Predicting Spatial Patterns of Sindbis Virus (SINV) Infection Risk in Finland Using Vector, Host and Environmental Data
Abstract
:1. Introduction
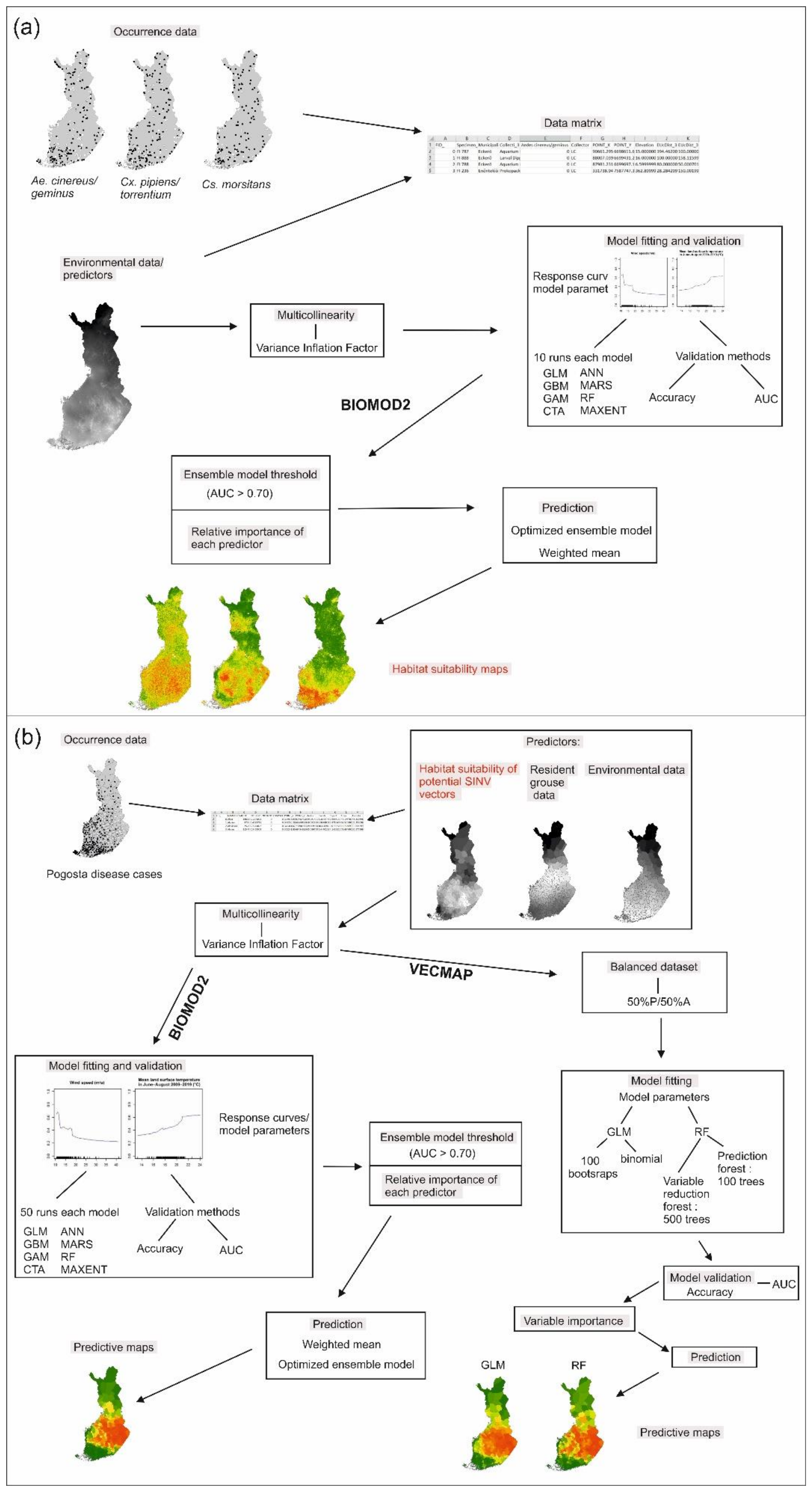
2. Materials and Methods
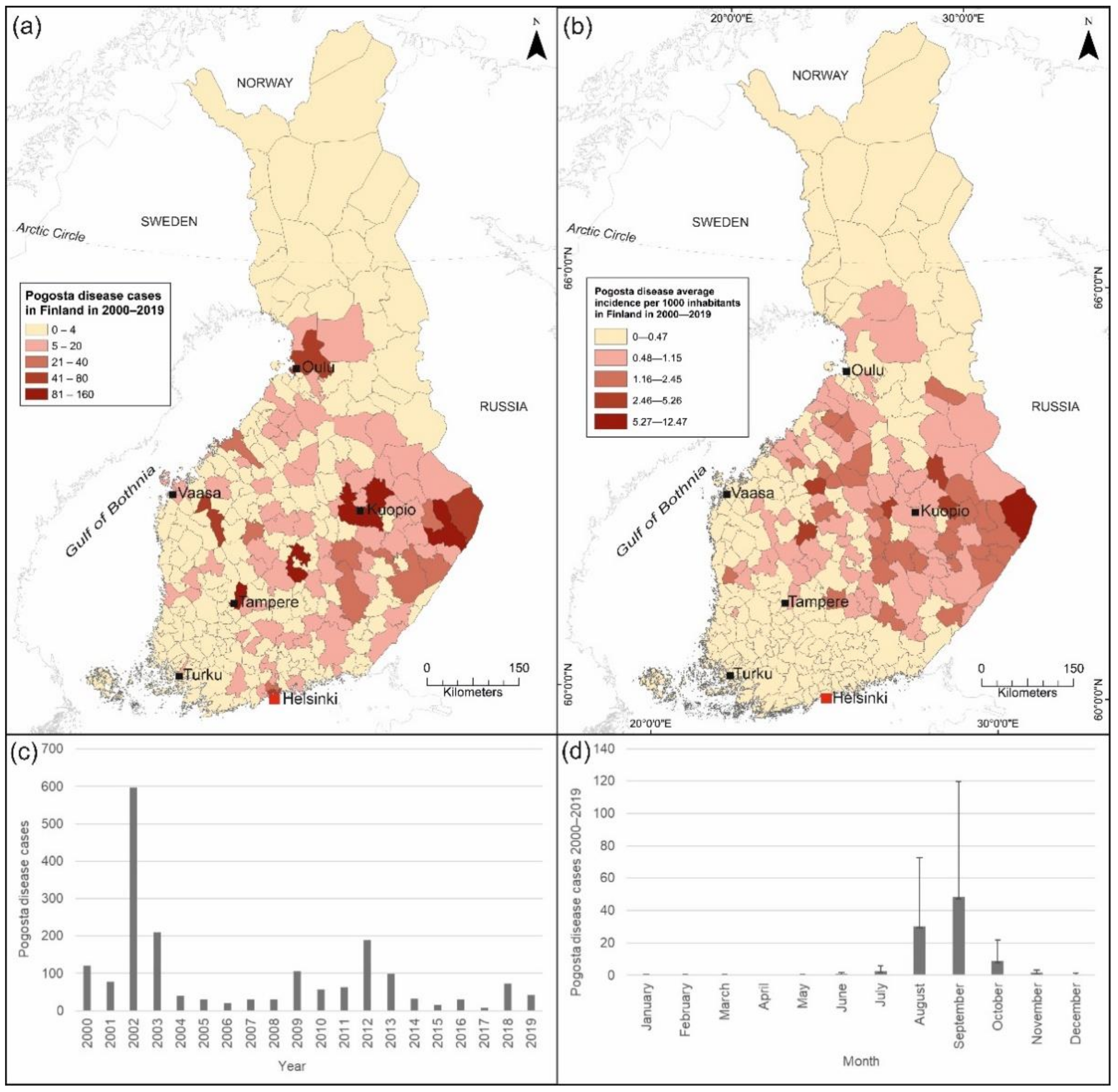
2.1. Pogosta Disease Data
2.2. Data of Potential SINV Vectors
2.3. SINV Host Species Data
2.4. Environmental and Other Predictors
2.5. Data Analysis
3. Results
3.1. Modeling of SINV Mosquito Vector Distributions
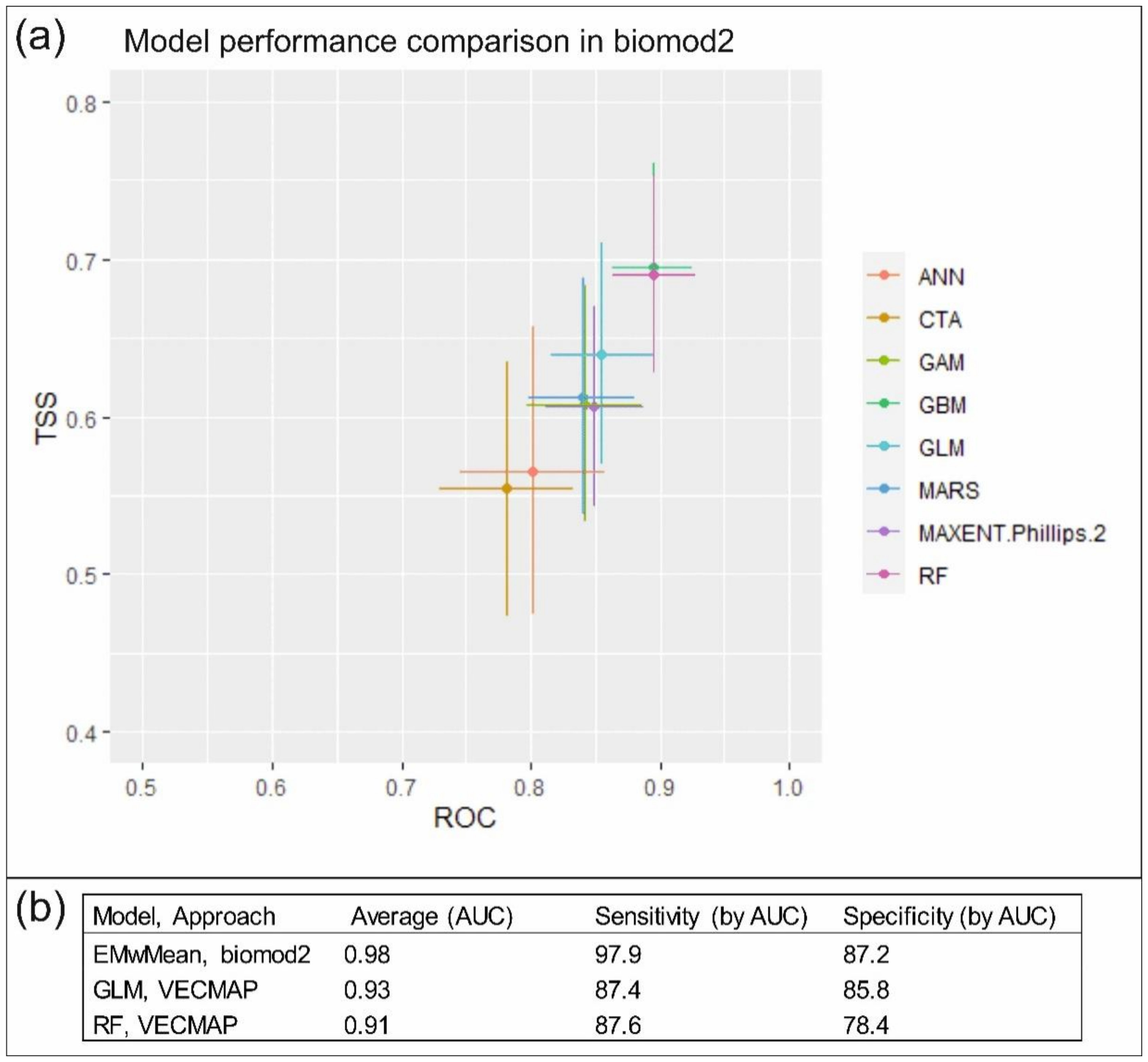
3.1.1. Predictive Performance
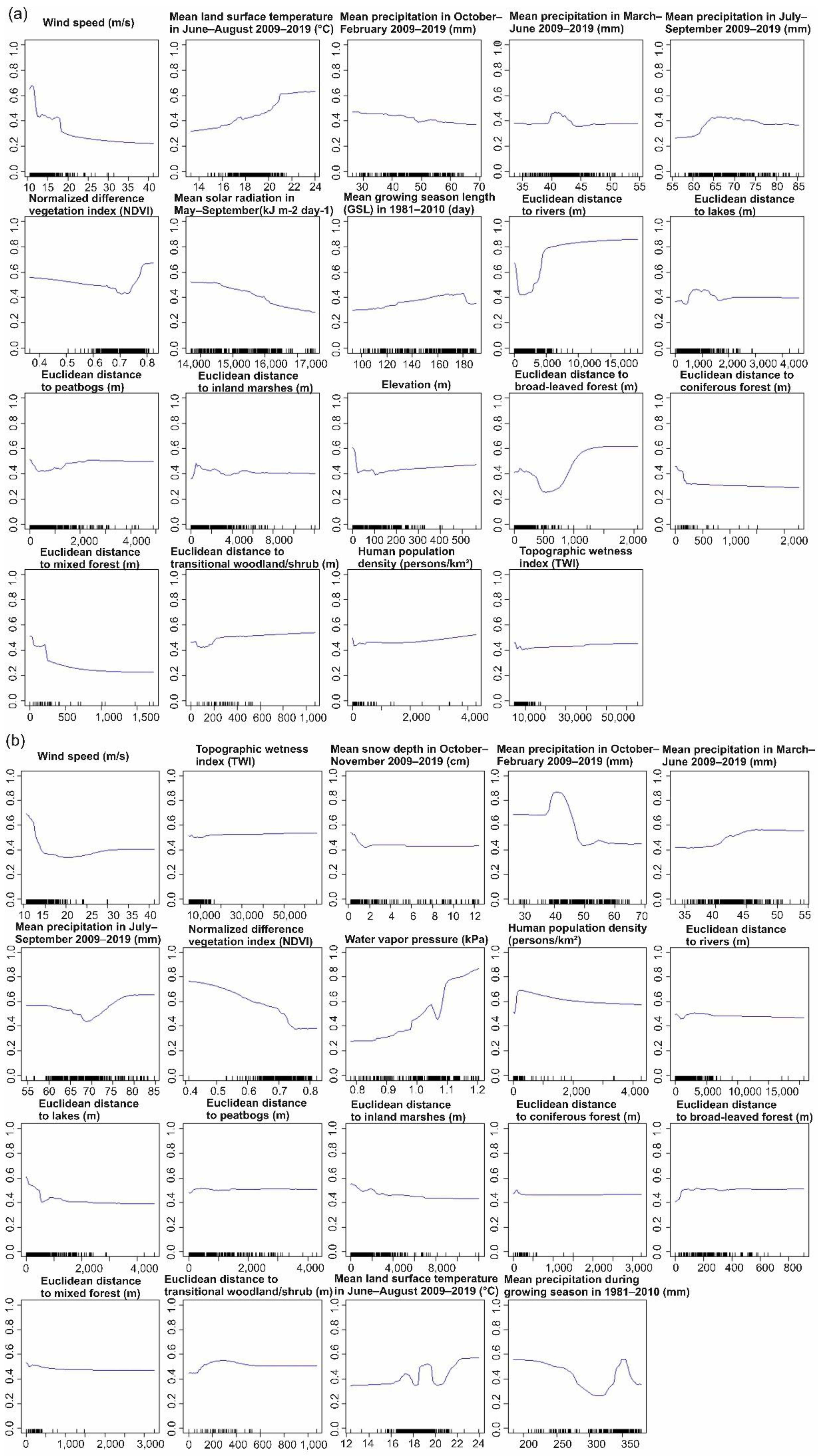
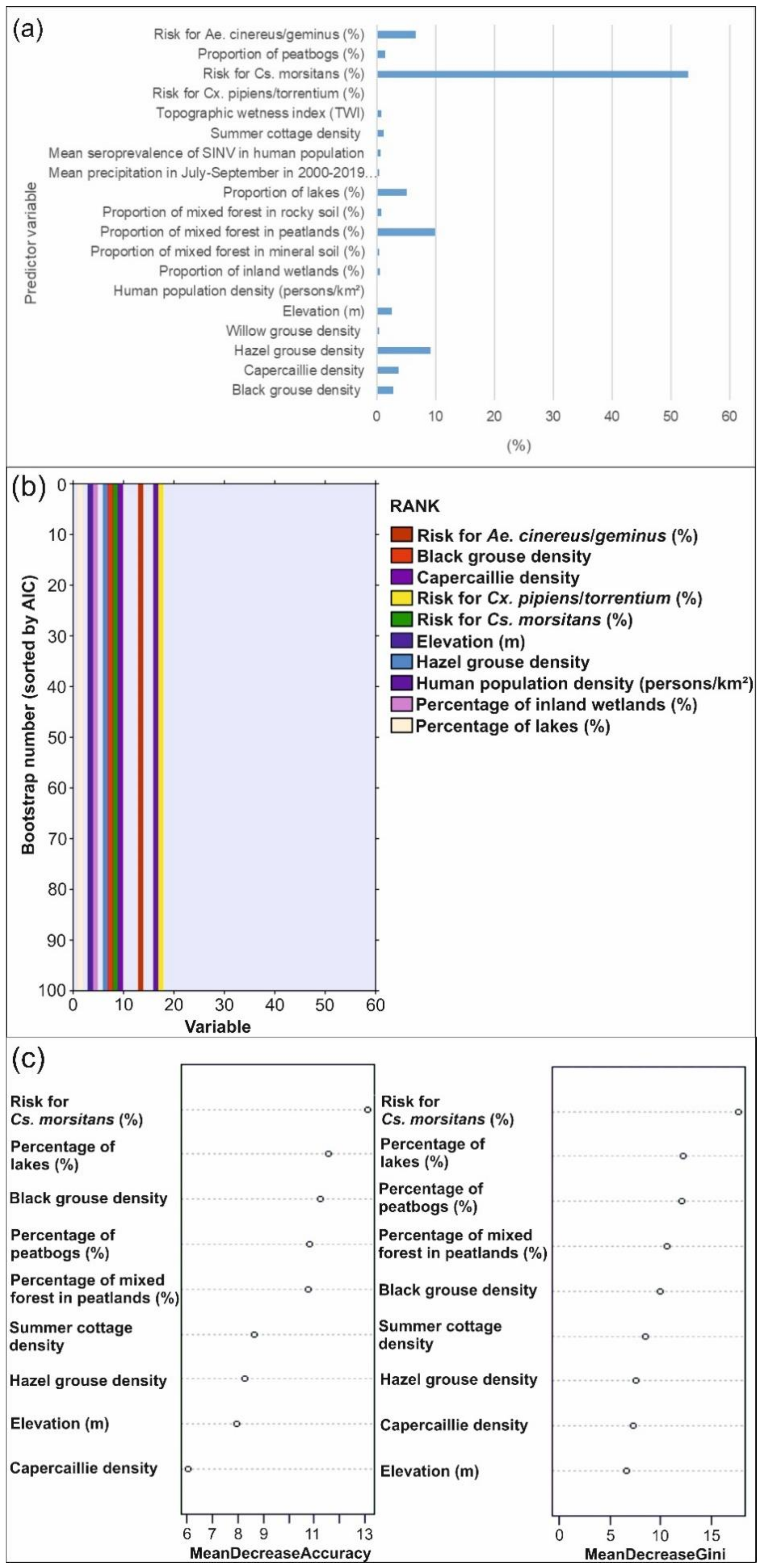
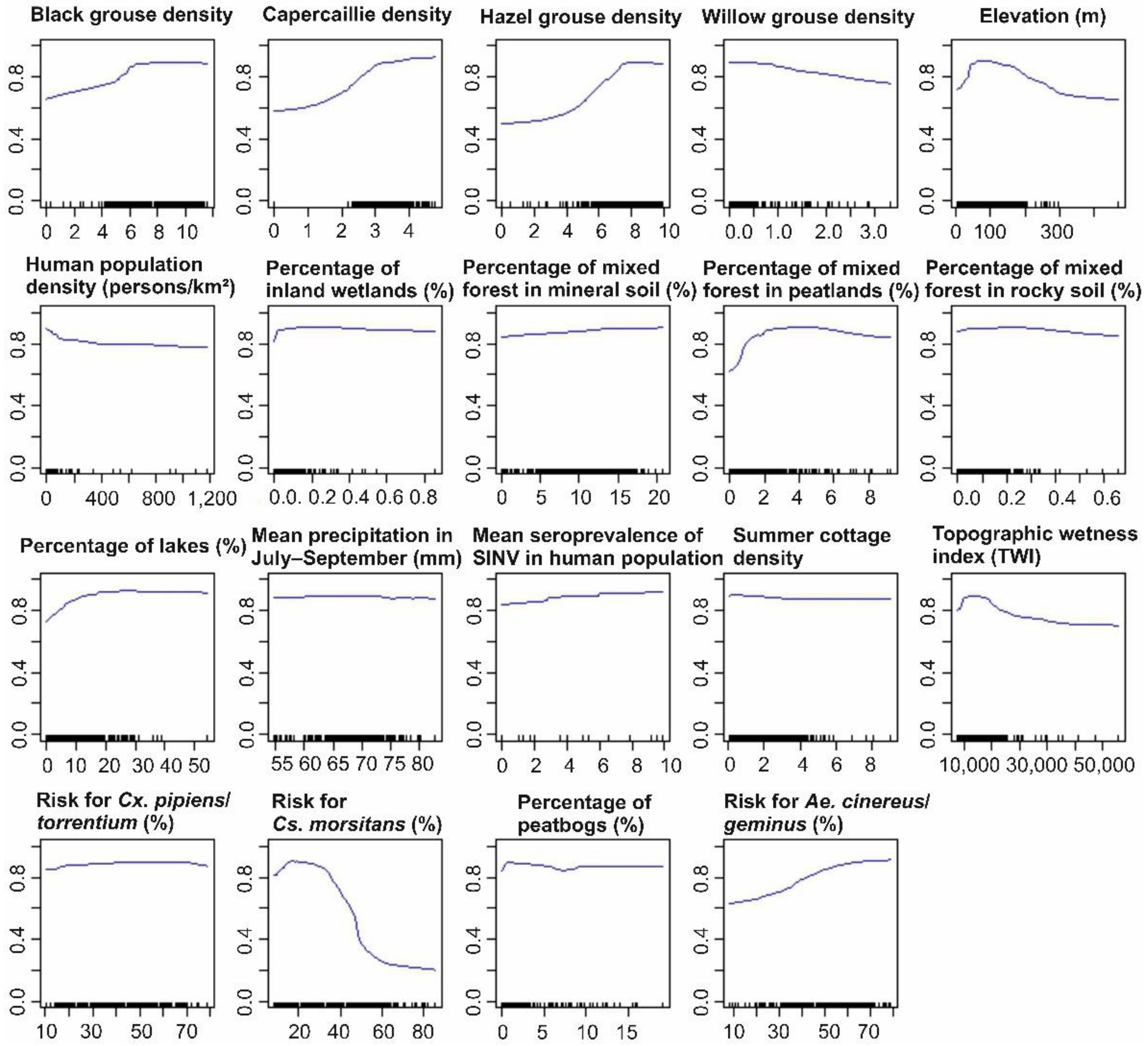
3.1.2. Predictor Contributions to the Distribution of SINV Vectors
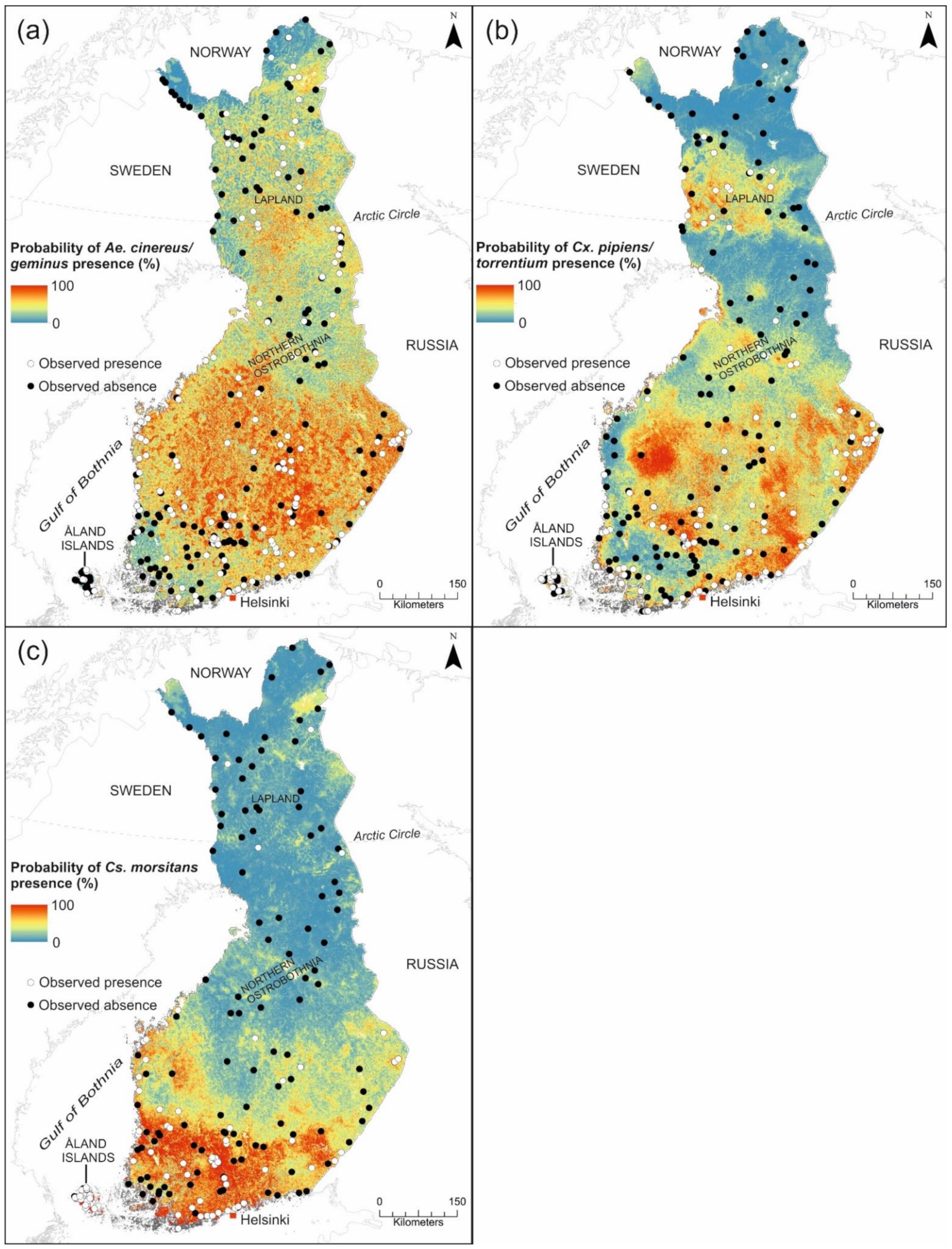
3.1.3. Prediction Maps for SINV Vectors
3.2. Pogosta Disease Modeling
3.2.1. Predictive Performance
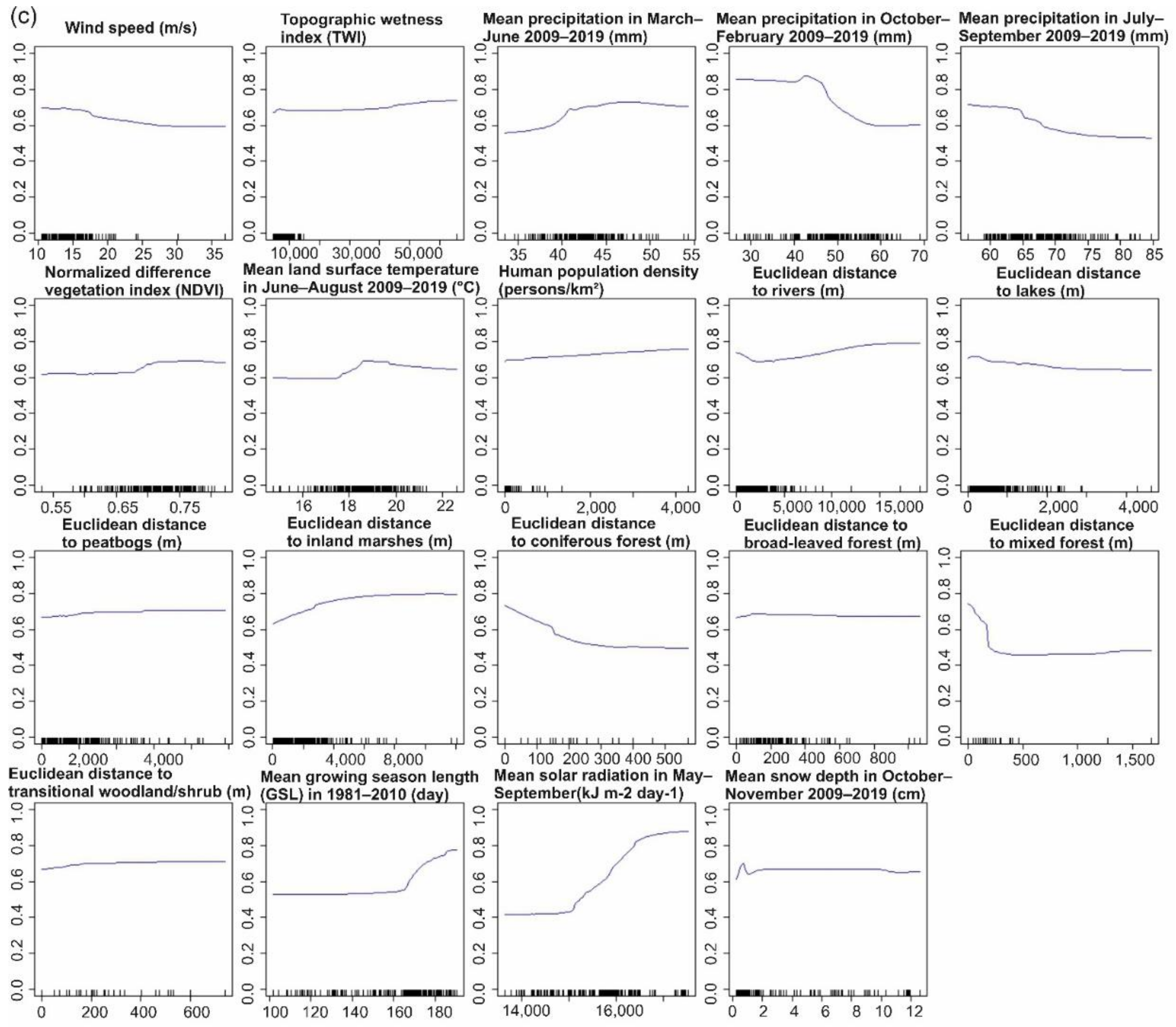
3.2.2. Predictor Contributions to the Distribution of Pogosta Disease Distribution
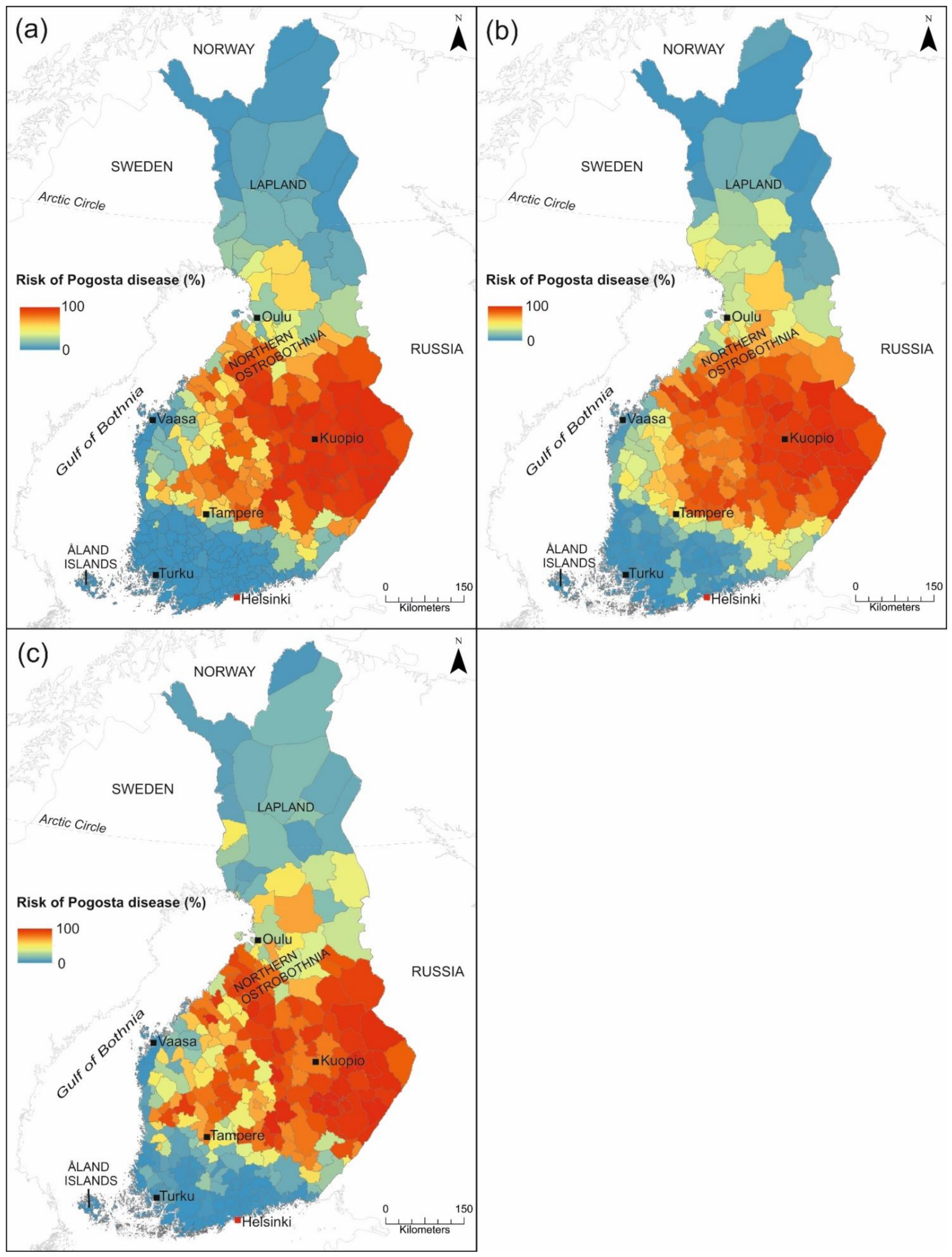
3.2.3. Pogosta Disease Risk Maps
4. Discussion
4.1. Validity of the Study
4.2. Influential Variables
4.3. The Suitability for Potential SINV Vectors and the Risk of Pogosta Disease in Finland
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Sutherst, R.W. Global change and human vulnerability to vector-borne diseases. Clin. Microbiol. Rev. 2004, 17, 136–173. [Google Scholar] [CrossRef] [Green Version]
- Lambin, E.F.; Tran, A.; Vanwambeke, S.O.; Linard, C.; Soti, V. Pathogenic landscapes: Interactions between land, people, disease vectors, and their animal hosts. Int. J. Health Geogr. 2010, 9, 54. [Google Scholar] [CrossRef] [Green Version]
- Eisen, R.J.; Eisen, L. Spatial modeling of human risk of exposure to vector-borne pathogens based on epidemiological versus arthropod vector data. J. Med. Entomol. 2008, 45, 181–192. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Talmoudi, K.; Bellali, H.; Ben-Alaya, N.; Saez, M.; Malouche, D.; Chahed, M.K. Modeling zoonotic cutaneous leishmaniasis incidence in Central Tunisia from 2009-2015: Forecasting models using climate variables as predictors. PLoS Negl. Trop. Dis. 2017, 11, e0005844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messina, J.P.; Brady, O.J.; Golding, N.; Kraemer, M.U.G.; Wint, G.R.W.; Ray, S.E.; Pigott, D.M.; Shearer, F.M.; Johnson, K.; Earl, L.; et al. The current and future global distribution and population at risk of dengue. Nat. Microbiol. 2019, 4, 1508–1515. [Google Scholar] [CrossRef]
- EEA. European Environment Agency. Vector-Borne Diseases. 2020. Available online: https://www.eea.europa.eu/data-and-maps/indicators/vector-borne-diseases-2/assessment (accessed on 18 December 2020).
- Lundström, J.O.; Hesson, J.C.; Schäfer, M.L.; Östman, Ö.; Semmler, T.; Bekaert, M.; Weidmann, M.; Lundkvist, Å.; Pfeffer, M. Sindbis virus polyarthritis outbreak signalled by virus prevalence in the mosquito vectors. PLoS Negl. Trop. Dis. 2019, 13, e0007702. [Google Scholar] [CrossRef] [Green Version]
- Musa, G.J.; Chiang, P.; Sylk, T.; Bavley, R.; Keating, W.; Lakew, B.; Tsou, H.; Hoven, C.W. Use of GIS mapping as a public health tool-from cholera to cancer. Health Serv. Insights 2013, 6, 111–116. [Google Scholar] [CrossRef]
- Carroll, L.N.; Au, A.P.; Detwiler, L.T.; Fu, T.; Painter, I.S.; Abernethy, N.F. Visualization and analytics tools for infectious disease epidemiology: A Systematic Review. J. Biomed. Inform. 2014, 51, 287–298. [Google Scholar] [CrossRef] [Green Version]
- Taylor, R.M.; Hurlbut, H.S.; Work, T.H.; Kingston, J.R.; Frothingham, T.E. Sindbis Virus: A newly recognized arthropod-transmitted virus. Am. J. Trop. Med. Hyg. 1955, 4, 844–862. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H.; Monath, T.P.; Mitchell, C.J.; Sabattini, M.S.; Cropp, C.B.; Kerschner, J.; Hunt, A.R.; Lazuick, J.S. Arbovirus investigations in Argentina, 1977–1980. Am. J. Trop. Med. Hyg. 1985, 34, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Hubálek, Z. Mosquito-borne viruses in Europe. Parasitol. Res. 2008, 103 (Suppl. 1), 29. [Google Scholar] [CrossRef]
- Laine, M.; Luukkainen, R.; Toivanen, A. Sindbis viruses and other alphaviruses as cause of human arthritic disease. J. Intern. Med. 2004, 256, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Kurkela, S.; Manni, T.; Myllynen, J.; Vaheri, A.; Vapalahti, O. Clinical and laboratory manifestations of sindbis virus infection: Prospective study, Finland, 2002–2003. J. Infect. Dis. 2005, 191, 1820–1829. [Google Scholar] [CrossRef]
- Kurkela, S. Sindbis Virus and Pogosta Disease in Finland. Ph.D. Thesis, University of Helsinki, Helsinki, Finland, 2007. Retrieved from HELDA. Available online: http://hdl.handle.net/10138/20554 (accessed on 20 September 2020).
- ECDC. European Centre for Disease Prevention and Control. Facts about Sindbis Fever. 2020. Available online: https://www.ecdc.europa.eu/en/sindbis-fever/facts (accessed on 18 December 2020).
- Go, Y.; Balasuriya, U.; Lee, C. Zoonotic encephalitides caused by arboviruses: Transmission and epidemiology of alphaviruses and flaviviruses. Clin. Exp. Vaccine Res. 2014, 3, 58–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jupp, P.G.; Blackburn, N.K.; Thompson, D.L.; Meenehan, G.M. Sindbis and West Nile virus infections in the Witwatersrand-Pretoria region. S. Afr. Med. J. 1986, 70, 218–220. [Google Scholar]
- Lundström, J.O.; Vene, S.; Espmark, A.; Engvall, M.; Niklasson, B. Geographical and temporal distribution of Ockelbo disease in Sweden. Epidemiol. Infect. 1991, 106, 567–574. [Google Scholar] [CrossRef]
- Brummer-Korvenkontio, M.; Vapalahti, O.; Kuusisto, P.; Saikku, P.; Manni, T.; Koskela, P.; Nygrén, T.; Brummer-Korvenkontio, H.; Vaheri, A. Epidemiology of Sindbis virus infections in Finland 1981–96: Possible factors explaining a peculiar disease pattern. Epidemiol. Infect. 2002, 129. [Google Scholar] [CrossRef]
- NIDR. Finnish National Infectious Diseases Register. 2021. Available online: https://thl.fi/en/web/infectious-diseases-and-vaccinations/surveillance-and-registers/finnish-national-infectious-diseases-register (accessed on 25 May 2021).
- Kurkela, S.; Rätti, O.; Huhtamo, E.; Uzcátegui, N.Y.; Nuorti, J.P.; Laakkonen, J.; Manni, T.; Helle, P.; Vaheri, A.; Vapalahti, O. Sindbis virus infection in resident birds, migratory birds, and humans, Finland. Emerg. Infect. Dis. 2008, 14, 41–47. [Google Scholar] [CrossRef]
- THL. Terveyden ja Hyvinvoinnin Laitos, National Institute for Health and Welfare. Infectious Diseases in Finland 1995–2009. Report 28/2010. 2010. Available online: http://urn.fi/URN:NBN:fi-fe201205085094 (accessed on 18 December 2020).
- Sane, J.; Guedes, S.; Kurkela, S.; Lyytikäinen, O.; Vapalahti, O. Epidemiological analysis of mosquito-borne Pogosta disease in Finland, 2009. Eurosurveillance 2010, 15, 19462. [Google Scholar] [CrossRef]
- Lundström, J.O.; Lindström, K.M.; Olsen, B.; Dufva, R.; Krakower, D.S. Prevalence of Sindbis virus neutralizing antibodies among swedish passerines indicates that thrushes are the main amplifying hosts. J. Med. Entomol. 2001, 38, 289–297. [Google Scholar] [CrossRef]
- Hesson, J.C.; Verner-Carlsson, J.; Larsson, A.; Ahmed, R.; Lundkvist, Å.; Lundström, J.O. Culex torrentium mosquito role as major enzootic vector defined by rate of Sindbis virus infection, Sweden, 2009. Emerg. Infect. Dis. 2015, 21, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Francy, D.B.; Jaenson, T.G.; Lundström, J.O.; Schildt, E.B.; Espmark, A.; Henriksson, B.; Niklasson, B. Ecologic studies of mosquitoes and birds as hosts of Ockelbo virus in Sweden and isolation of Inkoo and Batai viruses from mosquitoes. Am. J. Trop. Med. Hyg. 1989, 41, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Turell, M.; Lundström, J.; Niklasson, B. Transmission of Ockelbo virus by Aedes cinereus, A. communis, and A. excrucians (Diptera: Culicidae) collected in an enzootic area in central Sweden. J. Med. Entomol. 1990, 27, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Turell, M.J. Members of the Culex pipiens complex as vectors of viruses. J. Am. Mosq. Control Assoc. 2012, 28, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Sane, J.; Kurkela, S.; Putkuri, N.; Huhtamo, E.; Vaheri, A.; Vapalahti, O. Complete coding sequence and molecular epidemiological analysis of Sindbis virus isolates from mosquitoes and humans, Finland. J. Gen. Virol. 2012, 93, 1984–1990. [Google Scholar] [CrossRef]
- Tingström, O.; Lwande, O.; Näslund, J.; Spyckerelle, I.; Engdahl, C.; Von Schoenberg, P.; Ahlm, C.; Evander, M.; Bucht, G. Detection of Sindbis and Inkoo virus RNA in genetically typed mosquito larvae sampled in northern Sweden. Vector Borne Zoonotic Dis. 2016, 16. [Google Scholar] [CrossRef] [Green Version]
- Korhonen, E.M.; Suvanto, M.T.; Uusitalo, R.; Faolotto, G.; Smura, T.; Sane, J.; Vapalahti, O.; Huhtamo, E. Sindbis virus strains of divergent origin isolated from humans and mosquitoes during a recent outbreak in Finland. Vector Borne Zoonotic Dis. 2020, 20, 843–849. [Google Scholar] [CrossRef]
- Becker, N.; Petrić, D.; Zgomba, M.; Boase, C.; Madon, M.; Dahl, C.; Kaiser, A. Mosquitoes and Their Control, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3–8. [Google Scholar] [CrossRef]
- Lwande, O.W.; Näslund, J.; Lundmark, E.; Ahlm, K.; Ahlm, C.; Bucht, G.; Evander, M. Experimental infection and transmission competence of Sindbis virus in Culex torrentium and Culex pipiens mosquitoes from Northern Sweden. Vector Borne Zoonotic Dis. 2019, 19, 128–133. [Google Scholar] [CrossRef] [Green Version]
- Bergman, A.; Dahl, E.; Lundkvist, Å.; Hesson, J.C. Sindbis virus infection in non-blood-fed hibernating Culex pipiens mosquitoes in Sweden. Viruses 2020, 12, 1441. [Google Scholar] [CrossRef] [PubMed]
- Reiter, P. Weather, vector biology, and arboviral recrudescence. In The Arboviruses: Epidemiology and Ecology, 1st ed.; CRC Press: Boca Raton, FL, USA, 1988; pp. 245–256. [Google Scholar]
- Brady, O.J.; Johansson, M.A.; Guerra, C.A.; Bhatt, S.; Golding, N.; Pigott, D.M.; Delatte, H.; Grech, M.G.; Leisnham, P.T.; Maciel-de-Freitas, R.; et al. Modelling adult Aedes aegypti and Aedes albopictus survival at different temperatures in laboratory and field settings. Parasites Vectors 2013, 6, 351. [Google Scholar] [CrossRef] [Green Version]
- ECDC. European Centre for Disease Prevention and Control. Vector-Borne Diseases. 2020. Available online: https://www.ecdc.europa.eu/en/climate-change/climate-change-europe/vector-borne-diseases (accessed on 18 December 2020).
- Reisen, W.K.; Fang, Y.; Martinez, V.M. Effects of temperature on the transmission of West Nile virus by Culex tarsalis (Diptera: Culicidae). J. Med. Entomol. 2006, 43, 309–317. [Google Scholar] [CrossRef] [Green Version]
- Tian, H.; Bi, P.; Cazelles, B.; Zhou, S.; Huang, S.; Jing, Y.; Pei, Y.; Wu, X.; Fu, S.; Tong, S.; et al. How environmental conditions impact mosquito ecology and Japanese encephalitis: An eco-epidemiological approach. Environ. Int. 2015, 79, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Poh, K.C.; Chaves, L.F.; Reyna-Nava, M.; Roberts, C.M.; Fredregill, C.; Bueno, R.; Debboun, M.; Hamer, G.L. The influence of weather and weather variability on mosquito abundance and infection with West Nile virus in Harris County, Texas, USA. Sci. Total Environ. 2019, 675, 260–272. [Google Scholar] [CrossRef]
- Morin, C.W.; Comrie, A.C. Regional and seasonal response of a West Nile virus vector to climate change. Proc. Natl. Acad. Sci. USA 2013, 110, 15620–15625. [Google Scholar] [CrossRef] [Green Version]
- Trawinski, P.R.; Mackay, D.S. Identification of environmental covariates of West Nile virus vector mosquito population abundance. Vector Borne Zoonotic Dis. 2010, 10, 515–526. [Google Scholar] [CrossRef]
- Jalava, K.; Sane, J.; Ollgren, J.; Ruuhela, R.; Rätti, O.; Kurkela, S.; Helle, P.; Hartonen, S.; Pirinen, P.; Vapalahti, O.; et al. Climatic, ecological and socioeconomic factors as predictors of Sindbis virus infections in Finland. Epidemiol. Infect. 2012, 141, 1–10. [Google Scholar] [CrossRef]
- Hesson, J.; Rettich, F.; Merdic, E.U.H.; Vignjević, G.; Östman, Ö.; Schäfer, M.; Schaffner, F.; Foussadier, R.; Besnard, G.; Medlock, J.; et al. The arbovirus vector Culex torrentium is more prevalent than Culex pipiens in northern and central Europe. Med. Vet. Entomol. 2014, 28, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Culverwell, C.L.; Uusitalo, R.J.; Korhonen, E.M.; Vapalahti, O.P.; Huhtamo, E.; Harbach, R.E. The mosquitoes of Finland: Updated distributions and bionomics. Med. Vet. Entomol. 2021, 35, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Barbet-Massin, M.; Jiguet, F.; Albert, C.; Thuiller, W. Selecting pseudo-absences for species distribution models: How, where and how many? Methods Ecol. Evol. 2012, 3, 327–338. [Google Scholar] [CrossRef]
- Helle, P.; Ikonen, K.; Kantola, A. Wildlife monitoring in Finland-on-line information to game administration, hunters and the wider public. Can. J. For. Res. 2016, 46. [Google Scholar] [CrossRef]
- Venäläinen, A.; Laapas, M.; Pirinen, P.; Horttanainen, M.; Hyvönen, R.; Lehtonen, I.; Junila, P.; Hou, M.; Peltola, H. Estimation of the high-spatial-resolution variability in extreme wind speeds for forestry applications. Earth Syst. Dyn. 2017, 8, 529–545. [Google Scholar] [CrossRef] [Green Version]
- Aalto, J.; Pirinen, P.; Jylhä, K. New gridded daily climatology of Finland: Permutation-based uncertainty estimates and temporal trends in climate. J. Geophys. Res. Atmos. 2016, 121. [Google Scholar] [CrossRef] [Green Version]
- Pirinen, P.; Simola, H.; Aalto, J.; Kaukoranta, J.; Karlsson, P.; Ruuhela, R. Tilastoja Suomen Ilmastosta 1981–2010 (Climatological Statistics of Finland 1981–2010); Finnish Meteorological Institute: Helsinki, Finland, 2012. [Google Scholar]
- SYKE, Finnish Environment Institute. Corine Maanpeite 2018. 2020. Available online: https://ckan.ymparisto.fi/dataset/%7B0B4B2FAC-ADF1-43A1-A829-70F02BF0C0E5%7D (accessed on 17 December 2020).
- Statistics Finland. 2020. Available online: https://tilastokeskus-kartta.swgis.fi/# (accessed on 17 September 2020).
- Salmivaara, A.; Launiainen, S.; Tuominen, S.; Ala-Ilomäki, J.; Finér, L. Topographic Wetness Index for Finland; Natural Resources Institute Finland: Helsinki, Finland, 2017; Available online: http://urn.fi/urn:nbn:fi:csc-kata20170511114638598124 (accessed on 10 May 2020).
- NLS of Finland, National Land Survey of Finland. Digital Elevation Model 2019. 10 m x 10 m. Available online: http://urn.fi/urn:nbn:fi:csc-kata00001000000000000622 (accessed on 10 May 2020).
- Fick, S.; Hijmans, R. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37. [Google Scholar] [CrossRef]
- Didan, K. MOD13A3 MODIS/Terra Vegetation Indices Monthly L3 Global 1 km SIN Grid V006. NASA EOSDIS Land Processes DAAC. 2015. [Google Scholar]
- Wan, Z.; Hook, S.; Hulley, G. MYD11C3 MODIS/Aqua Land Surface Temperature/Emissivity Monthly L3 Global 0.05Deg CMG V006 [Data set]. NASA EOSDIS Land Processes DAAC. 2015. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: http://www.R-project.org/ (accessed on 7 June 2020).
- Thuiller, W.; Lafourcade, B.; Engler, R.; Araújo, M. BIOMOD-A platform for ensemble forecasting of species distributions. Ecography 2009, 32, 369–373. [Google Scholar] [CrossRef]
- Thuiller, W.; Georges, D.; Engler, R. Biomod2: Ensemble Platform for Species Distribution Modelling. 2014. R Package Version 3.1-64. Available online: https://mran.microsoft.com/snapshot/2014-10-20/web/packages/biomod2/biomod2.pdf (accessed on 2 February 2020).
- Kruijff, M.; Hendrickx, G.; Wint, W.; Ginati, A. Mapping habitats for vectors of infectious disease: VECMAP. In Proceedings of the International Astronautical Congress, Cape Town, South Africa, 3–7 October 2011; Available online: https://business.esa.int/sites/default/files/KruijffIAC-11-B5%201%2010%20-%20VECMAP%20-%20v2.2.pdf (accessed on 18 September 2020).
- Besley, D.A.; Kuh, E.; Welsh, R.E. Regression Diagnostics: Identifying Influential Data and Sources of Collinearity; Wiley & Sons, Inc.: New York, NY, USA, 1980. [Google Scholar]
- Naimi, B.; Hamm, N.A.S.; Groen, T.A.; Skidmore, A.K.; Toxopeus, A.G. Where is positional uncertainty a problem for species distribution modelling? Ecography 2014, 37, 191–203. [Google Scholar] [CrossRef]
- Dormann, C.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Diekötter, T.; García Márquez, J.; Gruber, B.; Lafourcade, B.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- McCullagh, P.; Nelder, J.A. Generalized Linear Models, 2nd ed.; Chapman & Hall: London, UK, 1989. [Google Scholar]
- Hastie, T.J.; Tibshirani, R. Generalized Additive Models, 1st ed.; Chapman and Hall: London, UK, 1990. [Google Scholar]
- Breiman, L.; Friedman, J.; Stone, C.J.; Olshen, R.A. Classification and Regression Trees, 1st ed.; Chapman and Hall: London, UK, 1984. [Google Scholar]
- Ripley, B.D. Pattern Recognition and Neural Networks; Cambridge University Press: Cambridge, UK, 1996. [Google Scholar] [CrossRef]
- Friedman, J.H. Multivariate adaptive regression splines. Ann. Statist. 1991, 19, 1–67. [Google Scholar] [CrossRef]
- Ridgeway, G. The state of boosting. Comput. Sci. Stat. 1999, 31, 172–181. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Phillips, S.; Dudík, M.; Schapire, R. A maximum entropy approach to species distribution modeling. In Proceedings of the Twenty-First International Conference on Machine Learning (ICML 2004), Banff, AB, Canada, 4–8 July 2004; p. 83. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Gao, F. A nested-loop fisher discriminant analysis algorithm. Chemom. Intellig. Lab. Syst. 2015, 146, 396–406. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.; Anderson, R.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.; Huettmann, F.; Leathwick, J.; Lehmann, A.; et al. Novel methods improve prediction of species’ distributions from occurence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef] [Green Version]
- Pearson, R.; Thuiller, W.; Araújo, M.; Martínez-Meyer, E.; Brotons, L.; Mcclean, C.; Miles, L.; Segurado, P.; Dawson, T.; Lees, D. Model-based uncertainty in species range prediction. J. Biogeogr. 2006, 33, 174–1711. [Google Scholar] [CrossRef]
- Wood, S.N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. Ser. B 2011, 73, 3–36. [Google Scholar] [CrossRef] [Green Version]
- Guisan, A.; Zimmermann, N. Predictive habitat distribution models in ecology. Ecol. Model. 2000, 135, 147–186. [Google Scholar] [CrossRef]
- Fielding, A.H.H. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. J. 1997, 24, 38–49. [Google Scholar] [CrossRef]
- Ruete, A.; Leynaud, G. Goal-oriented evaluation of species distribution models’ accuracy and precision: True skill statistic profile and uncertainty maps. PeerJ Prepr. 2015, 3, e1478. [Google Scholar] [CrossRef] [Green Version]
- Akhtar, M.; Kraemer, M.U.G.; Gardner, L.M. A dynamic neural network model for predicting risk of zika in real time. BMC Med. 2019, 17, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araújo, M.B.; New, M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Sintayehu, D.W.; Tassie, N.; De Boer, W.F. Present and future climatic suitability for dengue fever in Africa. Infect. Ecol. Epidemiol. 2020, 10, 1782042. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y. Applying various algorithms for species distribution modelling. Integr. Zool. 2013, 8, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Lippi, C.; Gaff, H.; White, A.; John, H.; Richards, A.; Ryan, S. Exploring the niche of Rickettsia montanensis (Rickettsiales: Rickettsiaceae) infection of the American dog tick (Acari: Ixodidae), using multiple species distribution model approaches. J. Med. Entomol. 2021, 58, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Pearce, J.L.; Boyce, M.S. Modelling distribution and abundance with presence-only data. J. Appl. Ecol. 2006, 43, 405–412. [Google Scholar] [CrossRef]
- Lundström, J.; Turell, M.; Niklasson, B. Viremia in three orders of birds (anseriformes, galliformes and passeriformes) inoculated with Ockelbo virus. J. Wildl. Dis. 1993, 29, 189–195. [Google Scholar] [CrossRef] [Green Version]
- Laurila, T.K.; Sinclair, V.A.; Gregow, H. Climatology, variability, and trends in near-surface wind speeds over the North Atlantic and Europe during 1979–2018 based on ERA5. Int. J. Climatol. 2021, 41, 2253–2278. [Google Scholar] [CrossRef]
- Lembrechts, J.J.; Lenoir, J.; Roth, N.; Hattab, T.; Milbau, A.; Haider, S.; Pellissier, L.; Pauchard, A.; Ratier Backes, A.; Dimarco, R.D.; et al. Comparing temperature data sources for use in species distribution models: From in-situ logging to remote sensing. Global Ecol. Biogeogr. 2019, 28, 1578–1596. [Google Scholar] [CrossRef]
- Mohrig, W. Die Culiciden Deutschlands. Untersuchungen zur taxonomie, biologie und ökologie der einheimischen stechmücken. Parasitol. Schr. 1969, 18, 1–260. [Google Scholar]
- Harbach, R.E. Culex pipiens: Species versus species complex taxonomic history and perspective. J. Am. Mosq. Control Assoc. 2012, 28, 10–23. [Google Scholar] [CrossRef]
- Medlock, J.M.; Snow, K.R.; Leach, S. Potential transmission of West Nile virus in the British Isles: An ecological review of candidate mosquito bridge vectors. Med. Vet. Entomol. 2005, 19, 2–21. [Google Scholar] [CrossRef] [PubMed]
- Medlock, J.M.; Vaux, A.G.C. Seasonal dynamics and habitat specificity of mosquitoes in an English wetland: Implications for UK wetland management and restoration. J. Vector Ecol. 2015, 40, 90–106. [Google Scholar] [CrossRef]
- Finnish Museum of Natural History. Finnish Breeding Bird Atlas. 2020. Available online: http://atlas3.lintuatlas.fi/results/species/willow%20grouse (accessed on 17 December 2020).
- NRIF. Natural Resources Institute of Finland. Metsäkanalintujen Kannat Edelleen Kasvussa. 2019. Available online: https://www.luke.fi/uutinen/metsakanalintujen-kannat-edelleen-kasvussa/ (accessed on 17 December 2020).
- Lindström, J.; Ranta, E.; Kaitala, V.; Lindén, H. The clockwork of Finnish tetraonid population dynamics. Oikos 1995, 74, 185–194. [Google Scholar] [CrossRef]
- Ernek, E.; Kožuch, O.; Nosek, J.; Labuda, M. Evidence for circulation of Sindbis virus and other arboviruses by using sentinel animals in western Slovakia. Intervirology 1973, 2, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Lundström, J.; Schäfer, M.; Hesson, J.; Blomgren, E.; Lindström, A.; Wahlqvist, P.; Halling, A.; Hagelin, A.; Ahlm, C.; Evander, M.; et al. The geographic distribution of mosquito species in Sweden. J. Am. Mosq. Control Assoc. 2013, 31, 35. [Google Scholar]
- Ciota, A.T.; Keyel, A.C. The role of temperature in transmission of zoonotic arboviruses. Viruses 2019, 11, 1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Data Source | Data Layer(s) | Modifications | Year | Spatial Resolution | References |
|---|---|---|---|---|---|
| FMI | Wind speed 50 years return interval (m/s) | Calculated mean wind speed 50 y interval per municipality. | 1979–2015 | 20 m | [50] |
| FMI | Mean monthly air temperature (°C) | Calculated mean monthly temperature per municipality in 2000–2019 in July–September, October–February and March–June. Air temperature measurement height was 2 m (FMI). | 2000–2019 | 1000 m | [51] |
| FMI | Mean monthly precipitation (mm) | Calculated mean monthly precipitation per municipality in 2000–2019 in July–September, October–February and March–June. | 2000–2019 | 1000 m | [51] |
| FMI | Mean monthly snow depth (cm) | Calculated mean monthly snow depth per municipality in 2000–2019 in October–November, December–February and March–April. | 2000–2019 | 1000 m | [51] |
| FMI | Mean precipitation during growing season (mm) | Calculated mean precipitation during growing season per municipality. | averages for 1981–2010 | 1000 m | [52] |
| FMI | Mean heat summation during growing season (°C day) | Calculated mean heat summation during growing season per municipality. | averages for 1981–2010 | 1000 m | [52] |
| FMI | Growing season length (GLS) (day) | Calculated growing season length (GLS) per municipality. | averages for 1981–2010 | 1000 m | [52] |
| LUKE | Density (individuals/km2) of willow grouse (Lagopus lagopus), black grouse (Lyrurus tetrix), capercaillie (Tetrao urogallus) and hazel grouse (Tetrastes bonasia) | Average annual densities at a 100 km radius from the municipality center, further averaged over the years. Based on wildlife triangle census. | 2000–2019 | Municipality | [49] |
| SYKE, EEA, EU/Copernicus programme | CORINE land cover 2018 | Euclidean distances to selected land cover types from mosquito species PA point were calculated in ArcGIS. Proportion (%) of chosen land cover types were derived by calculating percentage of each land cover type for municipality. | 2018 | 20 m | [53] |
| Statistics Finland | Human population density (persons/km2) | Calculated as sum per municipality. | 2019 | 1000 m | [54] |
| Statistics Finland | Summer cottage density (cottages/km2) | Calculated in ArcGIS in order to present summer cottages per area (km2) of municipality. | 2019 | Municipality | [54] |
| Kurkela et al. 2008 | Mean seroprevalence of SINV in human population in Finland | Seroprevalence rate was taken from the hospital district in which municipality belongs to. | 1999–2003 | Hospital districts | [23] |
| NLS of Finland | Topographic wetness index (TWI) | Calculated mean TWI per municipality. | 2016 | 16 × 16 m | [55] |
| NLS of Finland | Digital elevation model (m) | Calculated mean elevation per municipality. | 2019 | 10 × 10 m | [56] |
| WorldClim- Global climate data | Solar radiation (kJ m−2 day−1) | Calculated mean solar radiation in May–September. | averages for 1980–2000 | ~1000 m | [57] |
| WorldClim- Global climate data | Water vapor pressure (kPa) | Calculated mean water vapor pressure in May–September. | averages for 1980–2000 | ~1000 m | [57] |
| NASA Earthdata | Normalized Difference Vegetation Index (NDVI) (MOD13A3) | Calculated mean NDVI in June 2000–2019 per municipality. | in June, 2000–2019 | 1000 m | [58] |
| NASA Earthdata | Land surface temperature (°C) (MOD11C3) | Calculated mean monthly LST per municipality in 2000–2019 April–May, June–August and September–October | 2000–2019 | 5600 m | [59] |
| Lorna Culverwell, Jenny Hesson | The distribution data of Cs. morsitans, Cx.pipiens/torrentium and Ae. cinereus/geminus | The occurrence data of Cx. pipiens and Cx. torrentium, and Ae. cinereus and Ae. geminus, were unified to Cx. pipiens/torrentium and Ae. cinereus/geminus due to the lack of reliable identification methods to distinguish them to either of the species. | 2009–2018 | Location (Longitude, Latitude) | [46,47] |
| THL, NIDR | Patient Pogosta disease data | Pogosta disease average incidence per 1000 inhabitants during 2000–2019 per municipality was calculated in ArcGIS. | 2000–2019 | Municipality | [22] |
| (a) | Ae. cinereus/geminus | Cx. pipiens/torrentium | Cs. morsitans | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Predictor | Min. | Max. | Avg. | Min. | Max. | Avg. | Min. | Max. | Avg. |
| Wind speed 50 years interval | 10.5 | 41.3 | 14.4 | 10.5 | 14.6 | 14.6 | 10.5 | 36.9 | 14.6 |
| Topographic wetness index (TWI) | 4269 | 65,535 | 16,708 | 4617 | 65,535 | 15,261 | 4617 | 65,535 | 15,261 |
| Mean snow depth in October–November 2009–2019 | x | x | x | 0.2 | 12.4 | 2.3 | 0.2 | 12.6 | 2.3 |
| Mean precipitation in October–February 2009–2019 | 26.3 | 68.8 | 47.4 | 26.3 | 49 | 49 | 26.8 | 68.8 | 49 |
| Mean precipitation in March–June 2009–2019 | 33.5 | 54.7 | 42.2 | 33.5 | 54.3 | 41.9 | 33.5 | 54.3 | 41.9 |
| Mean precipitation in July–September 2009–2019 | 55.8 | 85.3 | 67.5 | 54.7 | 84.7 | 67.1 | 56.6 | 84.7 | 67.1 |
| Mean normalized difference vegetation index (NDVI) in June 2009–2019 | 0.4 | 0.8 | 0.7 | 0.4 | 0.8 | 0.7 | 0.5 | 0.8 | 0.7 |
| Mean water vapor pressure in May–September 2009–2019 | x | x | x | 0.8 | 1.21 | 1.04 | x | x | x |
| Human population density | 0 | 4281 | 141.9 | 0 | 4281 | 73.4 | 0 | 4281 | 73.4 |
| Euclidean distance to water courses | 0 | 19,194.3 | 2485.6 | 0 | 20,634.4 | 2884.2 | 0 | 19,194.3 | 2884.2 |
| Euclidean distance to water bodies | 0 | 4628.9 | 665.8 | 0 | 4628.9 | 757.1 | 0 | 4628.9 | 757.1 |
| Euclidean distance to peatbogs | 0 | 4891.8 | 1021.5 | 20 | 4303.6 | 1093.1 | 0 | 4891.8 | 1093.1 |
| Euclidean distance to inland marshes | 0 | 12,041.4 | 2155.2 | 0 | 12,041.4 | 2182.8 | 44.7 | 12,041.4 | 2182.8 |
| Euclidean distance to coniferous forest | 0 | 2272.7 | 98.2 | 0 | 3217.6 | 76.3 | 0 | 570.1 | 76.3 |
| Euclidean distance to broad-leaved forest | 0 | 2062.4 | 201.6 | 0 | 906.9 | 170.4 | 0 | 1063.2 | 170.4 |
| Euclidean distance to mixed forest | 0 | 1724.1 | 136.6 | 0 | 3265 | 119.3 | 0 | 1668.8 | 119.3 |
| Euclidean distance to transitional woodland/shrub | 0 | 1073.6 | 141.9 | 0 | 1073.6 | 135.3 | 0 | 738.2 | 135.3 |
| Mean land surface temperature in June–August 2009–2019 | 13.3 | 24 | 18.4 | 12.4 | 23.9 | 18.7 | 14.7 | 22.6 | 18.7 |
| Mean precipitation during growth season 1981–2010 | 93.1 | 190.3 | 157.8 | 184.1 | 378.5 | 326.5 | 101.9 | 190.3 | 162.6 |
| Mean solar radiation in May–September | 13,718 | 17,526 | 15,460 | x | x | x | 13,639 | 17,526 | 15,662 |
| Elevation | 0.1 | 116.5 | 564.6 | x | x | x | x | x | x |
| (b) | Pogosta disease | ||||||||
| Predictor | Min. | Max. | Avg. | ||||||
| Black grouse density | 0 | 11.6 | 6.9 | ||||||
| Capercaillie density | 0 | 4.9 | 3.2 | ||||||
| Hazel grouse density | 0 | 10 | 7.3 | ||||||
| Willow grouse density | 0 | 3.3 | 0.3 | ||||||
| Elevation (m) | 4.9 | 471.3 | 94.2 | ||||||
| Human population density (persons/km2) | 0 | 1177 | 36.7 | ||||||
| Percentage of inland wetlands (%) | 0 | 0.9 | 0.1 | ||||||
| Percentage of mixed forest in mineral soil (%) | 0 | 20.9 | 10.4 | ||||||
| Percentage of mixed forest in peatlands (%) | 0 | 9.3 | 2 | ||||||
| Percentage of mixed forest in rocky soil (%) | 0 | 0.7 | 0.1 | ||||||
| Percentage of lakes (%) | 0 | 54.4 | 8.4 | ||||||
| Mean precipitation in July–September in 2000–2019 (mm) | 54.8 | 82.8 | 68.2 | ||||||
| Mean seroprevalence of SINV in human population | 0 | 9.9 | 3.5 | ||||||
| Summer cottage density | 0.1 | 9.1 | 1.8 | ||||||
| Topographic wetness index (TWI) | 7751 | 65,535 | 17,062 | ||||||
| Suitability for Cx. pipiens/torrentium (%) | 10.6 | 78.7 | 41.3 | ||||||
| Risk for Cs. morsitans (%) | 8.3 | 85.9 | 36.2 | ||||||
| Percentage of peatbogs (%) | 0 | 19.1 | 2.1 | ||||||
| Risk for Ae. cinereus/geminus (%) | 8.5 | 78.7 | 50.9 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uusitalo, R.; Siljander, M.; Culverwell, C.L.; Hendrickx, G.; Lindén, A.; Dub, T.; Aalto, J.; Sane, J.; Marsboom, C.; Suvanto, M.T.; et al. Predicting Spatial Patterns of Sindbis Virus (SINV) Infection Risk in Finland Using Vector, Host and Environmental Data. Int. J. Environ. Res. Public Health 2021, 18, 7064. https://doi.org/10.3390/ijerph18137064
Uusitalo R, Siljander M, Culverwell CL, Hendrickx G, Lindén A, Dub T, Aalto J, Sane J, Marsboom C, Suvanto MT, et al. Predicting Spatial Patterns of Sindbis Virus (SINV) Infection Risk in Finland Using Vector, Host and Environmental Data. International Journal of Environmental Research and Public Health. 2021; 18(13):7064. https://doi.org/10.3390/ijerph18137064
Chicago/Turabian StyleUusitalo, Ruut, Mika Siljander, C. Lorna Culverwell, Guy Hendrickx, Andreas Lindén, Timothée Dub, Juha Aalto, Jussi Sane, Cedric Marsboom, Maija T. Suvanto, and et al. 2021. "Predicting Spatial Patterns of Sindbis Virus (SINV) Infection Risk in Finland Using Vector, Host and Environmental Data" International Journal of Environmental Research and Public Health 18, no. 13: 7064. https://doi.org/10.3390/ijerph18137064
APA StyleUusitalo, R., Siljander, M., Culverwell, C. L., Hendrickx, G., Lindén, A., Dub, T., Aalto, J., Sane, J., Marsboom, C., Suvanto, M. T., Vajda, A., Gregow, H., Korhonen, E. M., Huhtamo, E., Pellikka, P., & Vapalahti, O. (2021). Predicting Spatial Patterns of Sindbis Virus (SINV) Infection Risk in Finland Using Vector, Host and Environmental Data. International Journal of Environmental Research and Public Health, 18(13), 7064. https://doi.org/10.3390/ijerph18137064







