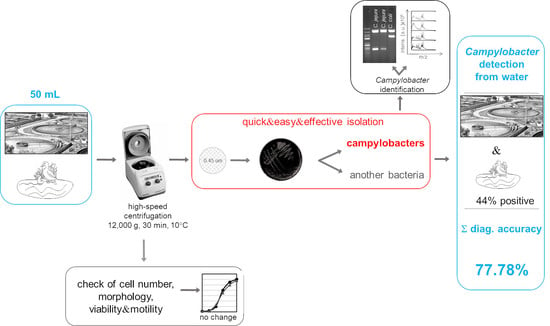
A Rapid Culture Method for the Detection of Campylobacter from Water Environments
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Campylobacters from Poorly Filterable Water Samples by the Standard Cultivation Method
2.3. Novel Cultivation Method for Campylobacters from Poorly Filterable Water Samples
2.4. Campylobacter Species Identification
2.5. Campylobacter Cell Numbers
2.6. Campylobacter Viability
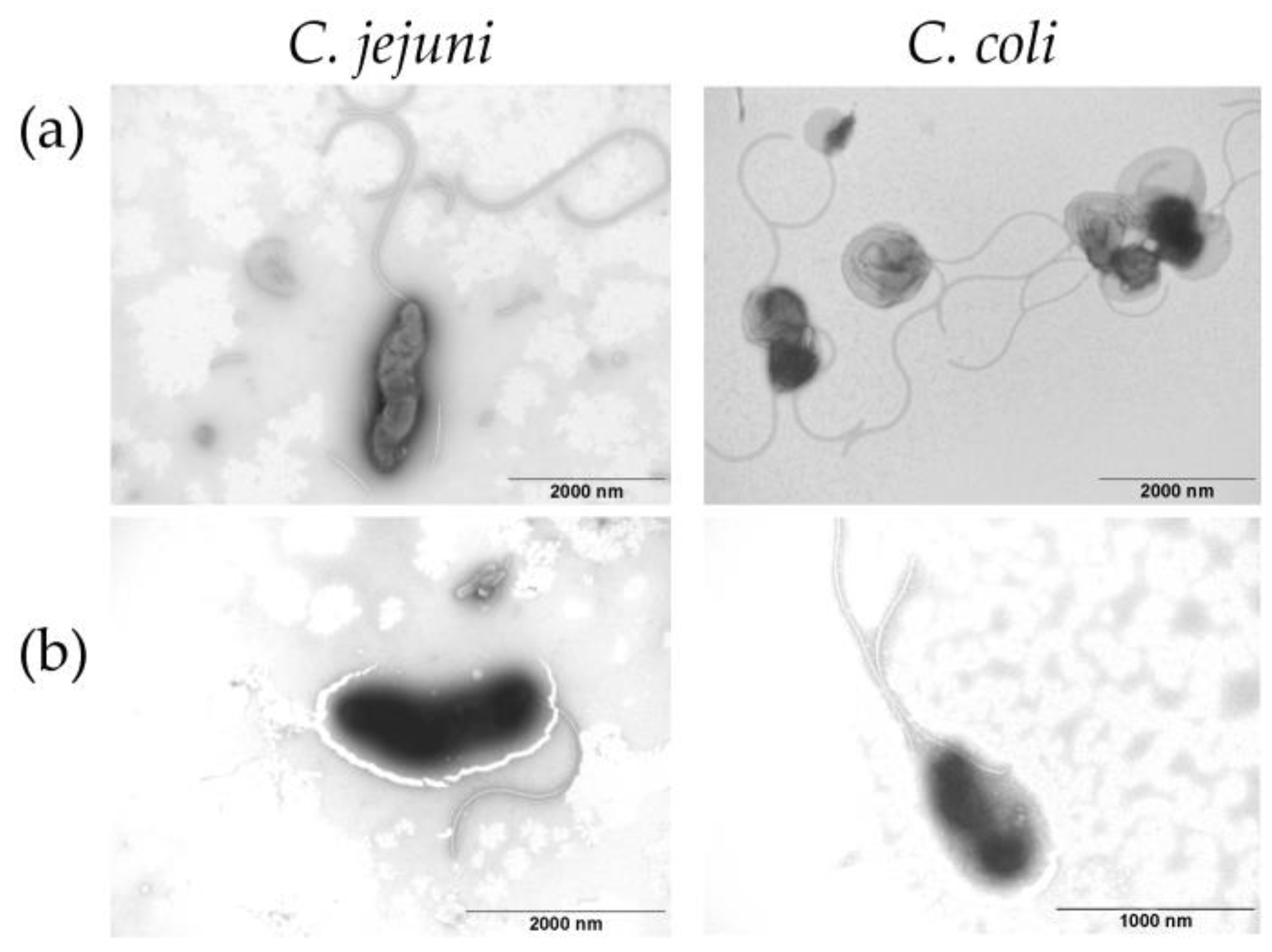
2.7. Campylobacter Morphology
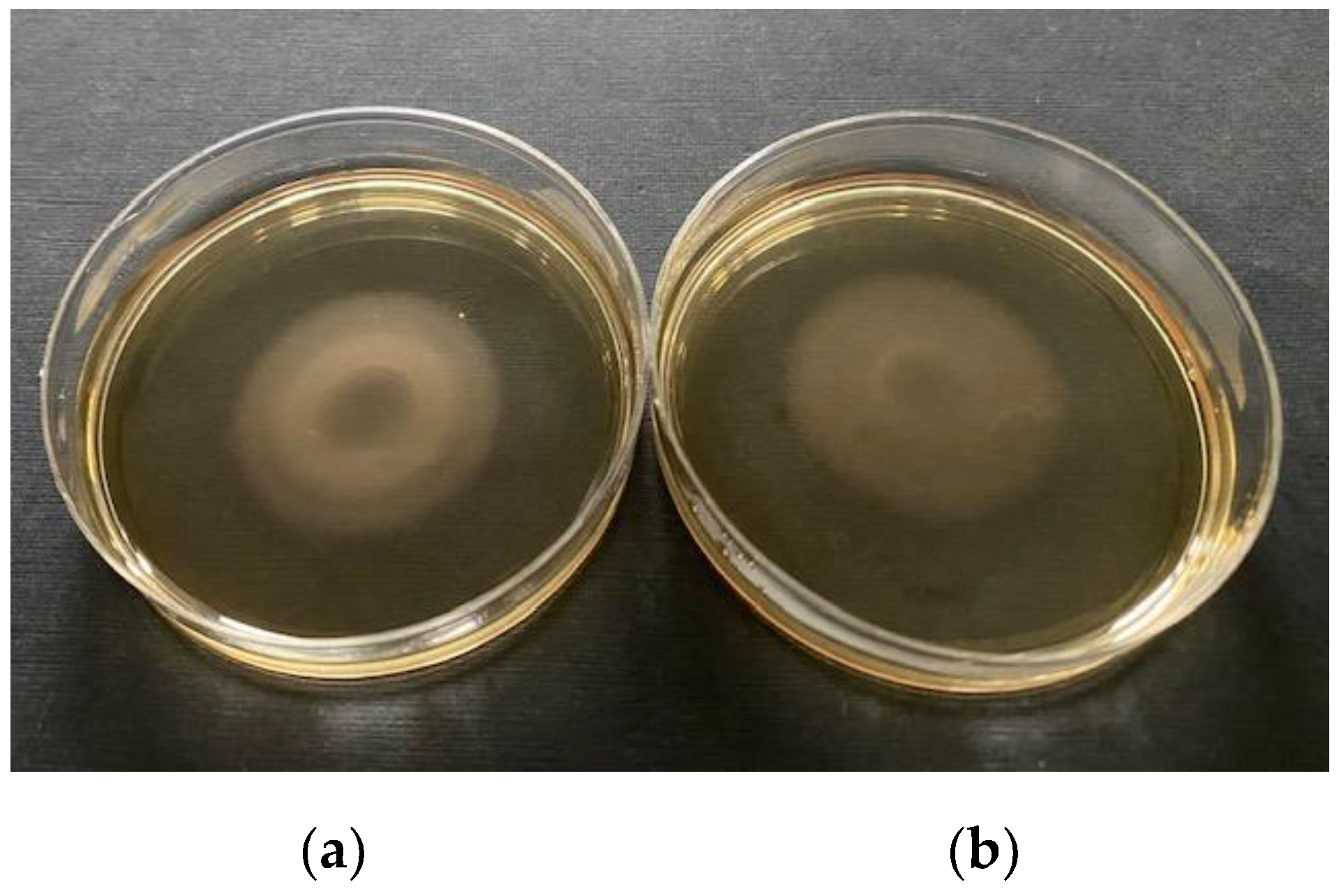
2.8. Campylobacter Motility
2.9. Evaluation of Selectivity of the Novel Culture-Based Method
2.10. Measurement of Diagnostic Accuracy of the Two Methods
3. Results
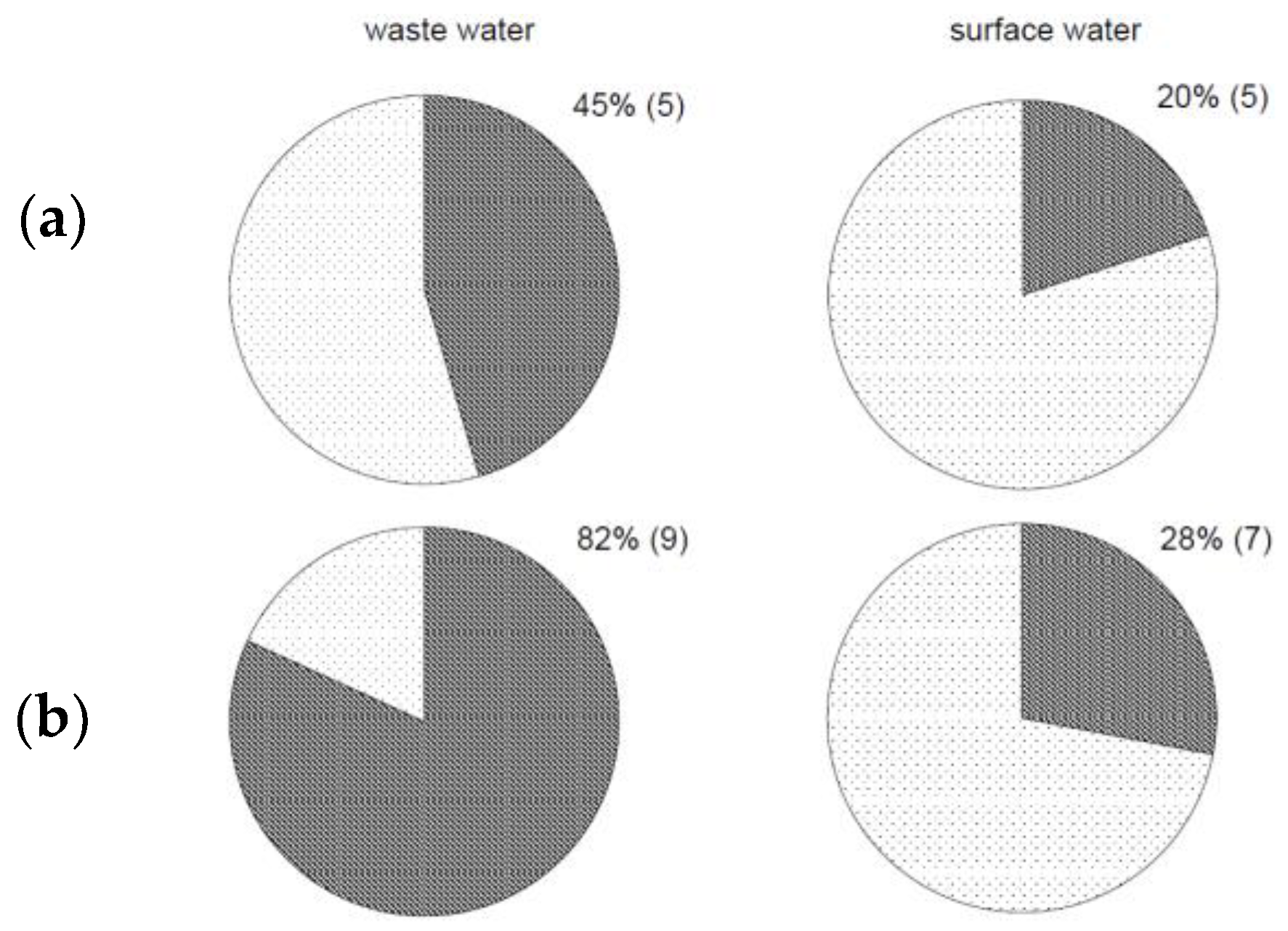
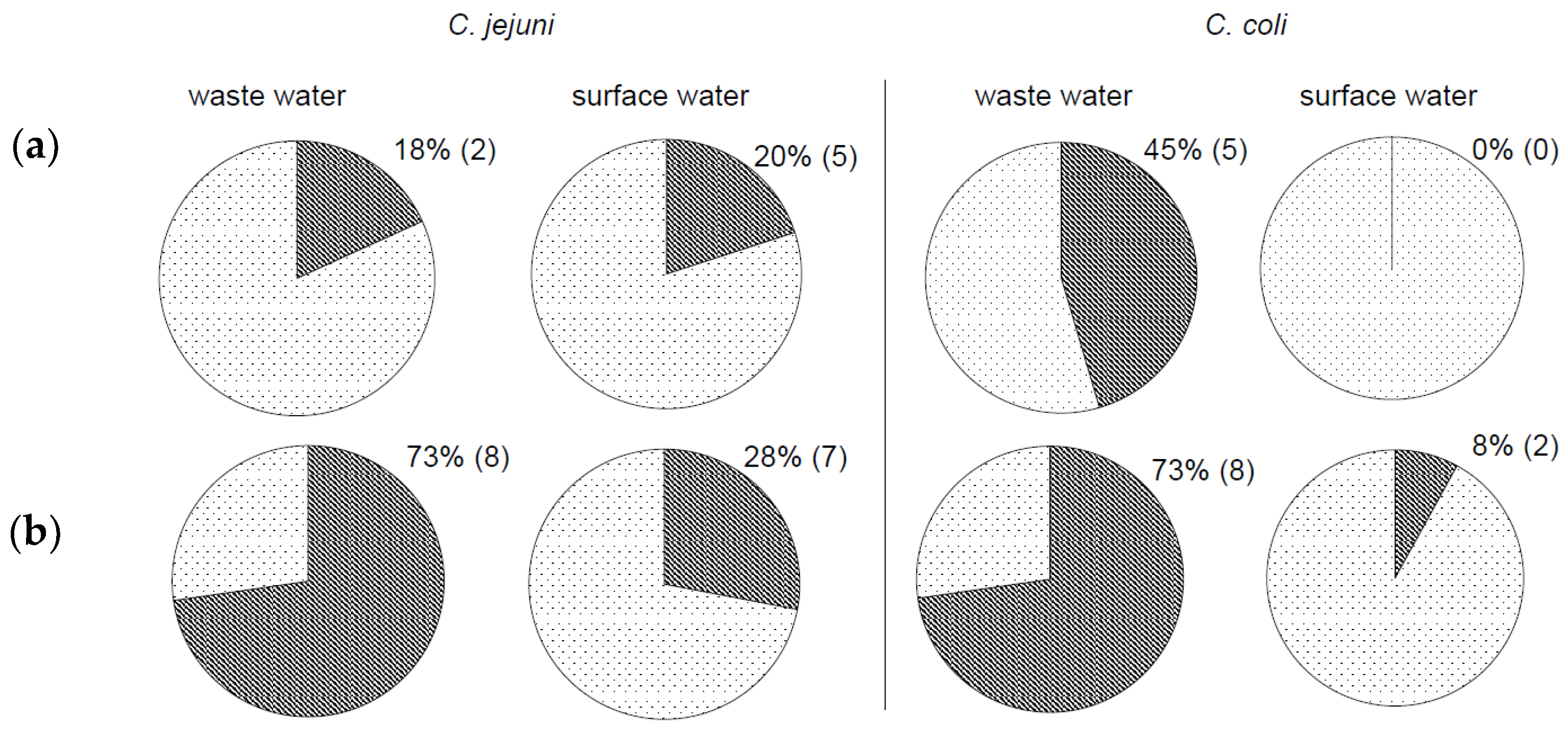
3.1. Detection of Campylobacter in Poorly Filterable Water Samples
3.2. Comparison of Effectiveness between Standard and Modified Methods
3.3. Evaluation of the Novel Method
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campylobacter Sebald and Véron 1963 (Approved Lists 1980). Available online: https://lpsn.dsmz.de/genus/campylobacter (accessed on 3 October 2020).
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef]
- Igwaran, A.; Okoh, A.I. Human campylobacteriosis: A public health concern of global importance. Heliyon 2019, 5, e02814. [Google Scholar] [CrossRef]
- Ghosh, R.; Uppal, B.; Aggarwal, P.; Chakravarti, A.; Jha, A.K.; Dubey, A.P. A comparative study of conventional and molecular techniques in diagnosis of Campylobacter gastroenteritis in children. Ann. Clin. Lab. Sci. 2014, 44, 42–48. [Google Scholar] [PubMed]
- Koga, M.; Ang, C.W.; Yuki, N.; Jacobs, C.; Herbrink, P.; Van Der Meché, G.A.; Hirata, K.; Van Doorn, P.A. Comparative study of preceding Campylobacter jejuni infection in Guillain-Barré syndrome in Japan and The Netherlands. J. Neurol. Neurosurg. Psychiatry 2001, 70, 693–695. [Google Scholar] [CrossRef]
- Nyati, K.K.; Nyati, R. Role of Campylobacter jejuni infection in the pathogenesis of Guillain-Barré syndrome: An update. BioMed Res. Int. 2013, 2013, 852195. [Google Scholar] [CrossRef] [PubMed]
- Young, K.Y.; Davis, L.M.; DiRita, V.J. Campylobacter jejuni: Molecular biology and pathogenesis. Nat. Rev. Microbiol. 2007, 5, 665–679. [Google Scholar] [CrossRef]
- Burnham, P.M.; Hendrixson, D.R. Campylobacter jejuni: Collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol. 2018, 16, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Pattis, I.; Moriarty, E.; Billington, C.; Gilpin, B.; Hodson, R.; Ward, N. Concentrations of Campylobacter spp., Escherichia coli, Enterococci, and Yersinia spp. in the Feces of Farmed Red Deer in New Zealand. J. Environ. Qual. 2017, 46, 819–827. [Google Scholar] [CrossRef]
- Hald, B.; Skov, M.N.; Nielsen, E.M.; Rahbek, C.; Madsen, J.J.; Wainø, M.; Chriél, M.; Nordentoft, S.; Baggesen, D.L.; Madsen, M. Campylobacter jejuni and Campylobacter coli in wild birds on Danish livestock farms. Acta Vet. Scand. 2016, 58, 11. [Google Scholar] [CrossRef]
- Moore, J.; Corcoran, D.; Dooley, J.; Fanning, S.; Lucey, B.; Matsuda, M.; McDowell, D.; Mégraud, F.; Millar, B.; O’Mahony, R.; et al. Campylobacter . Vet. Res. 2005, 36, 351–382. [Google Scholar] [CrossRef]
- Nag, R.; Whyte, P.; Markey, B.K.; O’Flaherty, V.; Bolton, D.; Fenton, O.; Richards, K.G.; Cummins, E. Ranking hazards pertaining to human health concerns from land application of anaerobic digestate. Sci. Total Environ. 2020, 710, 136297. [Google Scholar] [CrossRef]
- Ugarte-Ruiz, M.; Florez-Cuadrado, D.; Wassenaar, T.M.; Porrero, M.C.; Domínguez, L. Method Comparison for Enhanced Recovery, Isolation and Qualitative Detection of C. jejuni and C. coli from Wastewater Effluent Samples. Int. J. Environ. Res. Public Health 2015, 12, 2749–2764. [Google Scholar] [CrossRef] [PubMed]
- Koenraad, P.M.F.J.; Rombouts, F.M.; Notermans, S.H.W. Epidemiological aspects of thermophilic Campylobacter in water-related environments: A review. Water Environ. Res. 1997, 69, 52–63. [Google Scholar] [CrossRef]
- Jokinen, C.; Edge, T.A.; Ho, S.; Koning, W.; Laing, C.; Mauro, W.; Medeiros, D.; Miller, J.; Robertson, W.; Taboada, E.; et al. Molecular subtypes of Campylobacter spp., Salmonella enterica, and Escherichia coli O157:H7 isolated from faecal and surface water samples in the Oldman River watershed, Alberta, Canada. Water Res. 2011, 45, 1247–1257. [Google Scholar] [CrossRef]
- Pitkänen, T. Review of campylobacter spp. in drinking and environmental waters. J. Microbiol. Methods 2013, 95, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Itoh, S.; Zhou, L. Effect of unboiled water consumption data on sensitivity analysis in quantitative microbial risk assessment. NPJ Clean Water 2018, 1, 18. [Google Scholar] [CrossRef]
- Arimi, S.M.; Fricker, C.R.; Park, R.W.A. Occurrence of “thermophilic” campylobacters in sewage and their removal by treatment processes. Epidemiol. Infect. 1988, 101, 279–286. [Google Scholar] [CrossRef]
- Marti, E.; Jofre, J.; Balcazar, J.L. Prevalence of Antibiotic Resistance Genes and Bacterial Community Composition in a River Influenced by a Wastewater Treatment Plant. PLoS ONE 2013, 8, e78906. [Google Scholar] [CrossRef]
- Schönberg-Norio, D.; Takkinen, J.; Hänninen, M.-L.; Katila, M.-L.; Kaukoranta, S.-S.; Mattila, L.; Rautelin, H. Swimming and Campylobacter Infections. Emerg. Infect. Dis. 2004, 10, 1474–1477. [Google Scholar] [CrossRef] [PubMed]
- Rechenburg, A.; Kistemann, T. Sewage effluent as a source of Campylobacter spp. in a surface water catchment. Int. J. Environ. Health Res. 2009, 19, 239–249. [Google Scholar] [CrossRef]
- Abulreesh, H.H.; Paget, T.A.; Goulder, R. A pre-enrichment step is essential for detection of Campylobacter spp. in turbid pond water. Trop. Biomed. 2014, 31, 320–326. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardisation. Water Quality—Detection and Enumeration of Thermotolerant Campylobacter Species—ISO 17995; ISO: Geneva, Switzerland, 2005. [Google Scholar]
- Bang, D.D.; Wedderkopp, A.; Pedersen, K.; Madsen, M. Rapid PCR using nested primers of the 16s rRNA and the hippuricase (hipO) genes to detect Campylobacter jejuni and Campylobacter coli in environmental samples. Mol. Cell. Probes 2002, 16, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Linton, D.; Lawson, A.J.; Owen, R.J.; Stanley, J. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 1997, 35, 2568–2572. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Penny, C.; Ragimbeau, C.; Schets, F.M.; Blaak, H.; Duim, B.; Wagenaar, J.A.; de Boer, A.; Cauchie, H.M.; Mossong, J.; et al. Quantifying potential sources of surface water contamination with Campylobacter jejuni and Campylobacter coli. Water Res. 2016, 101, 36–45. [Google Scholar] [CrossRef]
- International Organization for Standardisation. Water Quality—Detection and Enumeration of Thermotolerant Campylobacter spp.—ISO 17995:2019; ISO: Geneva, Switzerland, 2019. [Google Scholar]
- Winters, D.K.; O’Leary, A.E.; Slavik, M.F. Polymerase chain reaction for rapid detection of Campylobacter jejuni in artificially contaminated foods. Lett. Appl. Microbiol. 1998, 27, 163–167. [Google Scholar] [CrossRef]
- Wolfe, A.J.; Berg, H.C. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. USA 1989, 86, 6973–6977. [Google Scholar] [CrossRef] [PubMed]
- Marini, E.; Magi, G.; Ferretti, G.; Bacchetti, T.; Giuliani, A.; Pugnaloni, A.; Rippo, M.R.; Facinelli, B. Attenuation of Listeria monocytogenes virulence by Cannabis sativa L. Essential oil. Front. Cell. Infect. Microbiol. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- International Organization for Standardisation. ISO 16140-4:2020: Microbiology of the Food Chain—Method Validation—Part 4: Protocol for Method Validation in a Single Laboratory; ISO: Geneva, Switzerland, 2020. [Google Scholar]
- Hanninen, M.; Haajanen, H.; Pummi, T.; Wermundsen, K.; Katila, M.; Sarkkinen, H.; Miettinen, I.; Rautelin, H. Detection and Typing of Campylobacter jejuni and Campylobacter coli and Analysis of Indicator Organisms in Three Waterborne Outbreaks in Finland. Appl. Environ. Microbiol. 2003, 69, 1391–1396. [Google Scholar] [CrossRef]
- Nygård, K.; Andersson, Y.; Røttingen, J.A.; Svensson, Å.; Lindbäck, J.; Kistemann, T.; Giesecke, J. Association between environmental risk factors and Campylobacter infections in Sweden. Epidemiol. Infect. 2004, 132, 317–325. [Google Scholar] [CrossRef]
- Vereen, E.; Lowrance, R.R.; Cole, D.J.; Lipp, E.K. Distribution and Ecology of Campylobacters in Coastal Plain Streams (Georgia, United States of America). Appl. Environ. Microbiol. 2007, 73, 1395–1403. [Google Scholar] [CrossRef]
- Black, R.E.; Levine, M.M.; Clements, M.L.; Hughes, T.P.; Blaser, M.J. Experimental Campylobacter jejuni Infection in Humans. J. Infect. Dis. 1988, 157, 472–479. [Google Scholar] [CrossRef]
- Sales-Ortells, H.; Agostini, G.; Medema, G. Quantification of Waterborne Pathogens and Associated Health Risks in Urban Water. Environ. Sci. Technol. 2015, 49, 6943–6952. [Google Scholar] [CrossRef]
- Phiri, B.J.; French, N.P.; Biggs, P.J.; Stevenson, M.A.; Reynolds, A.D.; Garcia-R, J.C.; Hayman, D.T.S. Microbial contamination in drinking water at public outdoor recreation facilities in New Zealand. J. Appl. Microbiol. 2021, 130, 302–312. [Google Scholar] [CrossRef]
- Li, J.; Zhao, X.; Tian, X.; Li, J.; Sjollema, J.; Wang, A. Retention in treated wastewater affects survival and deposition of Staphylococcus aureus and Escherichia coli in sand columns. Appl. Environ. Microbiol. 2015, 81, 2199–2205. [Google Scholar] [CrossRef]
- Nachamkin, I.; Szymanski, C.; Blaser, M.J. Campylobacter, 3rd ed.; Nachamkin, I., Szymanski, C., Blaser, M.J., Eds.; ASM Press: Washington, DC, USA, 2008; ISBN 9781119738039. [Google Scholar]
- Ferrari, S.; Frosth, S.; Svensson, L.; Fernström, L.L.; Skarin, H.; Hansson, I. Detection of Campylobacter spp. in water by dead-end ultrafiltration and application at farm level. J. Appl. Microbiol. 2019, 127, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- De Beer, D.M.; Botes, M.; Cloete, T.E. The microbial community of a biofilm contact reactor for the treatment of winery wastewater. J. Appl. Microbiol. 2018, 124, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Cui, Q.; Fang, T.; Huang, Y.; Wang, H. Occurrence of antibiotic resistance genes and bacterial pathogens in water and sediment in urban recreational water. J. Environ. Sci. 2019, 77, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Baffone, W.; Casaroli, A.; Citterio, B.; Pierfelici, L.; Campana, R.; Vittoria, E.; Guaglianone, E.; Donelli, G. Campylobacter jejuni loss of culturability in aqueous microcosms and ability to resuscitate in a mouse model. Int. J. Food Microbiol. 2006, 107, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.K.; Jørgensen, F.; Grogono-Thomas, R.; Humphrey, T.J. Enrichment culture for the isolation of Campylobacter spp.: Effects of incubation conditions and the inclusion of blood in selective broths. Int. J. Food Microbiol. 2009, 130, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Cody, H.J. Methods for isolating Campylobacter jejuni from low-turbidity water. Appl. Environ. Microbiol. 1986, 51, 312–315. [Google Scholar] [CrossRef]
- Vesga, F.J.; Moreno, Y.; Ferrús, M.A.; Campos, C.; Trespalacios, A.A. Detection of Helicobacter pylori in drinking water treatment plants in Bogotá Colombia, using cultural and molecular techniques. Int. J. Hyg. Environ. Health 2018, 221, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Carroll, Z.S.; Long, S.C.; Roa-Espinosa, A.; Runge, T. Centrifuge separation effect on bacterial indicator reduction in dairy manure. J. Environ. Manag. 2017, 191, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Malorny, B.; Hoorfar, J.; Hugas, M.; Heuvelink, A.; Fach, P.; Ellerbroek, L.; Bunge, C.; Dorn, C.; Helmuth, R. Interlaboratory diagnostic accuracy of a Salmonella specific PCR-based method. Int. J. Food Microbiol. 2003, 89, 241–249. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strakova, N.; Korena, K.; Gelbicova, T.; Kulich, P.; Karpiskova, R. A Rapid Culture Method for the Detection of Campylobacter from Water Environments. Int. J. Environ. Res. Public Health 2021, 18, 6098. https://doi.org/10.3390/ijerph18116098
Strakova N, Korena K, Gelbicova T, Kulich P, Karpiskova R. A Rapid Culture Method for the Detection of Campylobacter from Water Environments. International Journal of Environmental Research and Public Health. 2021; 18(11):6098. https://doi.org/10.3390/ijerph18116098
Chicago/Turabian StyleStrakova, Nicol, Kristyna Korena, Tereza Gelbicova, Pavel Kulich, and Renata Karpiskova. 2021. "A Rapid Culture Method for the Detection of Campylobacter from Water Environments" International Journal of Environmental Research and Public Health 18, no. 11: 6098. https://doi.org/10.3390/ijerph18116098
APA StyleStrakova, N., Korena, K., Gelbicova, T., Kulich, P., & Karpiskova, R. (2021). A Rapid Culture Method for the Detection of Campylobacter from Water Environments. International Journal of Environmental Research and Public Health, 18(11), 6098. https://doi.org/10.3390/ijerph18116098