Assessing Seasonality Variation with Harmonic Regression: Accommodations for Sharp Peaks
Abstract
1. Introduction
2. Methods
2.1. The Base Model
2.2. Model Extensions
2.3. Data
2.3.1. Example 1: Hospitalizations Due to Salmonellosis in U.S. Elderly
2.3.2. Example 2: Laboratory-Confirmed Cases of Shigellosis in Christian Medical College and Hospital, India
2.3.3. Example 3: Monthly Records of Pneumonia and Influenza Death in US
2.3.4. Example 4: Simulated Dataset
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chui, K.K.H.; Jagai, J.S.; Griffiths, J.K.; Naumova, E.N. Hospitalization of the Elderly in the United States for Nonspecific Gastrointestinal Diseases: A Search for Etiological Clues. Am. J. Public Health 2011, 101, 2082–2086. [Google Scholar] [CrossRef]
- Chui, K.K.; Webb, P.; Russell, R.M.; Naumova, E.N. Geographic variations and temporal trends of Salmonella-associated hospitalization in the U.S. elderly, 1991–2004: A time series analysis of the impact of HACCP regulation. BMC Public Health 2009, 9, 447. [Google Scholar] [CrossRef] [PubMed]
- Falconi, T.M.A.; Cruz, M.S.; Naumova, E.N. The shift in seasonality of legionellosis in the USA. Epidemiol. Infect. 2018, 146, 1824–1833. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, T.; Anandan, S.; Jeyaseelan, V.; Jeyaseelan, L.; Ramanathan, K.; Veeraraghavan, B. Role of seasonality and rainfall in Vibrio cholerae infections: A time series model for 11 years surveillance data. Clin. Epidemiol. Glob. Health 2015, 3, 144–148. [Google Scholar] [CrossRef]
- Wenger, J.B.; Naumova, E.N. Seasonal Synchronization of Influenza in the United States Older Adult Population. PLoS ONE 2010, 5, e10187. [Google Scholar] [CrossRef] [PubMed]
- Naumova, E.N.; Jagai, J.S.; Matyas, B.; DeMaria, A., Jr.; MacNeill, I.B.; Griffiths, J.K. Seasonality in six enterically transmitted diseases and ambient temperature. Epidemiol. Infect. 2007, 135, 281–292. [Google Scholar] [CrossRef]
- Grabowska, K.; Högberg, L.; Penttinen, P.; Svensson, A.; Ekdahl, K. Occurrence of invasive pneumococcal disease and number of excess cases due to influenza. BMC Infect. Dis. 2006, 6, 58. [Google Scholar] [CrossRef]
- Huq, A.; Sack, R.B.; Nizam, A.; Longini, I.M.; Nair, G.B.; Ali, A.; Morris, J.G.; Khan, M.N.H.; Siddique, A.K.; Yunus, M.; et al. Critical Factors Influencing the Occurrence of Vibrio cholerae in the Environment of Bangladesh. Appl. Environ. Microbiol. 2005, 71, 4645–4654. [Google Scholar] [CrossRef]
- Hu, W.; Tong, S.; Mengersen, K.; Connell, D. Weather Variability and the Incidence of Cryptosporidiosis: Comparison of Time Series Poisson Regression and SARIMA Models. Ann. Epidemiol. 2007, 17, 679–688. [Google Scholar] [CrossRef]
- Kinlin, L.M.; Spain, C.V.; Ng, V.; Johnson, C.C.; White, A.N.J.; Fisman, D.N. Environmental exposures and invasive meningococcal disease: An evaluation of effects on varying time scales. Am. J. Epidemiol. 2009, 169, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.E. The calendar of epidemics: Seasonal cycles of infectious diseases. PLOS Pathog. 2018, 14, e1007327. [Google Scholar] [CrossRef] [PubMed]
- Jagai, J.S.; Castronovo, D.A.; Monchak, J.; Naumova, E.N. Seasonality of cryptosporidiosis: A meta-analysis approach. Environ. Res. 2009, 109, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Lofgren, E.; Fefferman, N.H.; Naumov, Y.N.; Gorski, J.; Naumova, E.N. Influenza seasonality: Underlying causes and modeling theories. J. Virol. 2007, 81, 5429–5436. [Google Scholar] [CrossRef] [PubMed]
- Moorthy, M.; Castronovo, D.; Abraham, A.; Bhattacharyya, S.; Gradus, S.; Gorski, J.; Naumov, Y.N.; Fefferman, N.H.; Naumova, E.N. Deviations in influenza seasonality: Odd coincidence or obscure consequence? Clin. Microbiol. Infect. 2012, 18, 955–962. [Google Scholar] [CrossRef]
- Jagai, J.S.; Sarkar, R.; Castronovo, D.; Kattula, D.; McEntee, J.; Ward, H.; Kang, G.; Naumova, E.N. Seasonality of Rotavirus in South Asia: A Meta-Analysis Approach Assessing Associations with Temperature, Precipitation, and Vegetation Index. PLoS ONE 2012, 7, e38168. [Google Scholar] [CrossRef]
- Sarkar, R.; Kang, G.; Naumova, E.N. Rotavirus Seasonality and Age Effects in a Birth Cohort Study of Southern India. PLoS ONE 2013, 8, e71616. [Google Scholar] [CrossRef]
- Auget, J.L.; Balakrishnan, N.; Mesbah, M.; Molenberghs, G. Advances in Statistical Methods for the Health Sciences: Applications to Cancer and AIDS Studies, Genome Sequence Analysis, and Survival Analysis; Springer Science & Business Media: Boston, FL, USA, 2007; pp. 1–540. [Google Scholar]
- Shumway, R.H.; Stoffer, D.S. Characteristics of Time Series. In Time Series Analysis and Its Applications: With R Examples, Springer Text in Statistics; Springer: Cham, Switzerland, 2017; pp. 1–44. [Google Scholar]
- Nagpaul, P.S. Time Series Analysis in Win IDAMS. 2005. Available online: https://pdfs.semanticscholar.org/ddb0/14582fd074d682aec17151ff4d0833aa9b10.pdf?_ga=2.125368387.895857527.1575065250-760598625.1575065250 (accessed on 29 November 2019).
- Strickland, M.J.; Klein, M.; Correa, A.; Reller, M.D.; Mahle, W.T.; Riehle-Colarusso, T.J.; Botto, L.D.; Flanders, W.D.; Mulholland, J.A.; Siffel, C.; et al. Ambient air pollution and cardiovascular malformations in Atlanta, Georgia, 1986–2003. Am. J. Epidemiol. 2009, 169, 1004–1014. [Google Scholar] [CrossRef]
- Consonni, D.; Pesatori, A.C.; Zocchetti, C.; Sindaco, R.; D’Oro, L.C.; Rubagotti, M.; Bertazzi, P.A. Mortality in a Population Exposed to Dioxin after the Seveso, Italy, Accident in 1976: 25 Years of Follow-Up. Am. J. Epidemiol. 2008, 167, 847–858. [Google Scholar] [CrossRef]
- Lofgren, E.; Fefferman, N.H.; Doshi, M.; Naumova, E.N. Assessing Seasonal Variation in Multisource Surveillance Data: Annual Harmonic Regression; Springer: Berlin/Heidelberg, Germany, 2007; pp. 114–123. [Google Scholar]
- Stolwijk, A.M.; Straatman, H.; Zielhuis, G.A. Studying seasonality by using sine and cosine functions in regression analysis. J. Epidemiol. Community Health 1999, 53, 235–238. [Google Scholar] [CrossRef]
- Brownstein, J.S.; Kleinman, K.P.; Mandl, K.D. Identifying pediatric age groups for influenza vaccination using a real-time regional surveillance system. Am. J. Epidemiol. 2005, 162, 686–693. [Google Scholar] [CrossRef]
- Bliss, C.I. Periodic regression in biology and climatology. Conn. Agric. Exp. Stn. 1958, 615, 3–55. [Google Scholar]
- Alsova, O.K.; Loktev, V.B.; Naumova, E.N. Rotavirus Seasonality: An Application of Singular Spectrum Analysis and Polyharmonic Modeling. Int. J. Environ. Res. Public Health 2019, 16, 4309. [Google Scholar] [CrossRef] [PubMed]
- Stashevsky, P.S.; Yakovina, I.N.; Falconi, T.M.; Naumova, E.N. Agglomerative clustering of enteric infections and weather parameters to indentify seasonal outbreaks in cold climates. Int. J. Environ. Res. Public Health 2019, 16, 2083. [Google Scholar] [CrossRef] [PubMed]
- Naumova, E.N. Mystery of seasonality: Getting the rhythm of nature. J. Public Health Policy 2006, 27, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Eilers, P.H.C.; Gampe, J.; Marx, B.D.; Rau, R. Modulation models for seasonal time series and incidence tables. Stat. Med. 2008, 27, 3430–3441. [Google Scholar] [CrossRef] [PubMed]
- Chui, K.K.H.; Wenger, J.B.; Cohen, S.A.; Naumova, E.N. Visual Analytics for Epidemiologists: Understanding the Interactions Between Age, Time, and Disease with Multi-Panel Graphs. PLoS ONE 2011, 6, e14683. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chui, K.K.; Cohen, A.S.; Naumova, E.N. Snowbirds and infection—New phenomena in pneumonia and influenza hospitalizations from winter migration of older adults: A spatiotemporal analysis. BMC Public Health 2011, 11, 444. [Google Scholar] [CrossRef] [PubMed]
- Jutla, A.; Whitcombe, E.; Hasan, N.; Haley, B.; Akanda, A.; Huq, A.; Alam, M.; Sack, R.B.; Colwell, R. Environmental Factors Influencing Epidemic Cholera. Am. J. Trop. Med. Hyg. 2013, 89, 597–607. [Google Scholar] [CrossRef]
- Koelle, K. The impact of climate on the disease dynamics of cholera. Clin. Microbiol. Infect. 2009, 15, 29–31. [Google Scholar] [CrossRef]
- Longini, J.I.M.; Yunus, M.; Zaman, K.; Siddique, A.K.; Sack, R.B.; Nizam, A. Epidemic and Endemic Cholera Trends over a 33-Year Period in Bangladesh. J. Infect. Dis. 2002, 186, 246–251. [Google Scholar] [CrossRef][Green Version]
- Glass, R.I.; Becker, S.; Huq, M.I.; Stoll, B.J.; Khan, M.U.; Merson, M.H.; Lee, J.V.; Black, R.E. Endemic Cholera in Rural Bangladesh, 1966–1980. Am. J. Epidemiol. 1982, 116, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Moreno, D.; Pascual, M.; Bouma, M.; Dobson, A.; Cash, B. Cholera Seasonality in Madras (1901–1940): Dual Role for Rainfall in Endemic and Epidemic Regions. EcoHealth 2007, 4, 52–62. [Google Scholar] [CrossRef]
- Harboe, Z.B.; Benfield, T.L.; Valentiner-Branth, P.; Hjuler, T.; Lambertsen, L.; Kaltoft, M.; Krogfelt, K.A.; Slotved, H.C.; Christensen, J.J.; Konradsen, H.B. Temporal Trends in Invasive Pneumococcal Disease and Pneumococcal Serotypes over 7 Decades. Clin. Infect. Dis. 2010, 50, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Ajdacic-Gross, V.; Bopp, M.; Sansossio, R.; Lauber, C.; Gostynski, M.; Eich, M.; Gutzwiller, F.; Rössler, W. Diversity and change in suicide seasonality over 125 years. J. Epidemiol. Community Health 2005, 59, 967–972. [Google Scholar] [CrossRef]
- Bhaskaran, K.; Gasparrini, A.; Hajat, S.; Smeeth, L.; Armstrong, B. Time series regression studies in environmental epidemiology. Int. J. Epidemiol. 2013, 42, 1187–1195. [Google Scholar] [CrossRef]
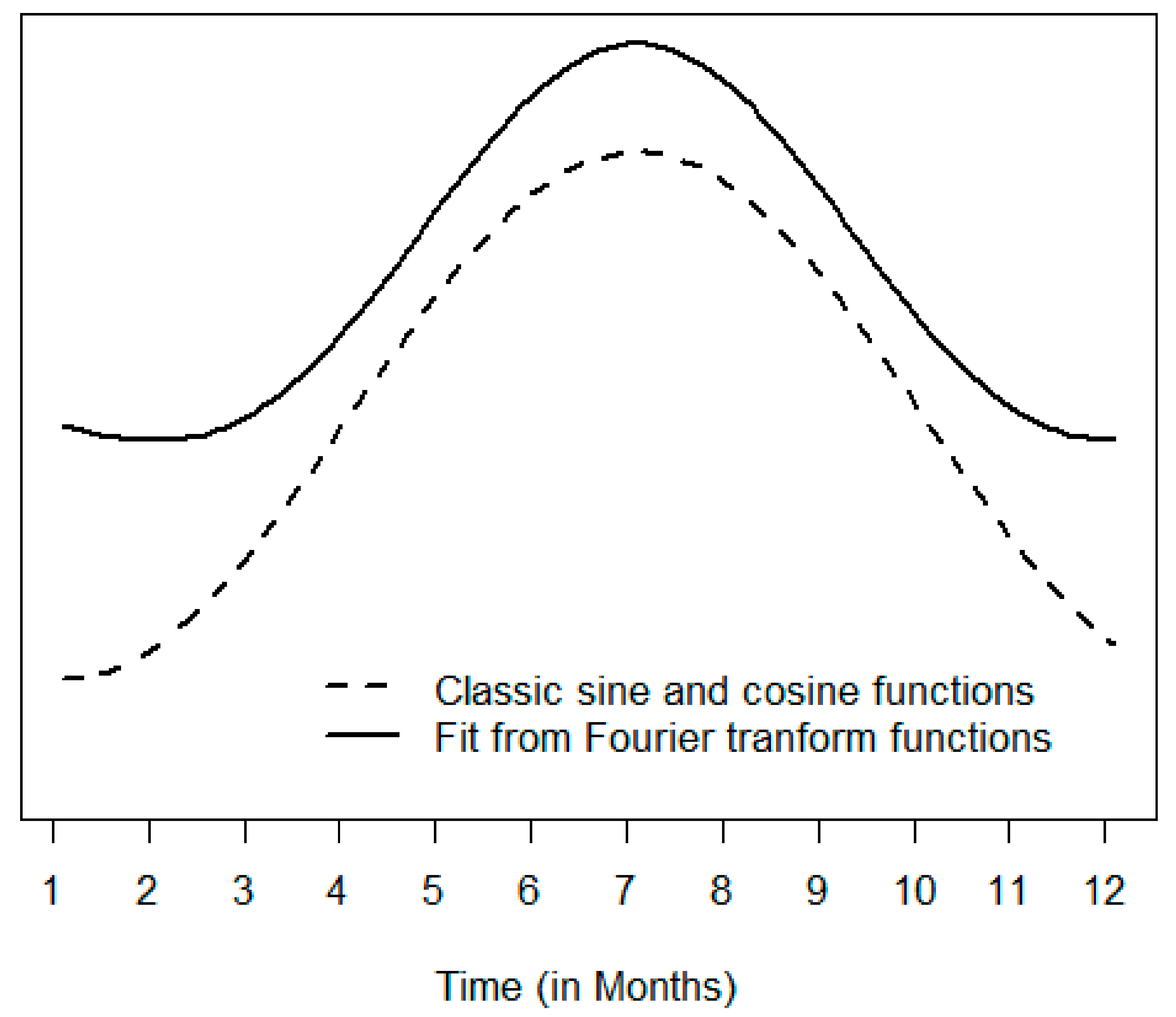
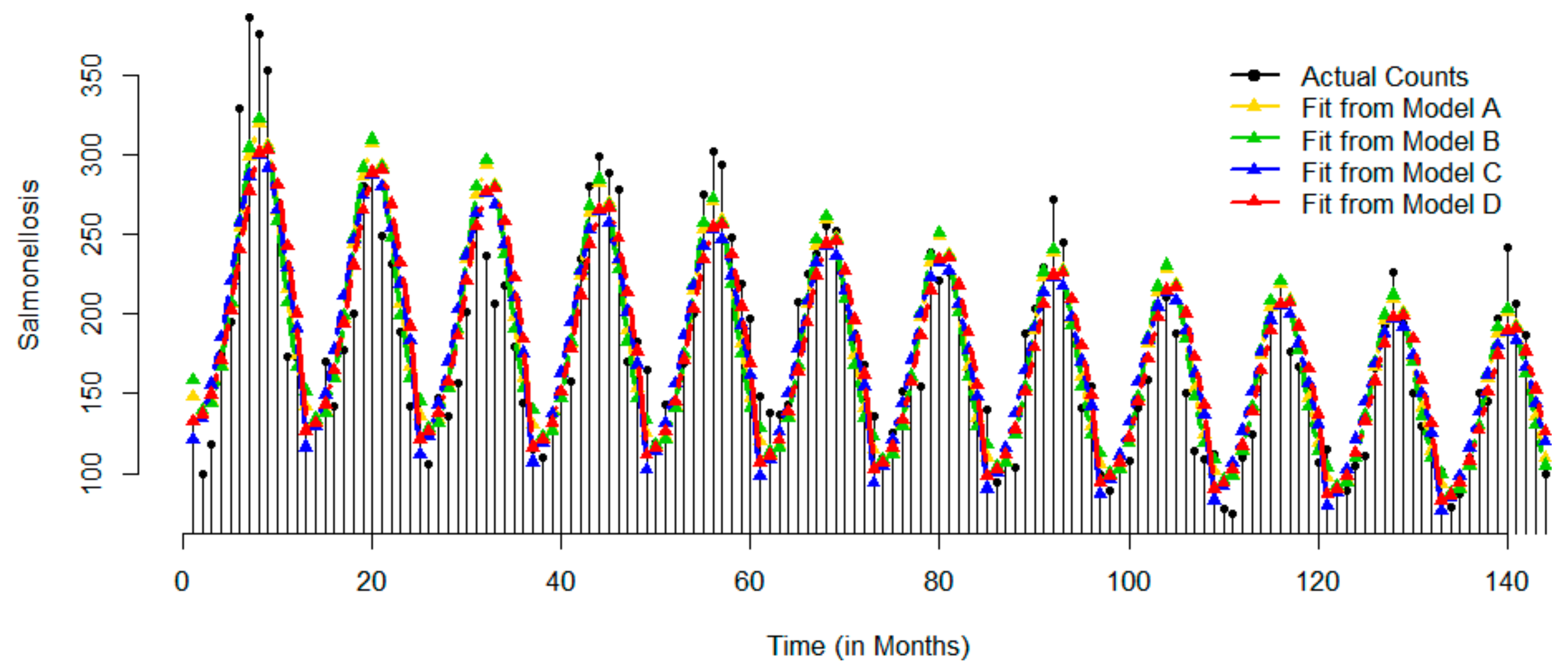
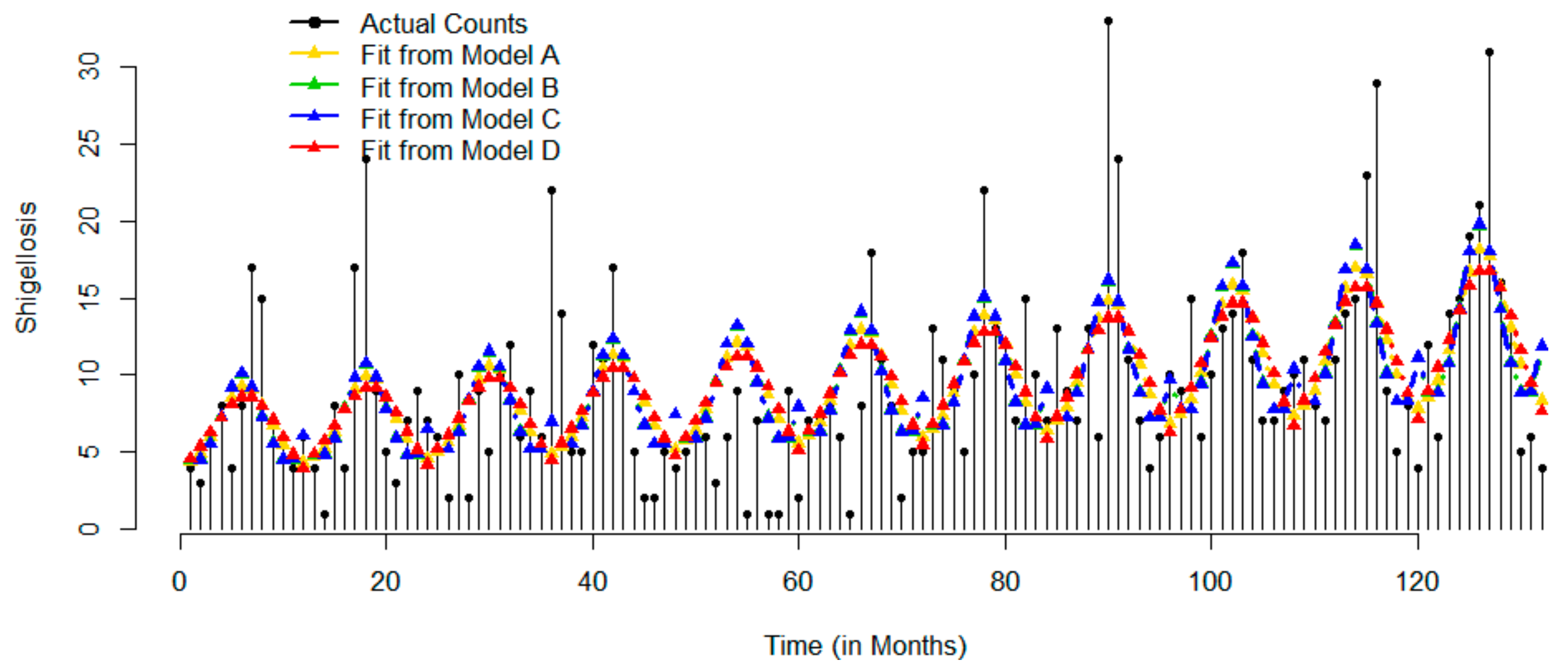


| Statistics | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug | Sep | Oct | Nov | Dec | Overall |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Example 1: Salmonellosis | |||||||||||||
| Mean | 129.3 | 103.5 | 120.5 | 131.1 | 161.3 | 198.5 | 249.8 | 262.5 | 239.9 | 209.7 | 163.6 | 144.4 | 176.2 |
| SD | 20.4 | 19.0 | 28.2 | 23.0 | 28.3 | 50.1 | 53.3 | 48.0 | 52.0 | 42.7 | 29.1 | 31.5 | 63.5 |
| Min | 99 | 78 | 75 | 104 | 111 | 145 | 193 | 211 | 176 | 150 | 114 | 100 | 75 |
| Max | 165 | 138 | 170 | 175 | 208 | 329 | 386 | 376 | 353 | 278 | 219 | 197 | 386 |
| 1st Qrt | 113.5 | 89.5 | 96.5 | 109 | 145.5 | 161.5 | 209 | 223.5 | 200 | 177 | 142 | 116.5 | 131 |
| 3rd Qrt | 143.5 | 114.5 | 140 | 145.5 | 182.5 | 214 | 277.5 | 294.5 | 270.5 | 239.5 | 183.5 | 170 | 216 |
| Example 2: Shigellosis | |||||||||||||
| Mean | 8.9 | 6.6 | 7.8 | 8.1 | 10.0 | 16.0 | 15.9 | 12.2 | 6.4 | 5.7 | 7.0 | 7.4 | 9.3 |
| SD | 3.9 | 4.1 | 2.5 | 4.4 | 5.5 | 8.5 | 8.3 | 6.6 | 3.4 | 3.9 | 2.1 | 5.5 | 6.1 |
| Min | 4 | 1 | 5 | 2 | 1 | 5 | 1 | 5 | 1 | 1 | 4 | 2 | 1 |
| Max | 14 | 15 | 14 | 15 | 19 | 33 | 31 | 29 | 13 | 15 | 10 | 22 | 33 |
| 1st Qrt | 5 | 3 | 6 | 4 | 6 | 8 | 10 | 7 | 3 | 2 | 5 | 4 | 5 |
| 3rd Qrt | 13 | 9 | 9 | 12 | 14 | 22 | 23 | 15 | 8 | 7 | 9 | 10 | 12 |
| Example 3: Pneumonia and Influenza | |||||||||||||
| Mean | 51.6 | 45.3 | 35.5 | 27.1 | 22.5 | 21.0 | 21.6 | 20.9 | 21.2 | 24.1 | 25.8 | 34.1 | 29.2 |
| SD | 19.4 | 9.1 | 10.0 | 2.7 | 2.0 | 2.3 | 2.4 | 2.2 | 1.9 | 2.0 | 3.6 | 13.4 | 12.6 |
| Min | 28 | 30 | 28 | 23 | 20 | 18 | 18 | 18 | 19 | 21 | 21 | 25 | 18 |
| Max | 82 | 57 | 64 | 31 | 26 | 25 | 25 | 25 | 24 | 27 | 32 | 73 | 82 |
| 1st Qrt | 31 | 37 | 30 | 25 | 21 | 19 | 19 | 19 | 19 | 22 | 23 | 26 | 22 |
| 3rd Qrt | 64 | 52 | 36 | 29 | 24 | 23 | 23 | 23 | 23 | 26 | 29 | 34 | 31 |
| Example 4: Simulated Data | |||||||||||||
| Mean | 47.5 | 50.5 | 56.5 | 55.1 | 51.3 | 46.2 | 41.3 | 40.8 | 38.6 | 36.4 | 43.6 | 47.7 | 46.3 |
| SD | 24.9 | 27.1 | 31.1 | 30.6 | 24.4 | 24.9 | 23.9 | 23.4 | 22.9 | 23.2 | 25.8 | 26.1 | 25.8 |
| Min | 9 | 9 | 9 | 10 | 10 | 10 | 8 | 7 | 6 | 4 | 6 | 7 | 4 |
| Max | 86 | 94 | 106 | 102 | 95 | 86 | 78 | 75 | 74 | 76 | 88 | 90 | 106 |
| 1st Qrt | 29 | 30 | 30 | 27 | 24 | 23 | 20 | 21 | 20 | 16 | 20 | 25 | 24.5 |
| 3rd Qrt | 69 | 75 | 84 | 82 | 78 | 69 | 64 | 62 | 58 | 55 | 62 | 68 | 66.5 |
| Statistics | Example 1: Salmonellosis | Example 2: Shigellosis | Example 3: Pneumonia | Example 4: Simulated Data |
|---|---|---|---|---|
| Skewness (SE) | 0.85 (0.20) | 1.44 (0.21) | 2.23 (0.21) | 0.25 (0.21) |
| Kurtosis (SE) | 0.58 (0.40) | 2.63 (0.42) | 5.17 (0.42) | −0.93 (0.42) |
| Peak timing (SE) | 8.09 (0.07) | 6.29 (0.47) | 1.47 (0.12) | 3.21 (0.26) |
| Amplitude (SE) | 0.42 (0.02) | 0.39 (0.08) | 0.43 (0.02) | 0.18 (0.03) |
| Estimator | Model A | Model B | Model C | Model D |
|---|---|---|---|---|
| Example 1: Salmonellosis | ||||
| Constant | 5.39 (5.37 to 5.42; < 0.001) | 7.07 (6.79 to 7.35; < 0.001) | 4.93 (4.90 to 4.97; < 0.001) | 4.79 (4.71 to 4.87; < 0.001) |
| Year | −0.04 (−0.05 to −0.04; < 0.001) | −0.04 (−0.05 to −0.04; < 0.001) | −0.04 (−0.05 to −0.04 < 0.001) | −0.04 (−0.05 to −0.04; < 0.001) |
| Sin | −0.37 (−0.39 to −0.36; < 0.001) | 3.91 (3.50 to 4.32; < 0.001) | 0.82 (0.78 to 0.86; < 0.001) | −0.18 (−0.21 to −0.15; < 0.001) |
| Cos | −0.19 (−0.21 to −0.17; < 0.001) | −5.16 (−5.84 to −4.48; < 0.001) | 0.02 (0.00 to 0.04; 0.047) | 0.80 (0.70 to 0.90; < 0.001) |
| RMSE | 24.47 | 24.28 | 28.58 | 29 |
| MAD | 18.76 | 18.66 | 22.67 | 22.41 |
| BIC | 1458.28 | 1451.72 | 1664.47 | 1643.73 |
| Example 2: Shigellosis | ||||
| Constant | 1.77 (1.63 to 1.90; < 0.001) | 7.92 (5.63 to 10.17; < 0.001) | 3.28 (2.60 to 3.95; < 0.001) | 0.76 (0.46 to 1.04; < 0.001) |
| Year | 0.07 (0.05 to 0.09; < 0.001) | 0.07 (0.05 to 0.09; < 0.001) | 0.07 (0.05 to 0.09; < 0.001) | 0.07 (0.05 to 0.09; < 0.001) |
| Sin | −0.06 (−0.14 to 0.02; 0.140) | 9.58 (6.52 to 12.60; < 0.001) | −2.58 (−3.71 to −1.43; < 0.001) | −0.06 (−0.14 to 0.02; 0.146) |
| Cos | −0.39 (−0.47 to −0.31; < 0.001) | −15.26 (−20.5 to −9.97; < 0.001) | −1.55 (−2.06 to −1.02; < 0.001) | 1.32 (1.01 to 1.64; < 0.001) |
| RMSE | 4.93 | 4.86 | 4.85 | 5.09 |
| MAD | 3.59 | 3.53 | 3.52 | 3.68 |
| BIC | 859.66 | 844.31 | 842.96 | 878.93 |
| Example 3: Pneumonia and Influenza | ||||
| Constant | 3.52 (3.45 to 3.58; < 0.001) | 3.92 (3.81 to 4.03; < 0.001) | 3.39 (3.32 to 3.46; < 0.001) | 3.42 (3.31 to 3.52; < 0.001) |
| Year | −0.03 (−0.04 to −0.02; < 0.001) | −0.03 (−0.04 to −0.02; < 0.001) | −0.03 (−0.04 to −0.02; < 0.001) | −0.03 (−0.04 to −0.02; < 0.001) |
| Sin | 0.30 (0.25 to 0.35; < 0.001) | 2.00 (1.73 to 2.27; < 0.001) | 0.48 (0.42 to 0.55; < 0.001) | 0.18 (0.10 to 0.27; < 0.001) |
| Cos | 0.31 (0.26 to 0.35; < 0.001) | −1.91 (−2.24 to −1.57; < 0.001) | 0.28 (0.23 to 0.32; < 0.001) | 0.25 (0.08 to 0.42; 0.004) |
| RMSE | 7.96 | 7.49 | 7.11 | 10.37 |
| MAD | 5.22 | 4.74 | 4.15 | 6.97 |
| BIC | 896.19 | 870.76 | 851.78 | 1064.65 |
| Example 4: Simulated Data | ||||
| Constant | 2.59 (2.52 to 2.66; < 0.001) | 3.06 (2.86 to 3.26; < 0.001) | 2.49 (2.41 to 2.56; < 0.001) | 2.63 (2.52 to 2.73; < 0.001) |
| Year | 0.18 (0.17 to 0.19; < 0.001) | 0.18 (0.17 to 0.19; < 0.001) | 0.18 (0.17 to 0.19; < 0.001) | 0.18 (0.17 to 0.19; < 0.001) |
| Sin | 0.18 (0.14 to 0.21; < 0.001) | 1.33 (0.96 to 1.70; < 0.001) | 0.24 (0.18 to 0.30; < 0.001) | 0.20 (0.14 to 0.25; < 0.001) |
| Cos | −0.02 (−0.05 to 0.02; 0.396) | −1.63 (−2.16 to −1.09; < 0.001) | 0.04 (0.01 to 0.08; 0.026) | −0.05 (−0.17 to 0.07; 0.383) |
| RMSE | 5.79 | 5.79 | 6.76 | 5.79 |
| MAD | 4.62 | 4.62 | 5.27 | 4.63 |
| BIC | 870.4 | 869.87 | 900.36 | 870.36 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramanathan, K.; Thenmozhi, M.; George, S.; Anandan, S.; Veeraraghavan, B.; Naumova, E.N.; Jeyaseelan, L. Assessing Seasonality Variation with Harmonic Regression: Accommodations for Sharp Peaks. Int. J. Environ. Res. Public Health 2020, 17, 1318. https://doi.org/10.3390/ijerph17041318
Ramanathan K, Thenmozhi M, George S, Anandan S, Veeraraghavan B, Naumova EN, Jeyaseelan L. Assessing Seasonality Variation with Harmonic Regression: Accommodations for Sharp Peaks. International Journal of Environmental Research and Public Health. 2020; 17(4):1318. https://doi.org/10.3390/ijerph17041318
Chicago/Turabian StyleRamanathan, Kavitha, Mani Thenmozhi, Sebastian George, Shalini Anandan, Balaji Veeraraghavan, Elena N. Naumova, and Lakshmanan Jeyaseelan. 2020. "Assessing Seasonality Variation with Harmonic Regression: Accommodations for Sharp Peaks" International Journal of Environmental Research and Public Health 17, no. 4: 1318. https://doi.org/10.3390/ijerph17041318
APA StyleRamanathan, K., Thenmozhi, M., George, S., Anandan, S., Veeraraghavan, B., Naumova, E. N., & Jeyaseelan, L. (2020). Assessing Seasonality Variation with Harmonic Regression: Accommodations for Sharp Peaks. International Journal of Environmental Research and Public Health, 17(4), 1318. https://doi.org/10.3390/ijerph17041318





