A Gated Dilated Convolution with Attention Model for Clinical Cloze-Style Reading Comprehension
Abstract
1. Introduction
2. Related Work
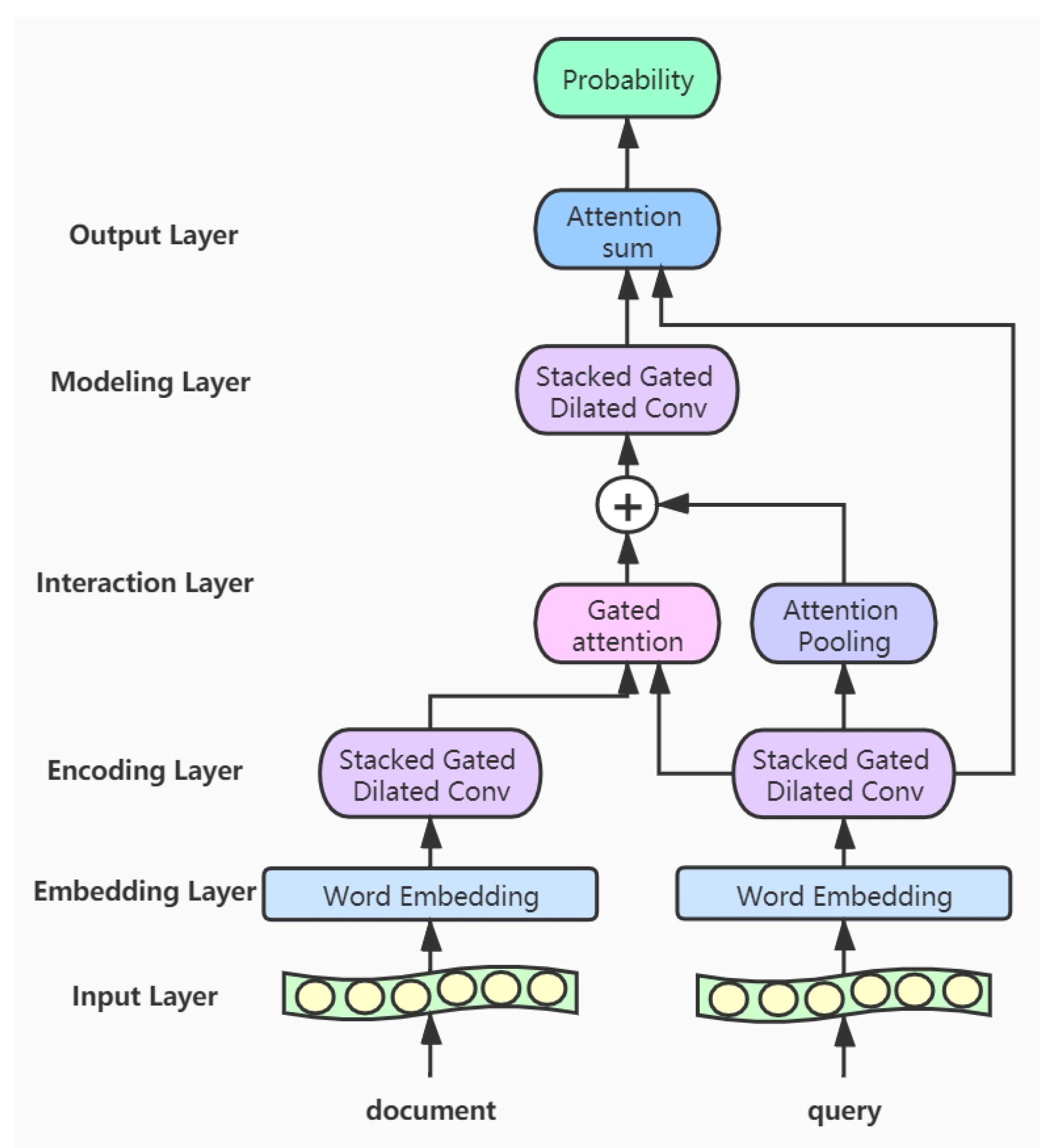
3. Model
- Input layer: This layer inputs the documents and queries into the model, and a variable-length approach is adopted so that the input text won’t be truncated.
- Embedding layer: In this layer, we convert the words of the input texts into word vector representations. The corpus for the word embedding is only from the CliCR training set and is learned by GloVe [35].
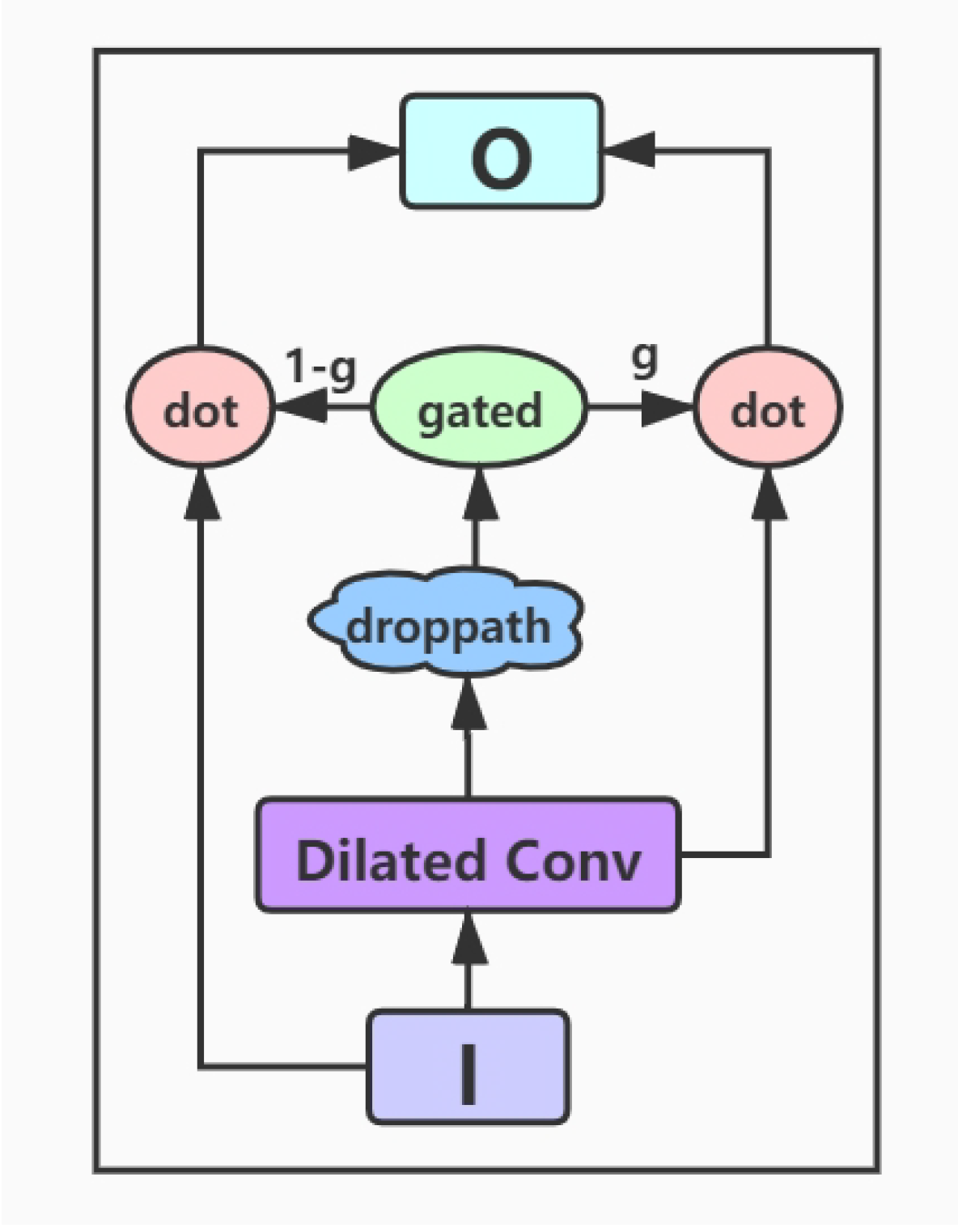
- Encoding layer: The encoding layer is a stack of gated dilated convolution modules, whose structure is similar to the Gated Linear Unit(GLU) [34,36]. The GLU structure is proposed by Facebook, and its advantage is that it hardly has to worry about the gradient disappearing because part of it is without any activation function. And dilated convolution can capture farther distances than conventional convolutions without causing any increase in parameters [37]. Once the convolution kernel and the step size are determined, the receptive field of the conventional convolution has a linear relationship with the number of layers of convolution, and the dilated convolution is an exponential relationship. The structure of the Gated Dilated Convolution module(GDConv) is shown in Figure 3.Assuming the document sequence , where k is the number of sentences in the document, the query sequence , where n is the number of sentences in the query. We input them into the dilated convolution layer and get a single output element and , respectively, with the dimension of :We divide the above outputs into two equal parts X and Y, both with the dimension d, which can be expressed as , . Then, we use the activation function sigmoid on Y to control which inputs X of the current context are relevant to, and perform the element-wise multiplicative operation with X. The formula is as follows:In order to solve the gradient disappearance problem and make the information transmit through multiple channels, the residual structure is used here, and the input sequence is also added. The formula is as follows:Then we use a droppath-like regularization method to make the model more robust:
- Interaction layer: Here we use the gated attention module proposed by Dhingra et al. [15], which obtains q by the soft attention, and then performs the element-wise multiplicative operation with the document representation vector d. The formula is as follows:Here we use the additive attention mechanism instead of simple pooling to complete the integration of the sequence information [38], namely, to encode the vector sequence of the query into a total query vector. Its formula is as follows:We concatenate the total query vector into the document representation and get the new document representation vector
- Modeling layer: The input to this layer is , which encodes the new representation of document words. Unlike the coding layer, since the representation of these words contain information about query’s integration, it can capture the words that are more relevant to the query in the document. We use five layers of Gated Dilated convolution(GDConv). Moreover, the of each layer is almost doubled, with the aim of establishing a farther relationship between words
- Output layer: In this layer, we calculate the inner product of the resulting document representation and the query representation and pass them through a softmax layer as the normalizing weightsThe vector s represents the probability of a word in the document. Then we integrate the probability of all the same words in the document for candidate set C. And this operation is the same as that in the AS model [28]:where indicates that the set of candidate c appears in document d.Finally, we calculate the candidate answer c with the highest probability as the final predicted answer:
4. Experiments and Results Analysis
4.1. Data Set Description
4.2. Experiment Setting
4.3. Results Analysis
4.4. Ablation Study for Model Components
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Liu, S.; Zhang, X.; Zhang, S.; Wang, H.; Zhang, W. Neural machine reading comprehension: Methods and trends. Appl. Sci. 2019, 9, 3698. [Google Scholar] [CrossRef]
- Tian, Y.; Ma, W.; Xia, F.; Song, Y. ChiMed: A Chinese Medical Corpus for Question Answering. In Proceedings of the 18th BioNLP Workshop and Shared Task, Florence, Italy, 1 August 2019; pp. 250–260. [Google Scholar]
- Pampari, A.; Raghavan, P.; Liang, J.; Peng, J. emrQA: A large corpus for question answering on electronic medical records. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium, 31 October–4 November 2018; pp. 2357–2368. [Google Scholar]
- Abacha, A.B.; Shivade, C.; Demner-Fushman, D. Overview of the mediqa 2019 shared task on textual inference, question entailment and question answering. In Proceedings of the 18th BioNLP Workshop and Shared Task, Florence, Italy, 1 August 2019; pp. 370–379. [Google Scholar]
- Nentidis, A.; Krithara, A.; Bougiatiotis, K.; Paliouras, G.; Kakadiaris, I. Results of the sixth edition of the BioASQ challenge. In Proceedings of the 6th BioASQ Workshop A Challenge on Large-Scale Biomedical Semantic Indexing and Question Answering, Brussels, Belgium, 31 October–1 November 2018; pp. 1–10. [Google Scholar]
- Wu, S.; Roberts, K.; Datta, S.; Du, J.; Ji, Z.; Si, Y.; Soni, S.; Wang, Q.; Wei, Q.; Xiang, Y.; et al. Deep learning in clinical natural language processing: A methodical review. J. Am. Med. Inform. Assoc. 2019. [Google Scholar] [CrossRef] [PubMed]
- Suster, S.; Daelemans, W. Clicr: A dataset of clinical case reports for machine reading comprehension. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), New Orleans, LA, USA, 1–6 June 2018; pp. 1551–1563. [Google Scholar]
- Jin, Q.; Dhingra, B.; Liu, Z.; Cohen, W.W.; Lu, X. PubMedQA: A Dataset for Biomedical Research Question Answering. arXiv 2019, arXiv:1909.06146. [Google Scholar]
- Smalheiser, N.R.; Luo, M.; Addepalli, S.; Cui, X. A manual corpus of annotated main findings of clinical case reports. Database 2019, 2019, bay143. [Google Scholar] [CrossRef] [PubMed]
- Smalheiser, N.R.; Shao, W.; Yu, P.S. Nuggets: Findings shared in multiple clinical case reports. Med. Libr. Assoc. 2015, 103, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Nye, B.; Li, J.J.; Patel, R.; Yang, Y.; Marshall, I.J.; Nenkova, A.; Wallace, B.C. A corpus with multi-level annotations of patients, interventions and outcomes to support language processing for medical literature. In Proceedings of the Conference Association for Computational Linguistics, Melbourne, Australia, 15–20 July 2018; pp. 197–207. [Google Scholar]
- Shardlow, M.; Batista-Navarro, R.; Thompson, P.; Nawaz, R.; McNaught, J.; Ananiadou, S. Identification of research hypotheses and new knowledge from scientific literature. BMC Med. 2018, 18, 46. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, D.I. What makes a case report publishable? Skeletal Radiol. 2006, 35, 627–628. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Bolton, J.; Manning, C.D. A Thorough Examination of the CNN/Daily Mail Reading Comprehension Task. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin, Germany, 7–12 August 2016; pp. 2358–2367. [Google Scholar]
- Dhingra, B.; Liu, H.; Yang, Z.; Cohen, W.; Salakhutdinov, R. Gated-Attention Readers for Text Comprehension. In Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Vancouver, BC, Canada, 30 July–4 August 2017; pp. 1832–1846. [Google Scholar]
- Yu, A.W.; Dohan, D.; Luong, M.T.; Zhao, R.; Chen, K.; Norouzi, M.; Le, Q.V. QANet: Combining Local Convolution with Global Self-Attention for Reading Comprehension. arXiv 2018, arXiv:1804.09541. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Chen, D. Neural Reading Comprehension and Beyond. Ph.D. Thesis, Stanford University, Stanford, CA, USA, 2018. [Google Scholar]
- Qiu, B.; Chen, X.; Xu, J.; Sun, Y. A Survey on Neural Machine Reading Comprehension. arXiv 2019, arXiv:1906.03824. [Google Scholar]
- Hermann, K.M.; Koisk, T.; Grefenstette, E.; Espeholt, L.; Kay, W.; Suleyman, M.; Blunsom, P. Teaching Machines to Read and Comprehend. In Proceedings of the 28th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; MIT Press: Cambridge, MA, USA, 2015; Volume 1, pp. 1693–1701. [Google Scholar]
- Hill, F.; Bordes, A.; Chopra, S.; Weston, J. The Goldilocks Principle: Reading Children’s Books with Explicit Memory Representations. arXiv 2015, arXiv:1511.02301. [Google Scholar]
- Bajgar, O.; Kadlec, R.; Kleindienst, J. Embracing Data Abundance: Booktest Dataset for Reading Comprehension. arXiv 2016, arXiv:1610.00956. [Google Scholar]
- Paperno, D.; Kruszewski, G.; Lazaridou, A.; Pham, N.Q.; Bernardi, R.; Pezzelle, S.; Baroni, M.; Boleda, G.; Fernandez, R. The LAMBADA Dataset: Word Prediction Requiring a Broad Discourse Context. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin, Germany, 7–12 August 2016; pp. 1525–1534. [Google Scholar]
- Onishi, T.; Wang, H.; Bansal, M.; Gimpel, K.; McAllester, D. Who did What: A Large-Scale Person-Centered Cloze Dataset. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Austin, TX, USA, 1–5 November 2016; pp. 2230–2235. [Google Scholar]
- Xie, Q.; Lai, G.; Dai, Z.; Hovy, E. Large-Scale Cloze Test Dataset Created by Teachers. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium, 31 October–4 November 2018; pp. 2344–2356. [Google Scholar]
- Weston, J.; Chopra, S.; Bordes, A. Memory Networks. arXiv 2014, arXiv:1410.3916. [Google Scholar]
- Trischler, A.; Ye, Z.; Yuan, X.; Bachman, P.; Sordoni, A.; Suleman, K. Natural Language Comprehension with the EpiReader. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Austin, TX, USA, 1–5 November 2016; pp. 128–137. [Google Scholar]
- Kadlec, R.; Schmid, M.; Bajgar, O.; Kleindienst, J. Text Understanding with the Attention Sum Reader Network. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin, Germany, 7–12 August 2016; pp. 908–918. [Google Scholar]
- Cui, Y.; Chen, Z.; Wei, S.; Wang, S.; Liu, T.; Hu, G. Attention-over-Attention Neural Networks for Reading Comprehension. In Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Vancouver, BC, Canada, 30 July–4 August 2017; pp. 593–602. [Google Scholar]
- Sordoni, A.; Bachman, P.; Trischler, A.; Bengio, Y. Iterative Alternating Neural Attention for Machine Reading. arXiv 2016, arXiv:1606.02245. [Google Scholar]
- Shen, Y.; Huang, P.S.; Gao, J.; Chen, W. Reasonet: Learning to Stop Reading in Machine Comprehension. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Halifax, NS, Canada, 13–17 August 2017; ACM: New York, NY, USA, 2017; pp. 1047–1055. [Google Scholar]
- Seo, M.; Kembhavi, A.; Farhadi, A.; Hajishirzi, H. Bidirectional Attention Flow for Machine Comprehension. arXiv 2016, arXiv:1611.01603. [Google Scholar]
- Wang, J.; Yu, L.; Lai, K.; Zhang, X. Community-based Weighted Graph Model for Valence-Arousal Prediction of Affective Words. IEEE-ACM Trans. Audio Spe. 2016, 24, 1957–1968. [Google Scholar] [CrossRef]
- Gehring, J.; Auli, M.; Grangier, D.; Yarats, D.; Dauphin, Y.N. Convolutional sequence to sequence learning. In Proceedings of the 34th International Conference on Machine Learning-Volume 70, Sydney, Australia, 6–11 August 2017; pp. 1243–1252. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C. Glove: Global Vectors for Word Representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Wang, J.; Peng, B.; Zhang, X. Using a Stacked Residual LSTM Model for Sentiment Intensity Prediction. Neurocomputing 2018, 322, 93–101. [Google Scholar] [CrossRef]
- Yu, F.; Koltun, V.; Funkhouser, T. Dilated residual networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Hawaii, HI, USA, 21–26 July 2017; pp. 472–480. [Google Scholar]
- Wang, W.; Yang, N.; Wei, F.; Chang, B.; Zhou, M. Gated self-matching networks for reading comprehension and question answering. In Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Vancouver, BC, Canada, 30 July–4 August 2017; pp. 189–198. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]

| Category | Number |
|---|---|
| Cases | 11,846 |
| Queries in train/dev/test | 91,344/6391/7184 |
| Tokens in documents | 16,544,217 |
| Distinct answers | 56,093 |
| Distinct answers(extended) | 288,211 |
| Entity types in documents | 591,960 |
| Model | EM | F1 | B-2 | B-4 |
|---|---|---|---|---|
| 35 | 53.7 | 0.46 | 0.23 | |
| 31 | 45.1 | 0.43 | 0.24 | |
| rand-entity | 1.4 | 5.1 | 0.03 | 0.01 |
| maxfreq-entity | 8.5 | 12.6 | 0.10 | 0.05 |
| lang-model | 2.1 | 3.5 | 0.00 | 0.00 |
| sim-entity | 20.8 | 29.4 | 0.22 | 0.15 |
| SA-Anonym | 19.6 | 27.2 | 0.22 | 0.16 |
| SA-Ent | 6.1 | 11.4 | 0.07 | 0.05 |
| GA-Anonym | 24.5 | 33.2 | 0.28 | 0.20 |
| GA-Ent | 22.2 | 30.2 | 0.25 | 0.18 |
| GA-NoEnt | 14.9 | 33.9 | 0.21 | 0.11 |
| Our model | 25.7 | 35.3 | 0.29 | 0.21 |
| Model | Time per epoch |
|---|---|
| GA Reader | 6 h 40 min |
| our model | 50 min |
| Embedding | EM | F1 |
|---|---|---|
| Pre-trained | 25.4 | 34.7 |
| CliCR training set | 25.7 | 35.3 |
| Method | EM | F1 |
|---|---|---|
| Convolution | 24.6 | 34.0 |
| Dilated convolution | 25.7 | 35.3 |
| Method | EM | F1 |
|---|---|---|
| Attention pooling/o | 25.0 | 34.9 |
| Attention pooling/w | 25.7 | 35.3 |
| Method | EM | F1 |
|---|---|---|
| Gated attention/o | 19.2 | 27.3 |
| Gated attention/w | 25.7 | 35.3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Zhang, X.; Zhou, X.; Li, J. A Gated Dilated Convolution with Attention Model for Clinical Cloze-Style Reading Comprehension. Int. J. Environ. Res. Public Health 2020, 17, 1323. https://doi.org/10.3390/ijerph17041323
Wang B, Zhang X, Zhou X, Li J. A Gated Dilated Convolution with Attention Model for Clinical Cloze-Style Reading Comprehension. International Journal of Environmental Research and Public Health. 2020; 17(4):1323. https://doi.org/10.3390/ijerph17041323
Chicago/Turabian StyleWang, Bin, Xuejie Zhang, Xiaobing Zhou, and Junyi Li. 2020. "A Gated Dilated Convolution with Attention Model for Clinical Cloze-Style Reading Comprehension" International Journal of Environmental Research and Public Health 17, no. 4: 1323. https://doi.org/10.3390/ijerph17041323
APA StyleWang, B., Zhang, X., Zhou, X., & Li, J. (2020). A Gated Dilated Convolution with Attention Model for Clinical Cloze-Style Reading Comprehension. International Journal of Environmental Research and Public Health, 17(4), 1323. https://doi.org/10.3390/ijerph17041323





