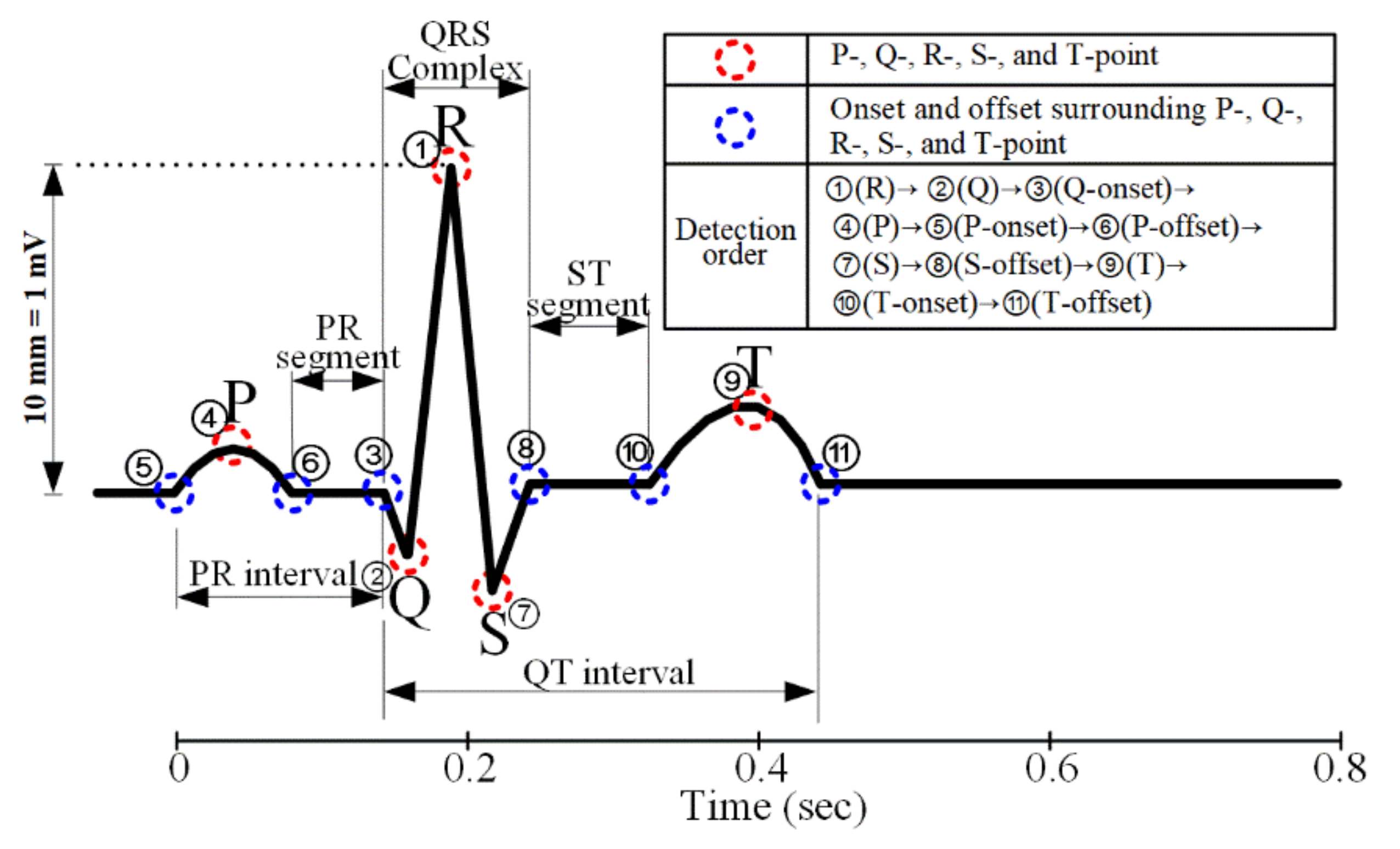
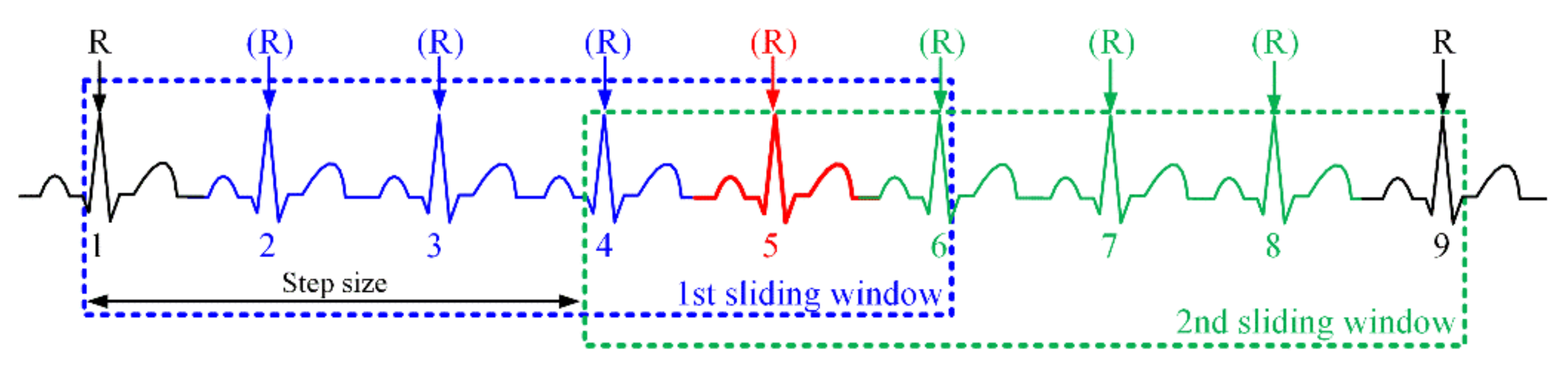
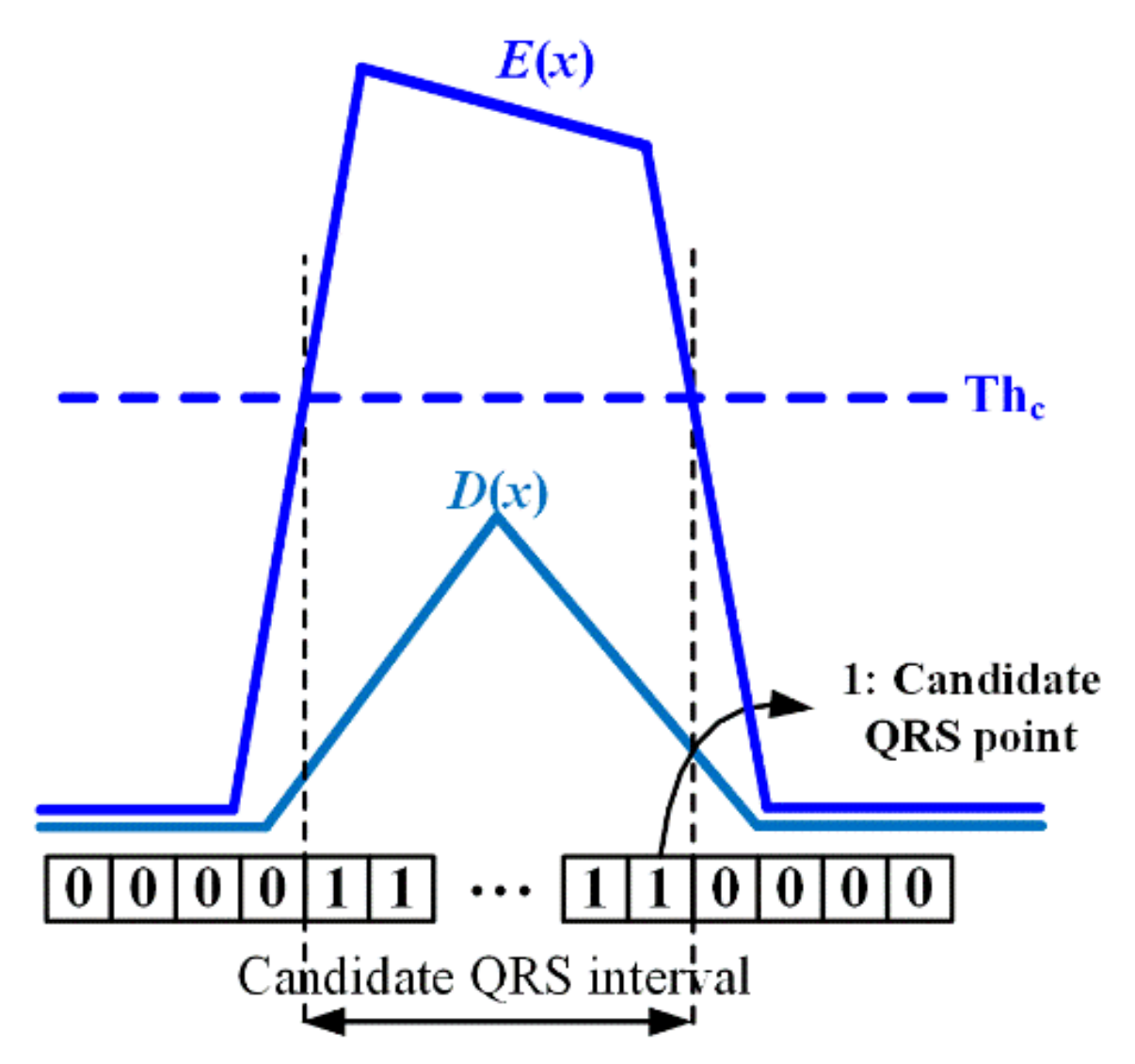
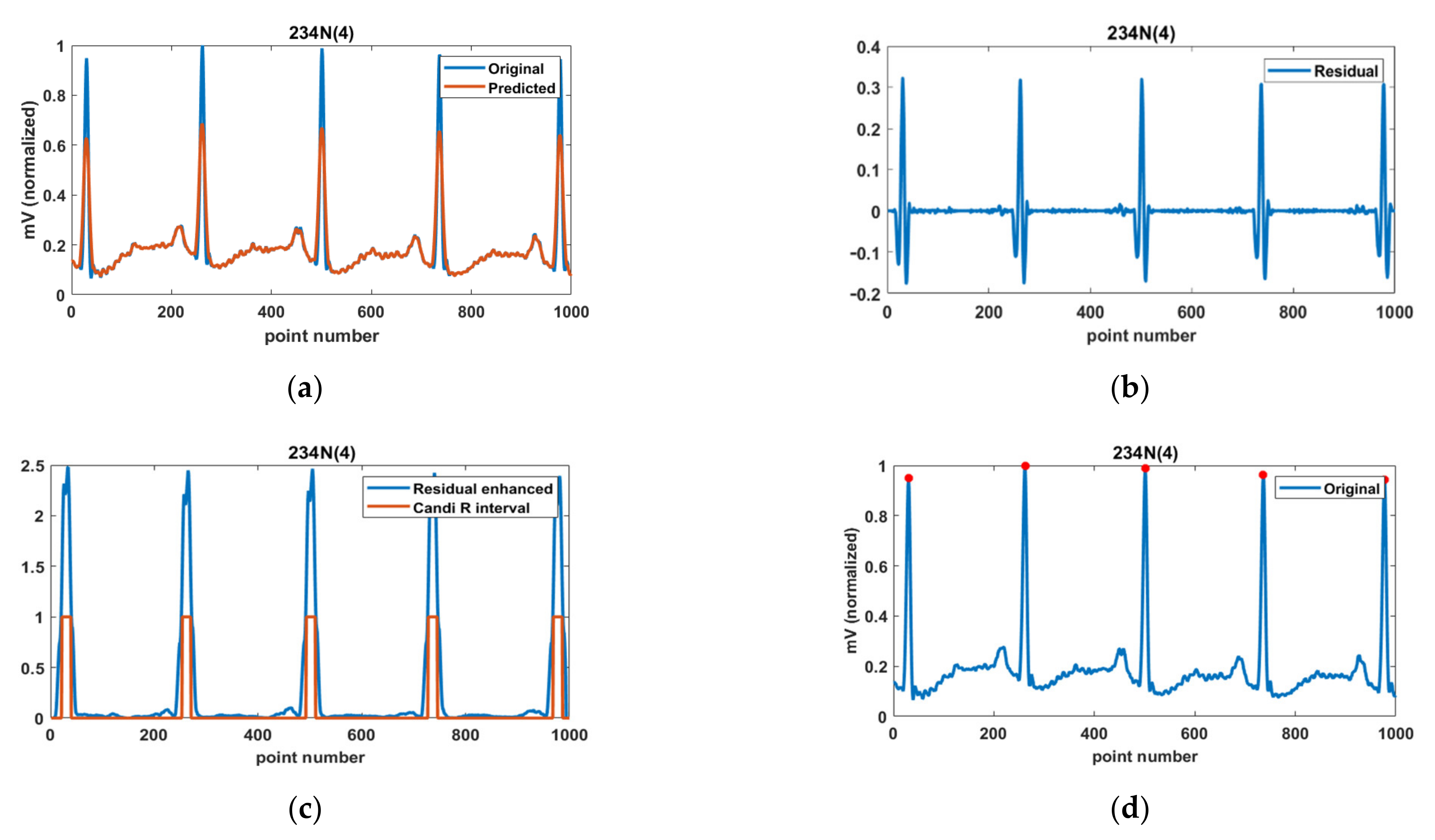
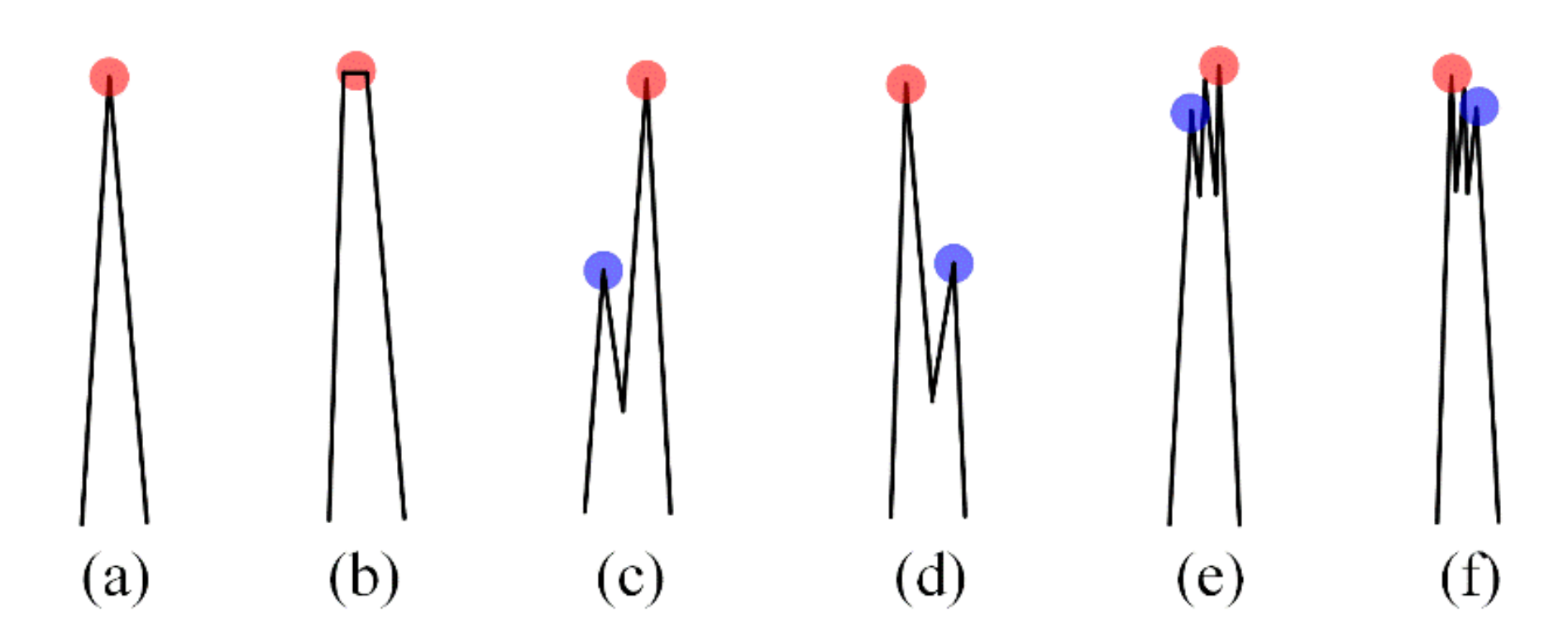
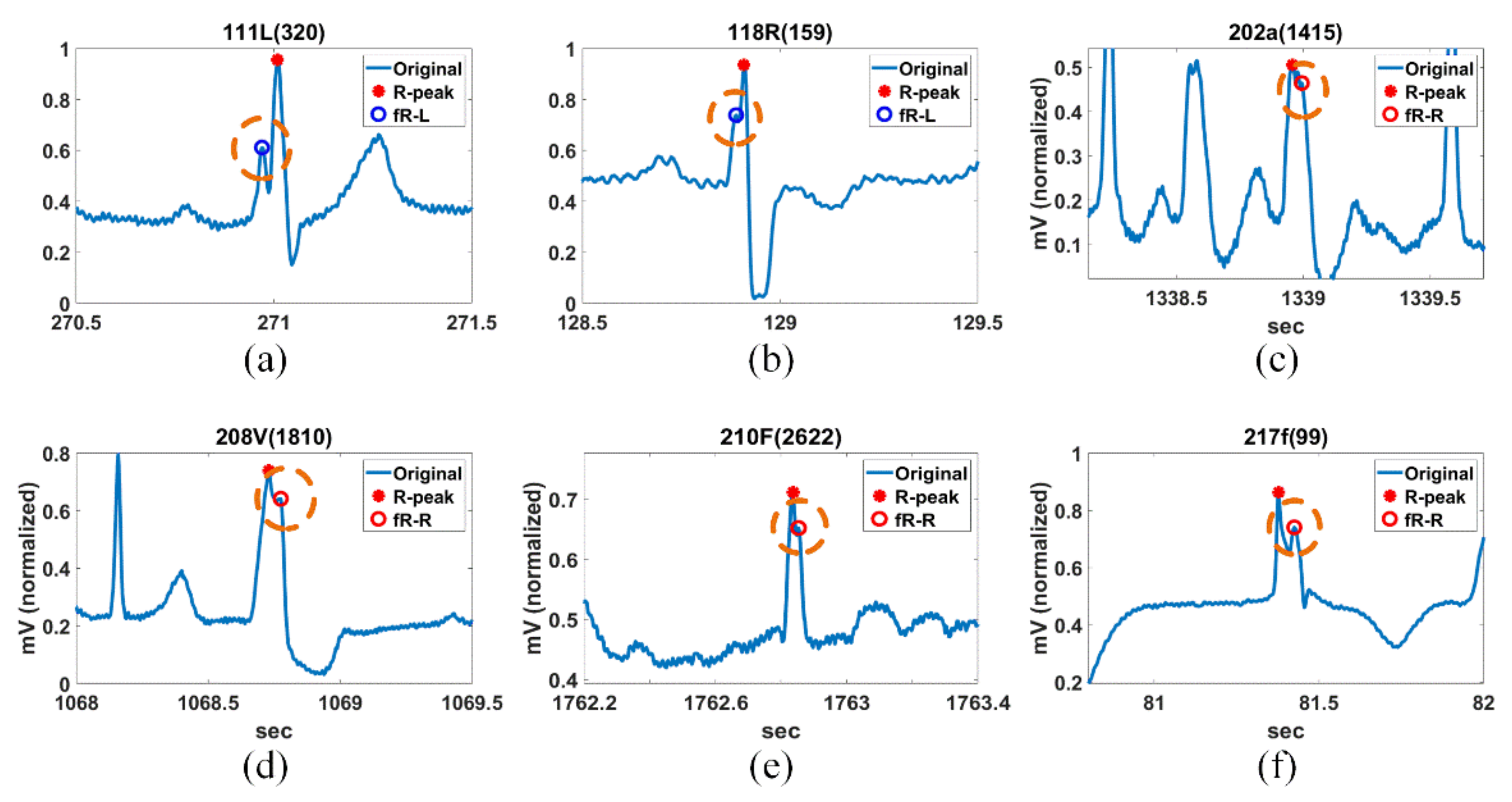
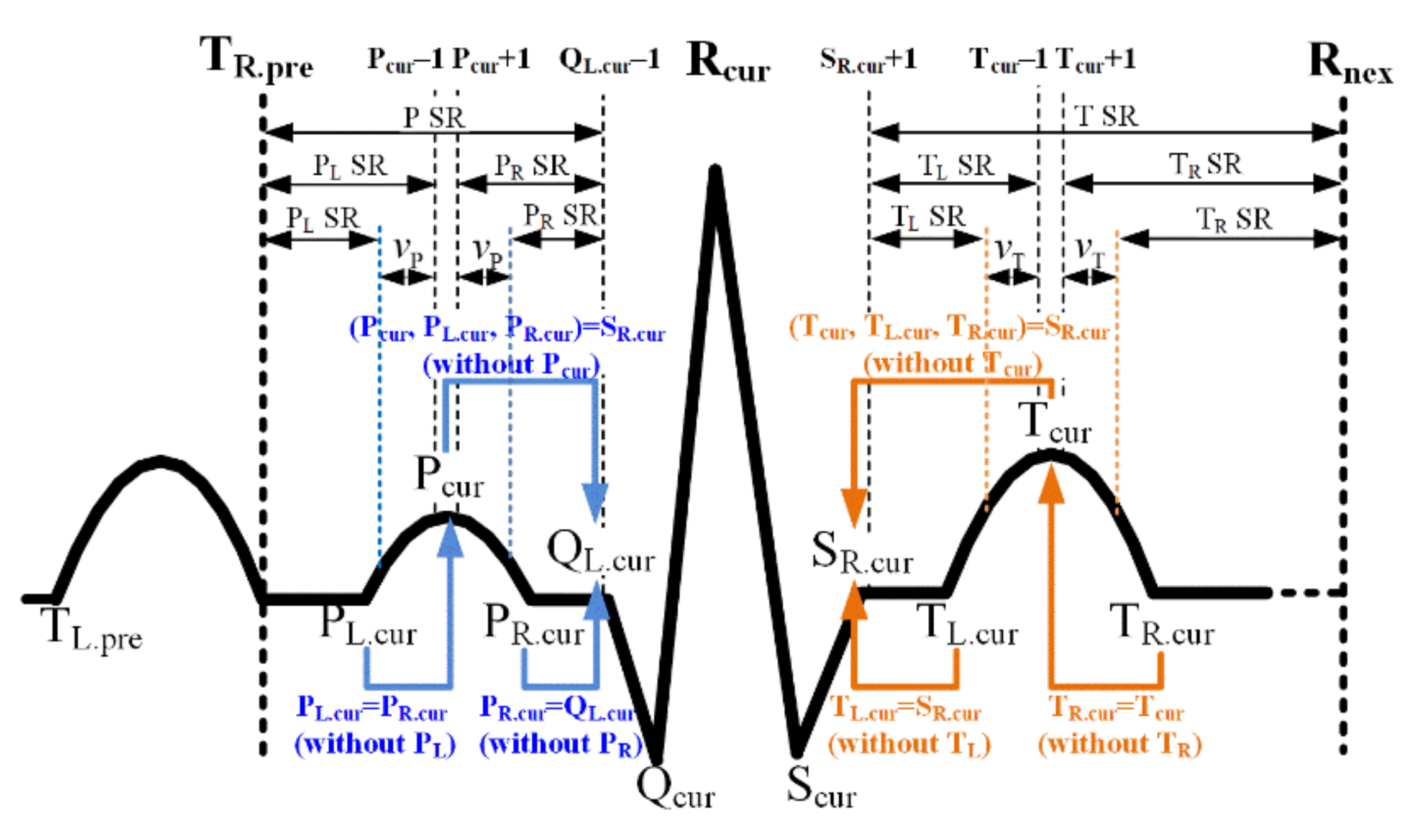
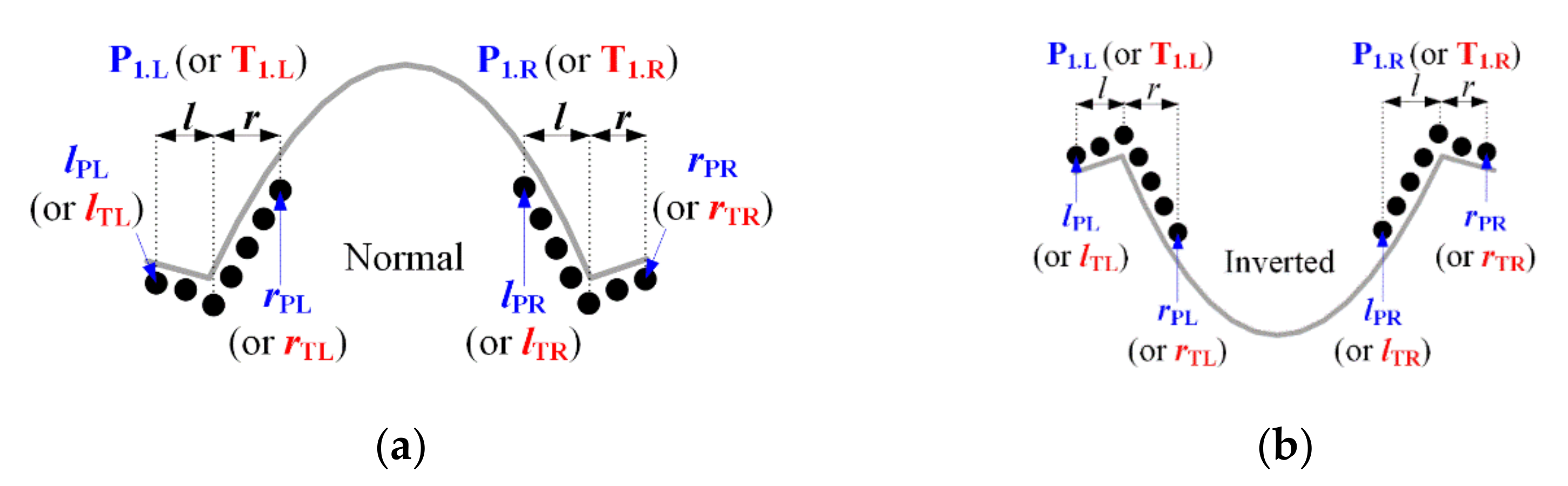
3.2. Performance Evaluation of R-Peak Detection
To assess the R-peak detection efficiency of the proposed method, several performance indexes are used. The indexes include the false positive (FP) and false negative (FN). The evaluation indices are as follows: sensitivity, Se = TP/(TP + FN), positive prediction, +P[%] = TP(TP + FP), and detection error, DER[%] = (FP + FN)/(TP + FN), respectively. TP represents the total number of accurately detected QRS [
52]. The annotated R-point location in each ECG data was used for beat-by-beat comparison with the detected R-peak location based on the WFDB library [
53]. If the difference between the annotated position and the detected R-point position is less than 50 ms, the proposed method is considered to have correctly detected the point [
54].
Table 2 shows the R-peak detection results by the proposed method for the MIT-DB. The proposed method accomplished Se = 99.83%, +P = 99.82%, and DER = 0.34% for the database.
The proposed R peak detection performance for noises is evaluated in
Table 3. Tape 102 is a relatively clean signal with no noise, while date 105 contains large induced noises. For evaluation, the MATLAB function ‘awgn’, which adds white Gaussian noise to the signal, was used for a signal-to-noise ratio (SNR) of 0.5 to 80. The proposed method for noisy signals of 5 dB SNR or higher showed relatively good sensitivity and positive prediction performance, but the performance deteriorated rapidly for noise signals of 1 dB SNR or less.
Table 4 compares the R-peak performance as well as supplemental function for several methods. The proposed method could obtain superior results similar to the existing methods, indicating a sensitivity of 99.83% on 109,510 annotated beats. As shown in
Table 4, while all six methods ([
5,
7,
17,
19,
30], and proposed) indicate the QRS detection technique, only two methods ([
11] and proposed) described the QRS-onset and -offset detection technique.
Table 4 shows that [
11,
19,
25] accomplished outstanding FP and FN as well as the best DER; thus, they represent fascinating techniques. However, they are not attractive for the detection of the QRS-onset, -offset, and fR-peak. In addition, only the proposed method could detect the fR-peak, which is important for detecting adverse cardiac events. Among the stated methods, only the proposed method can recognize the QRS-onset and -offset as well as the fR-peak. The comparison indicates that the proposed method has relatively high sensitivity, positive prediction as well as low DER by comparison with the previous methods. This demonstrates that the proposed method features superior trade-off between the QRS detection and identification of the fR-peak, QRS-onset, and -offset.
3.3. Detection Performance of PQRST Complex Using QT-DB
The QT-DB is used to assess the PQRST detection performance of the proposed method. It includes 105 recordings of 15 min length with 250 Hz, and each recording has a peak and onset and offset annotation information. The annotation for each record was manually performed by two cardiologists and its number reaches 3622 [
55,
56]. In case the detected S-offset and T-onset are very close (less than about 0.05 s, 13 sample), the detected T-offset was excluded in the evaluation.
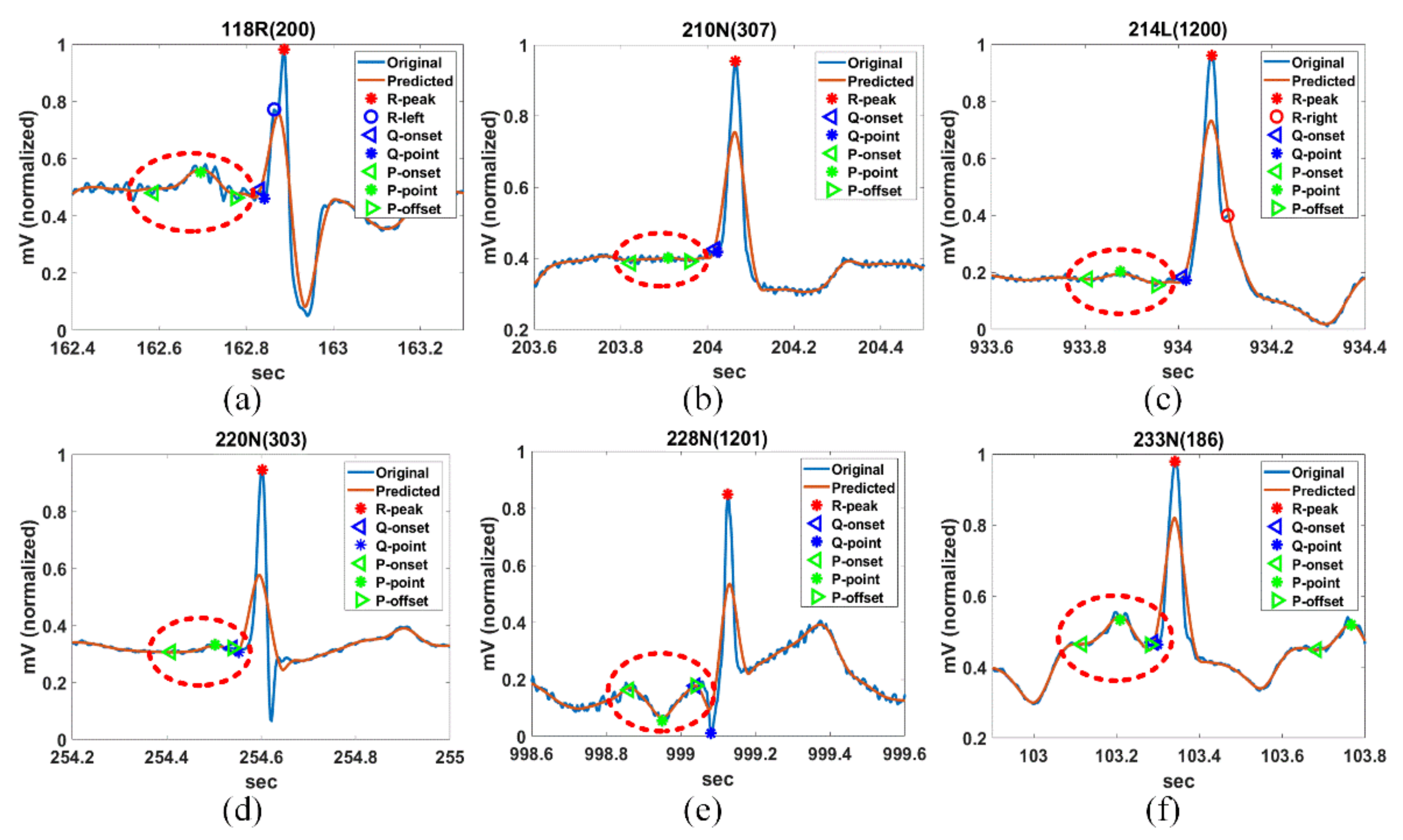
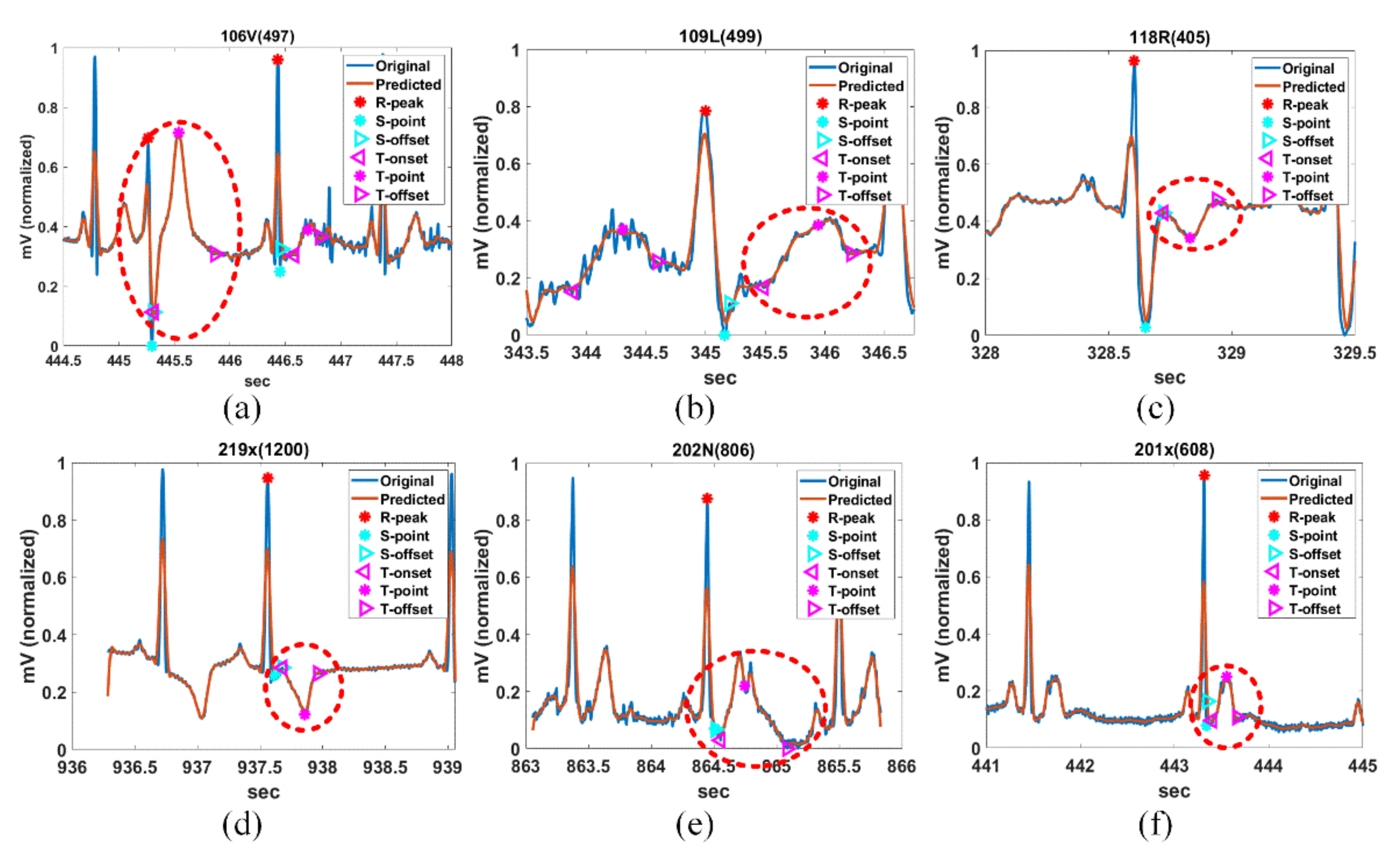
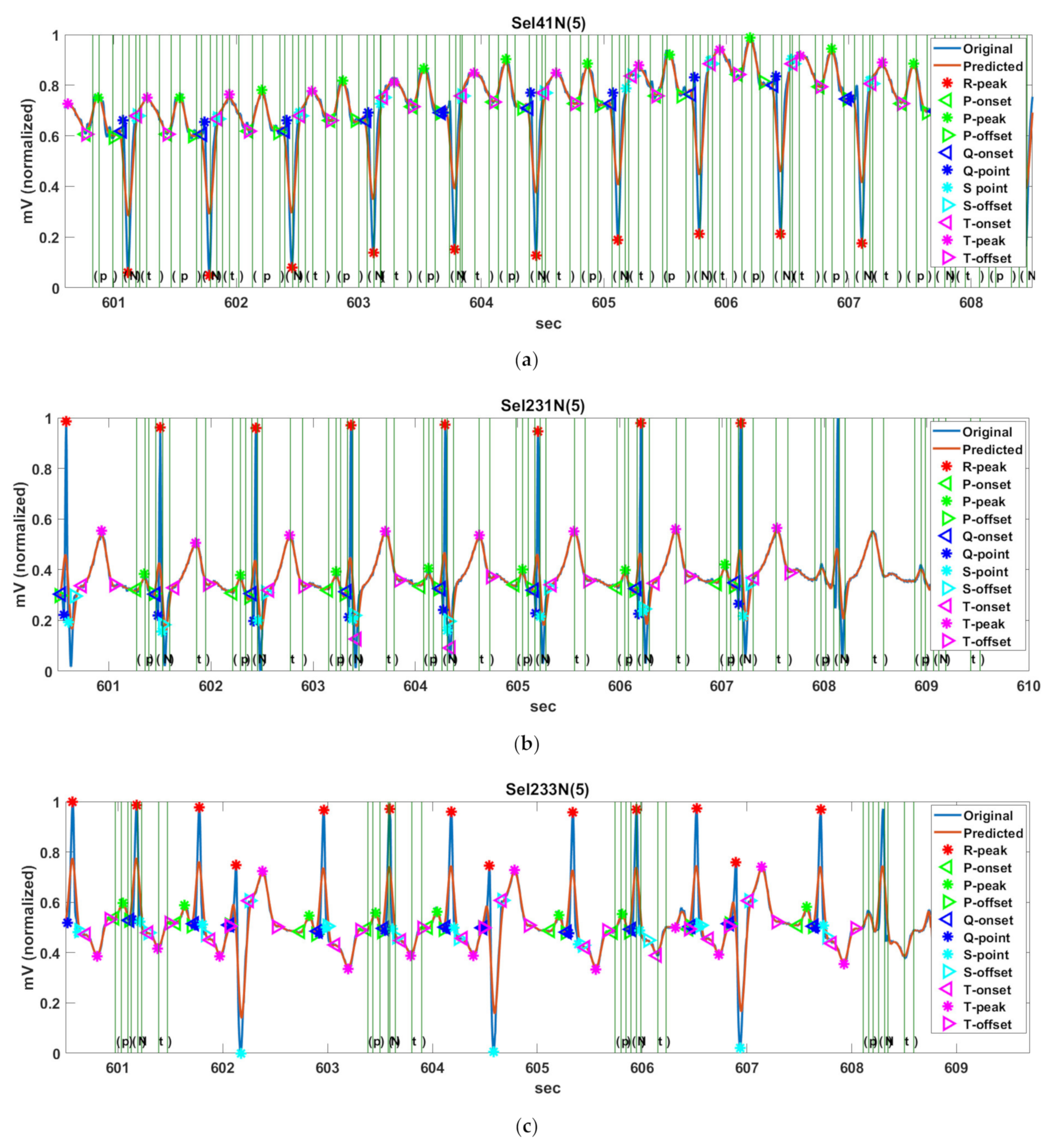
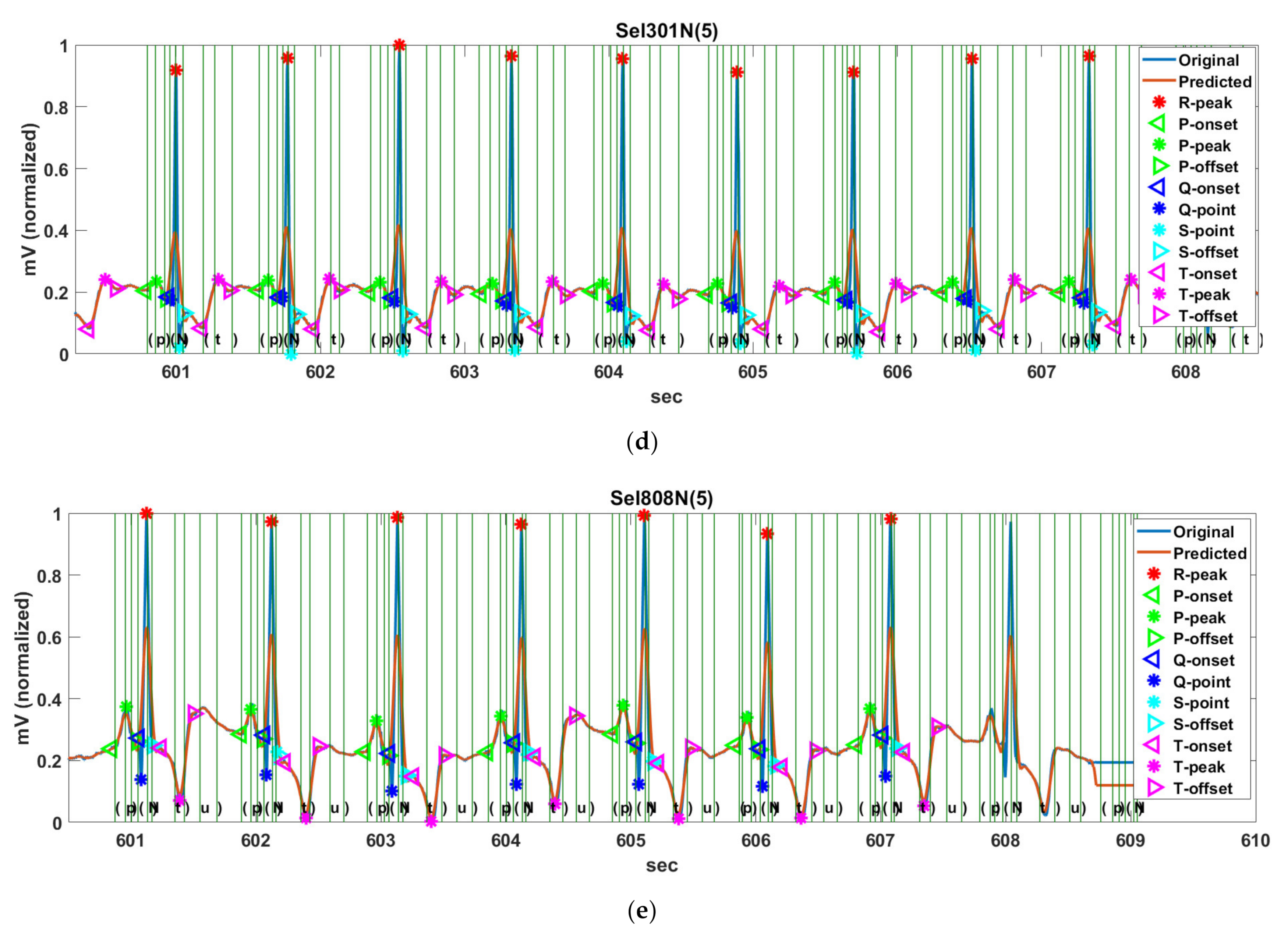
Figure 14 shows the PQRST complex detection results of Record Sel41N(5), Sel231N(5), Sel233N(5), Sel301N(5), and Sel808N(5) by the proposed method for QT-DB. The vertical green solid lines show the originally annotated positions in the QT-DB.
Figure 14a shows the locations of FPs detected by the proposed method for Sel41N(5), which has an inverted (negative) R-wave, bigger P- and T-waves, and is related to a sudden death. In this connection, the P-peak may be higher than the T-peak, and the interval between the current P-wave and the previous T-wave may be very close. We can see that the proposed method precisely detects the annotated locations.
Figure 14b shows the detection result for Sel231N(5), including hyperacute T-waves in a normal sinus rhythm. In the hyperacute T-wave, it is not easy to discriminate between the S-offset and T-onset, so the T-onset has no annotations. Nevertheless, the proposed method detects the T-onset used in HRV.
Figure 14c shows the detection result for Sel233N(5), which corresponds to an arrhythmia rhythm and has inverted T-waves, deep S-waves, and a hyperacute T-wave. Because the inverted T-wave appears immediately after the S-wave similar to Sel231N(5), the boundary between the S-offset and T-onset is unclear and the T-onset has no annotation. However, the proposed method estimates the T-onset used on HRV. As a result, we can see that the proposed method detects the peak and offset positions of the inverted T-waves. In addition, the beats with deep S-waves and hyperacute T-waves were not included in the original annotations. Therefore, these beats were excluded from the performance experiment.
Figure 14d shows the detection result of Sel301N(5), which corresponds to an ST change rhythm and has a biphasic T-wave. The positive peak was considered as the peak annotated by the cardiologist. The proposed method detects the peak and offset location of T-waves precisely.
Figure 14e shows the detection result for Sel808N(5), which is equivalent to a supraventricular rhythm, including inverted T-waves. As in the description of Sel233N(5), since the T-onset is indistinguishable from the S-offset in the inverted T-wave, it was ruled out from the annotations. We can see that the proposed method accurately estimates the original annotation locations for the normal P-wave as well as the inverted T-wave.
Table 5 shows the PQRST complex detection result of the proposed method and existing methods (LPD [
33], WT [
41], PCGS [
44], HFEA [
42], MHMM [
47]) on 27 Records (19 Records except for MHMM from the original paper) from the QT-DB. However, the result of TLBLDT was obtained by the LU database [
48]. Two-standard-deviation tolerances have been included in the last row [
57]. The mean (
) and standard deviation (
) between the detected positions by respective methods and the originally annotated positions are also listed. The root mean square error (
) is used for quantitative evaluation of the respective methods [
47]. The blue and green numbers in the table represent the smallest and the second smallest RMSE of the location difference of the FPs. The proposed method has the smallest RMSE value in the P-onset, P-offset, and T-wave, and the second lowest RMSE value in the P-peak, R-peak, and QRS-onset among all methods. The standard deviation of the proposed method is lower than the CSE tolerance in the P-offset, QRS-offset, and T-offset, and is close to the CSE tolerance in the P-onset and QRS-onset. Additionally, the proposed method also satisfies the criteria limit of the CSE tolerance for the P-offset with LPD and MHMM. Unfortunately, no method yet satisfies the severe criteria of the QRS-onset. The mean and standard deviation of all FPs are evaluated for LPD, WT, PCGS, HFEA, MHMM, TLBLDT, and the proposed method as 2.9 ± 13.5, 1.6 ± 12.7, 2.9 ± 15.1, −29 ± 17.4, −1.0 ± 8.5, 0.4 ± 7.4, and 0.4 ± 7.4 ms, respectively. The RMSE values across all FPs for the above-mentioned methods are evaluated to be 15.3, 13.1, 15.7, 19.8, 8.9, 9.5, and 8.3, respectively.
3.4. HRV Analysis for MIT-DB
The heart is continually regulated by the autonomic nervous system [
58]. HRV denotes the time variation between successive heartbeats [
58,
59]. HRV can estimate the sympathovagal balance through time- and frequency-domain methods [
58]. In the time domain, HRV is assessed by the variability of heartbeats observed in the range of 5 min to 24 h [
60]. These methods are calculated based on the beat-to-beat (or NN) intervals. The statistical time-domain measures (or metrics) include the mean HR, HR standard deviation (std.), RR-mean, NN50, pNN50, RMSSD, HRV triangular index (HTI), and TINN, which are listed in
Table 6 [
59,
61,
62,
63].
For the HR, variation in certain frequency bands is a very useful method to assess the sympathetic-parasympathetic balance of the cardiovascular system. Additionally, the high frequency band (HF) is also denoted as respiratory arrhythmia because it is associated with HRV associated with the breathing cycle and is controlled by the activity of the parasympathetic nervous system (PNS). The low frequency band (LF) is regulated by both the sympathetic nervous system (SNS) and PNS. The very low frequency band (VLF) is a variable dependent on the Lenin-Angiotensin system and is also affected in both SNS and PNS. A reduction in the ultra-low-frequency and VLF in 24-h ECG is the strongest predictor of acute heart attack and death from arrhythmia. Spectrum analysis is a useful method for evaluating the effects of the SNS and PNS at the same time. However, because individual deviations vary, it is difficult to obtain a normal number that can be used as a test tool. Therefore, it should always be investigated to see if it has decreased or increased statistically compared with the control group. The reduction in HRV mentioned above is a very powerful prognostic factor in all diseases with cardiovascular abnormalities.
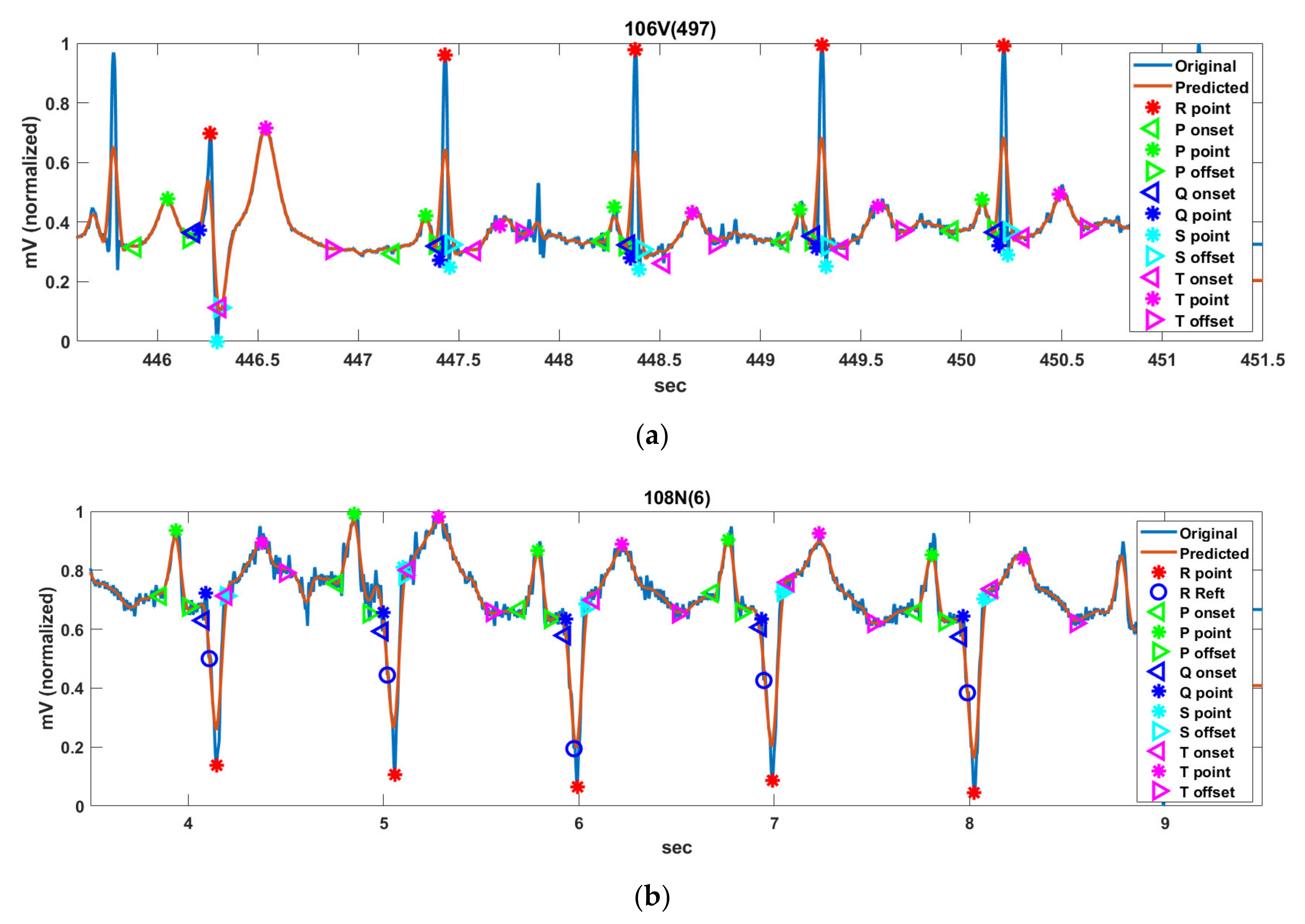
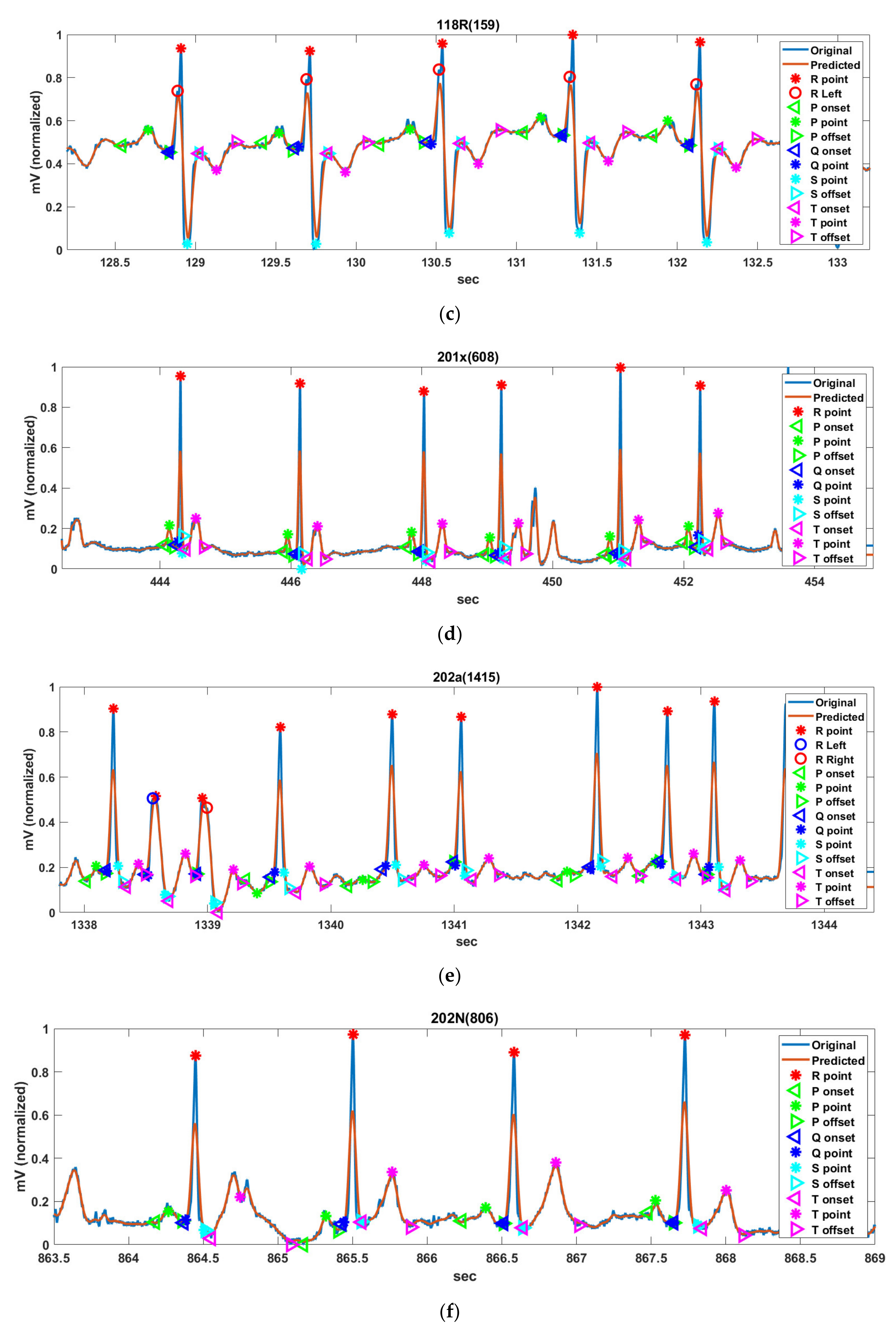
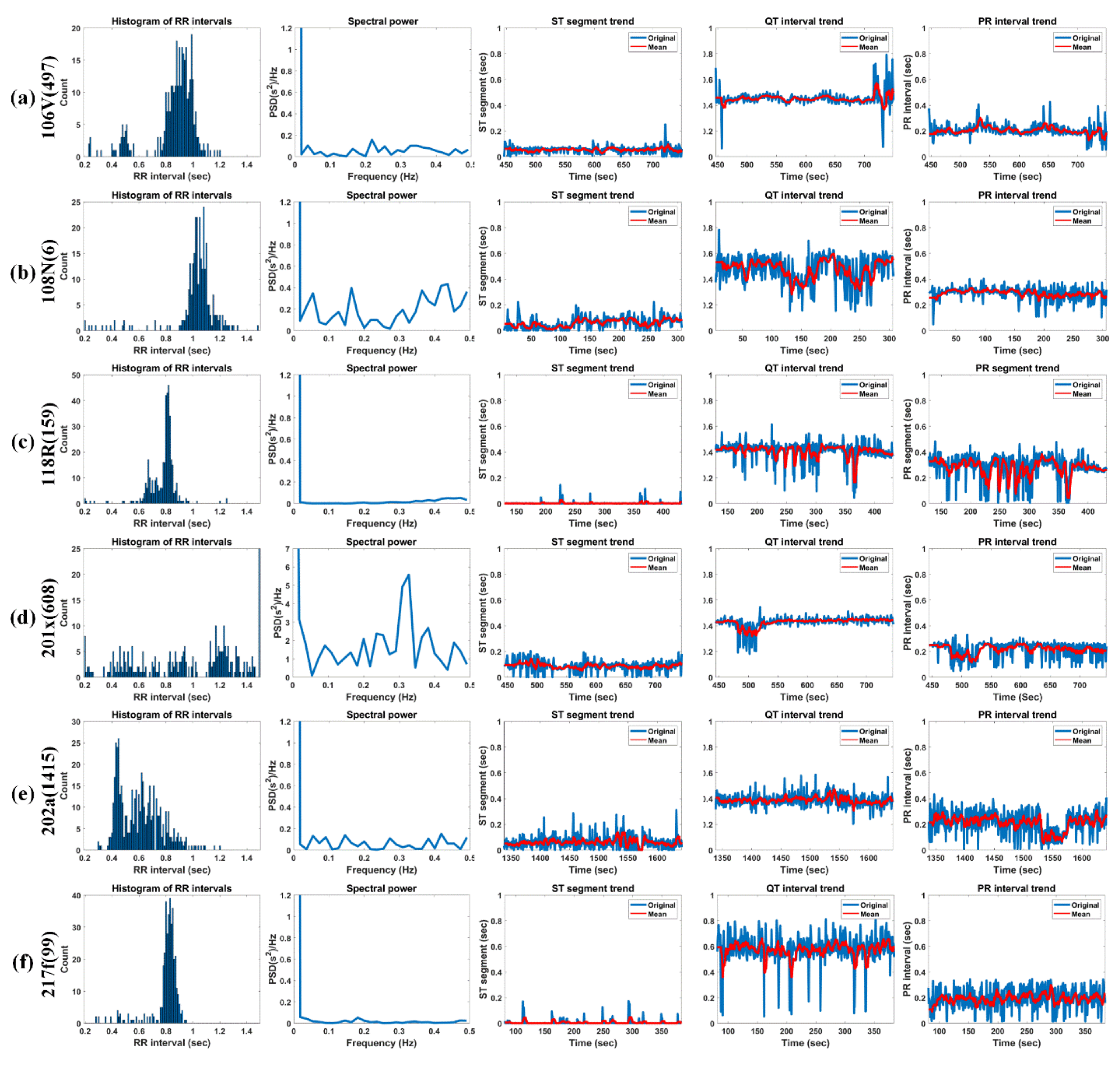
Figure 15 shows the histogram of RRIs, spectral power, ST-segment, QT-interval, and PR-interval for 5 min of Record 106V(497), 108N(6), 118R(159), 201x(608), 202a(1415), and 217f(99) in MIT-DB.
The ST-segment progresses from the S-offset to the T-onset of the following T-wave. This is the sluggish depolarization term of the ventricles after the contractions of the two ventricles [
64]. The baseline of the ST-segment typically is near the isoelectric line (IL) by height. The IL corresponds to the baseline of the whole ECG wave, typically located at 0 mV. Abnormal cardiac activities make the baseline of the ST-segment usually rise or fall from the IL. Elevation of the ST-segment represents myocardial infraction. On the other hand, depression of the ST-segment is typically relevant to hypokalemia [
65].
The typical term of the ST-segment corresponds to 0.005 to 0.15 s. It can be seen that all test signals have a normal range of ST-segments. Furthermore, 106V(497) (female, age 24) shows a relatively constant ST-segment of about 0.1 s or less. Moreover, 108N(6) has a somewhat irregular ST-segment, which is close to zero up to 100 s due to the effects of asymmetrically peaked (or hyperacute) T-waves. However, after 100 s, the normal T-waves are observed, but an unstable ST-segment distribution is observed due to the delay of the T-wave (ST-segment above 0.15 s) and noise (ST-segment close to zero). Additionally, 118R(159) has RBBB’s inverse T-waves but shows a very constant ST-segment distribution near zero and 201x(608) (male, age 68) shows a slightly higher ST-segment of 0.2 s due to the slowly rising T-waves up to 510 s and shows an ST-segment close to 0 s because of the deformation of the T-waves by the annotation “x” (Non-conducted P-wave, blocked APB) and the annotation “a” (Aberrared atrial premature) [
66]. However, from 510 s, it frequently shows ST-segments close to zero due to the frequent appearance of the annotation “V” (PVC). 202a(1415) (male, age 68) basically exhibits a relatively normal range of ST-segments but shows an unstable ST-segment distribution due to the gentle T-waves or T-wave delay and biphasic T-waves. 217f(99) shows an ST-segment distribution close to zero due to asymmetrically peaked (or hyperacute) T-waves. We know that the ST-segment is not easy to distinguish whether the detected T-wave is normal, an hyperacute T-wave, or a biphasic T-wave with a large falling curve.
The PR-interval represents the term from the P-onset to the Q-onset. It indicates the conduction by means of the AV node. In case the PR-interval is longer than 0.2 s, it can be considered that a first-degree heart block exists [
67]. The prolonged PR-interval arouses delayed conduction by the AV node. Furthermore, in case the PR-interval is less than 0.12 s, it is considered to manifest a pre-excitation or AV nodal rhythm.
The typical term of the PR-interval corresponds to the range of 0.12 to 0.2 s. 106V(497), 108N(6), 201x(608), and 217f(99) show a normal distribution, whereas 118R(159) and 202a(1415) show a somewhat unstable distribution. 106V(497) (female, age 24) has normal PR-intervals overall due to very clean P-waves. 108N(6) (female, age 87) also has a normal PR-interval and has a PR-interval of about 0.3 s because of the P-waves, which gradually increases in height. 118R(159) (male, age 69) has a notch (left atrial enlargement symptom) at the P-peaks, which is due to longer atrial depolarization times than normal. Therefore, the signal has a rather long PR-interval (about 0.37 secs). At 201 × 608 (male, age 68), the onset of AF with aberrated beats occurred at approximately 472 s. The ventricular fibrillation (AFIB) is the most severe from 480 to 520 s, and during this time, the P-waves are so weak that they cannot be detected; thus, the signal shows very low PR-intervals. After 550, 650, and 700 s, the annotation “V” (PVC) occurs frequently with little visible P-waves (“x” (paced) is very rarely present), resulting in very low PR-intervals. At 202a (1415) (male, age 68), AF with aberrated beats occurred at 1333 s, and then, atrial flutter with 2: 1 conduction occurred at 1558 s. This caused a momentary PR-interval drop near 1550 s. For the entire signal, the P-wave was very weak and accompanied by noise, which shows that the PR-interval is unstable. 217f (99) (male, age 65) shows relatively constant PR-intervals between 0.2 and 0.3 s, although the P-waves are very weak due to “/” (paced). However, because of the intermittent “f” (Fusion of paced and normal) or “V” (PVC), there are some intervals without P-waves; thus, the PR-interval with zero is partially visible.
The QT-interval represents the Q-onset to the T-offset. Specifically, it indicates the time from ventricle depolarization to ventricle repolarization. The unusually shortened or prolonged interval increases the danger of revelation of ventricular arrhythmia [
68]. In special cases, a prolonged QT-interval can cause a life-threatening ventricular tachycardia.
The typical term of the QT-interval corresponds to the range of 0.32 to 0.44 s. 106V(497) has a very constant QT-interval of 0.4 s, except for the long QT-interval (>0.6 s), due to the very high and wide T-waves of “V” (PVC) that appear around 450 s and after 710 s. At 108N(6), QT-intervals (more than 0.6 secs) beyond the normal range are observed because of the low but slowly increasing and then decreasing T-waves in the interval of less than 100 s and more than 160 s. In addition, the QT-interval was miscalculated because of severe noise near 150 and 250 s. 118R(159) has somewhat constant QT-intervals of 0.4 s because of clean inverse T-waves with a very constant ST-segment of 0.1 to 0.2 s. In the case of 201x(608), AFIB occurred at approximately 500 s due to the onset of AF with aberrated beats, which occurred at 472 s. Therefore, P- and T-waves were hardly seen and were not detected. Furthermore, the QT-interval near 500 s was momentarily reduced to 0.2 s. 202a(1415) has a QT-interval in the normal range of 0.4 s overall but exhibits a somewhat variable QT-interval distribution due to the gentle T-wave, T-wave delay, and biphasic T-wave as mentioned in the ST-segment experiments above. 217f(99) has a rather high QT-interval of approximately 0.6 s. 217f(99) has intermittently increasing QT-intervals due to a very tall asymmetrically peaked (or hyperacute) T-waves.
Table 7 shows the HRV analysis result of the test signals in MIT-DB. Bradycardia represents the HR of 45 beats/min or less while tachycardia denotes the HR of 100 beats/min or more at rest [
69]. All the test signals corresponded to a regular HR. 201x(608) with high RR std. and HR std. exhibited irregular RRI and HR, whereas 202a(1415) with a high HR std. has an irregular HR but showed a regular RRI. In fact, the high RR std. and HR of 201x(608) are due to fast and irregular “N” beats, “V”, “A”, and “x” (actually AFIB), and the irregular HR of 202a(1415) is due to irregular “N” beats per minute (actually AFIB). NN50, pNN50, and RMSSD indicate short-term cardiac variability and are associated with parasympathetic nerve activity. Additionally, those variables are deeply related to AF and sudden death [
70]. 201x(608) shows the highest RMSSD, NN50, and pNN50 among the tested signals and was actually annotated with AFIB at about 8:00. The patient was prescribed hypertension medication [
71]. 202a (1415) had a relatively high RMSNN but relatively high NN50 and pNN50 values, and was diagnosed with AFIB annotated at about 26:20; this patient was also prescribed medication for hypertension [
72]. In a previous study, when HTI > 20.42, the pattern was considered arrhythmic [
73]. As a criterion, 202a(1415) (HTI = 18.9) close to the threshold is classified as arrhythmia. On the other hand, since the remaining test signals show an HTI of 20 or less, those signals can be regarded as normal. However, specifically, 201x(608) had a very low HTI (2.693) but had AFIB. It can be seen that it is difficult to determine whether a signal is normal or not by means of only HTI. 201x(608) with TINN <15 can be estimated to have severely decreased HRV [
74].
HRV variables with a lower value indicate greater cardiac risk. VLF power is more deeply related to all-cause mortality compared to LF and HF power [
73]. Low VLF power has been researched to be related to arrhythmic death and is also associated with high inflammation [
73,
74]. The VLF rhythm appears to be influenced by the intrinsic nervous system of the heart, and the SNS affects the amplitude and frequency of its variations.
The power of most test signals is converged in DC to constant RRIs. Because the power spectral density only deals with the frequency of the RRIs, it is not directly related to BPM. In other words, in case the BPM is constant regardless of whether it is bradycardia (or tachycardia), it may show a high VLF value. It can be seen that the 108N(6) has a lower than average BPM than other test signals but have as much power as other signals in the VLF band. 201x(608) has more HF power compared to other test signals, which indicates that the RRI is unstable. In addition, in fact, 201x(608) contains numerous “V” and some “a” and “j” annotation beats with AFIB. 202a(1415) also contains AFIB and atrial flutter (AFL); however, no difference was observed in the HF power. As explained above, while LF works through sympathetic nerves, HF is affected by parasympathetic nerves [
73]. A low LF/HF, such as those for 106V(497), and 118R(159), denotes when to conserve energy, whereas a high LF/HF, such as those 108N(6), 202a(1415), and 217f(99), exhibits fight-or-flight behavior.