The Prevalence and Characterization of Extended-Spectrum β-Lactamase- and Carbapenemase-Producing Bacteria from Hospital Sewage, Treated Effluents and Receiving Rivers
Abstract
1. Introduction
2. Materials and Methods
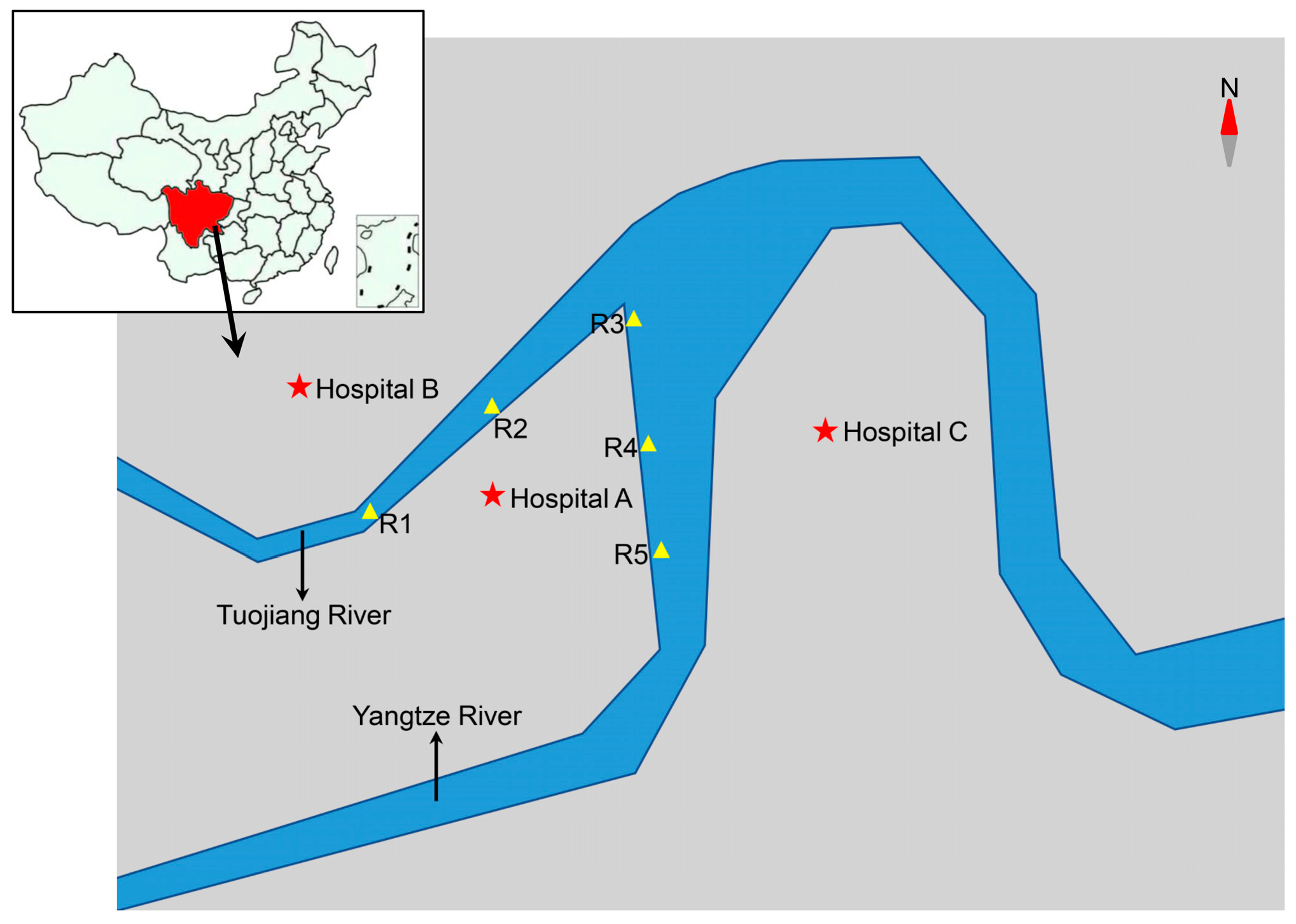
2.1. Sample Collection
2.2. Isolation of ESBL- and Carbapenem-Resistant Gram-Negative Bacteria
2.3. Bacterial Species Identification
2.4. Detection of Antibiotic Resistance Genes
2.5. Antimicrobial Susceptibility Testing
3. Results
3.1. Bacterial Isolation
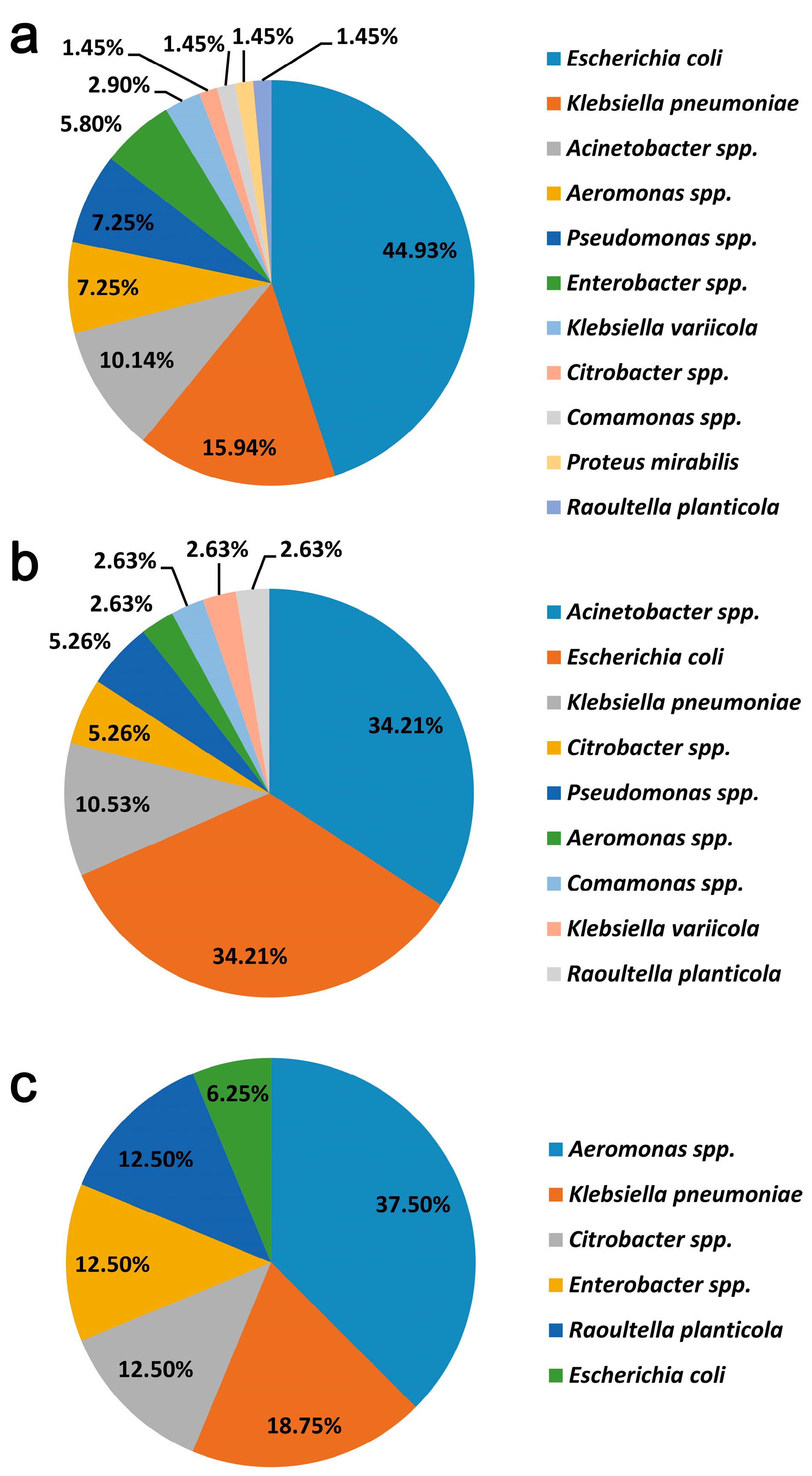
3.2. Bacterial Identification
3.3. Detection and Molecular Characterization of Resistance Genes
3.4. Antimicrobial Susceptibility of Enterobacteriaceae and Acinetobacter Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Laupland, K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef]
- Bonomo, R.A. Beta-Lactamases: A Focus on Current Challenges. Cold Spring Harb. Perspect. Med. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 2018, 62, e01076-18. [Google Scholar] [CrossRef]
- Schwaber, M.J.; Klarfeld-Lidji, S.; Navon-Venezia, S.; Schwartz, D.; Leavitt, A.; Carmeli, Y. Predictors of Carbapenem-Resistant Klebsiella pneumoniae Acquisition among Hospitalized Adults and Effect of Acquisition on Mortality. Antimicrob. Agents Chemother. 2008, 52, 1028–1033. [Google Scholar] [CrossRef]
- Martin, A.; Fahrbach, K.; Zhao, Q.; Lodise, T. Association Between Carbapenem Resistance and Mortality Among Adult, Hospitalized Patients With Serious Infections Due to Enterobacteriaceae: Results of a Systematic Literature Review and Meta-analysis. Open Forum Infect. Dis. 2018, 5, ofy150. [Google Scholar] [CrossRef]
- Potter, R.F.; D’Souza, A.W.; Dantas, G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Updated 2016, 29, 30–46. [Google Scholar] [CrossRef]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Maravic, A.; Skocibusic, M.; Fredotovic, Z.; Samanic, I.; Cvjetan, S.; Knezovic, M.; Puizina, J. Urban riverine environment is a source of multidrug-resistant and ESBL-producing clinically important Acinetobacter spp. Environ. Sci. Pollut. Res. Int. 2016, 23, 3525–3535. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-Resistance Genes in Waste Water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Weingarten, R.A.; Johnson, R.C.; Conlan, S.; Ramsburg, A.M.; Dekker, J.P.; Lau, A.F.; Khil, P.; Odom, R.T.; Deming, C.; Park, M.; et al. Genomic Analysis of Hospital Plumbing Reveals Diverse Reservoir of Bacterial Plasmids Conferring Carbapenem Resistance. mBio 2018, 9, e02011–e02017. [Google Scholar] [CrossRef] [PubMed]
- Lamba, M.; Graham, D.W.; Ahammad, S.Z. Hospital Wastewater Releases of Carbapenem-Resistance Pathogens and Genes in Urban India. Environ. Sci. Technol. 2017, 51, 13906–13912. [Google Scholar] [CrossRef]
- Parvez, S.; Khan, A.U. Hospital sewage water: A reservoir for variants of New Delhi metallo-beta-lactamase (NDM)- and extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2018, 51, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Nasri, E.; Subirats, J.; Sanchez-Melsio, A.; Mansour, H.B.; Borrego, C.M.; Balcazar, J.L. Abundance of carbapenemase genes (blaKPC, blaNDM and blaOXA-48) in wastewater effluents from Tunisian hospitals. Environ. Pollut. 2017, 229, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Cahill, N.; O’Connor, L.; Mahon, B.; Varley, A.; McGrath, E.; Ryan, P.; Cormican, M.; Brehony, C.; Jolley, K.A.; Maiden, M.C.; et al. Hospital effluent: A reservoir for carbapenemase-producing Enterobacterales? Sci. Total Environ. 2019, 672, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Haller, L.; Chen, H.; Ng, C.; Le, T.H.; Koh, T.H.; Barkham, T.; Sobsey, M.; Gin, K.Y. Occurrence and characteristics of extended-spectrum beta-lactamase- and carbapenemase- producing bacteria from hospital effluents in Singapore. Sci. Total Environ. 2018, 615, 1119–1125. [Google Scholar] [CrossRef]
- Zhang, X.; Lü, X.; Zong, Z. Enterobacteriaceae producing the KPC-2 carbapenemase from hospital sewage. Diagn. Microbiol. Infect. Dis. 2012, 73, 204–206. [Google Scholar] [CrossRef]
- Zhang, C.; Qiu, S.; Wang, Y.; Qi, L.; Hao, R.; Liu, X.; Shi, Y.; Hu, X.; An, D.; Li, Z.; et al. Higher isolation of NDM-1 producing Acinetobacter baumannii from the sewage of the hospitals in Beijing. PLoS ONE 2014, 8, e64857. [Google Scholar] [CrossRef]
- Jin, L.; Wang, R.; Wang, X.; Wang, Q.; Zhang, Y.; Yin, Y.; Wang, H. Emergence of mcr-1 and carbapenemase genes in hospital sewage water in Beijing, China. J. Antimicrob. Chemother. 2018, 73, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Zong, Z.; Zhang, X. blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J. Antimicrob. Chemother. 2013, 68, 1007–1010. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Zhang, X.; Zhou, Y.; Zhang, Z.; Zheng, X. Whole-genome sequencing of an NDM-1- and OXA-58-producing Acinetobacter towneri isolate from hospital sewage in Sichuan Province, China. J. Glob. Antimicrob. Resist. 2019, 16, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, X.; Yu, X.; Zhang, J.; Guo, L.; Huang, C.; Jiang, X.; Li, X.; Feng, Y.; Zheng, B. First detection and genomics analysis of KPC-2-producing Citrobacter isolates from river sediments. Environ. Pollut. 2018, 235, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Tafoukt, R.; Touati, A.; Leangapichart, T.; Bakour, S.; Rolain, J.M. Characterization of OXA-48-like-producing Enterobacteriaceae isolated from river water in Algeria. Water Res. 2017, 120, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Di, D.Y.W.; Jang, J.; Unno, T.; Hur, H.-G. Emergence of Klebsiella variicola positive for NDM-9, a variant of New Delhi metallo-β-lactamase, in an urban river in South Korea. J. Antimicrob. Chemother. 2017, 72, 1063–1067. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Azuma, T.; Otomo, K.; Kunitou, M.; Shimizu, M.; Hosomaru, K.; Mikata, S.; Ishida, M.; Hisamatsu, K.; Yunoki, A.; Mino, Y.; et al. Environmental fate of pharmaceutical compounds and antimicrobial-resistant bacteria in hospital effluents, and contributions to pollutant loads in the surface waters in Japan. Sci. Total Environ. 2019, 657, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.A.; Hellmark, B.; Ehricht, R.; Söderquist, B.; Jass, J. Related carbapenemase-producing Klebsiella isolates detected in both a hospital and associated aquatic environment in Sweden. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2241–2251. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Wiley: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Sivalingam, P.; Pote, J.; Prabakar, K. Environmental Prevalence of Carbapenem Resistance Enterobacteriaceae (CRE) in a Tropical Ecosystem in India: Human Health Perspectives and Future Directives. Pathogens 2019, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Marathe, N.P.; Berglund, F.; Razavi, M.; Pal, C.; Droge, J.; Samant, S.; Kristiansson, E.; Larsson, D.G.J. Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 2019, 7, 97. [Google Scholar] [CrossRef]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef]
- Yang, F.; Mao, D.; Zhou, H.; Wang, X.; Luo, Y. Propagation of New Delhi Metallo-β-lactamase Genes (blaNDM-1) from a Wastewater Treatment Plant to Its Receiving River. Environ. Sci. Technol. Lett. 2016, 3, 138–143. [Google Scholar] [CrossRef]
- Wang, R.N.; Zhang, Y.; Cao, Z.H.; Wang, X.Y.; Ma, B.; Wu, W.B.; Hu, N.; Huo, Z.Y.; Yuan, Q.B. Occurrence of super antibiotic resistance genes in the downstream of the Yangtze River in China: Prevalence and antibiotic resistance profiles. Sci. Total Environ. 2019, 651, 1946–1957. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huang, L.; Li, L.; Yang, Y.; Mao, D.; Luo, Y. Discharge of KPC-2 genes from the WWTPs contributed to their enriched abundance in the receiving river. Sci. Total Environ. 2017, 581, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Jiang, Y.; An, W.; Wang, H.; Zhang, X. Emergence of KPC-2-producing Escherichia coli isolates in an urban river in Harbin, China. World J. Microbiol. Biotechnol. 2015, 31, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, M.; Przygodzinska, D.; Matyjewicz, K.; Popowska, M. Occurrence and Variety of beta-Lactamase Genes among Aeromonas spp. Isolated from Urban Wastewater Treatment Plant. Front. Microbiol. 2017, 8, 863. [Google Scholar] [CrossRef] [PubMed]
- Harnisz, M.; Korzeniewska, E. The prevalence of multidrug-resistant Aeromonas spp. in the municipal wastewater system and their dissemination in the environment. Sci. Total Environ. 2018, 626, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Sekizuka, T.; Inamine, Y.; Segawa, T.; Hashino, M.; Yatsu, K.; Kuroda, M. Potential KPC-2 carbapenemase reservoir of environmental Aeromonas hydrophila and Aeromonas caviae isolates from the effluent of an urban wastewater treatment plant in Japan. Environ. Microbiol. Rep. 2019, 11, 589–597. [Google Scholar] [CrossRef]
- Jang, H.M.; Kim, Y.B.; Choi, S.; Lee, Y.; Shin, S.G.; Unno, T.; Kim, Y.M. Prevalence of antibiotic resistance genes from effluent of coastal aquaculture, South Korea. Environ. Pollut. 2018, 233, 1049–1057. [Google Scholar] [CrossRef]
| Isolation No. a | Species | Isolation Source | AMP b | CTX | FOX | MEM | AMC | CN | S | C | TET | CIP | SXT | NDM | KPC | CTX-M |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FA8C | Klebsiella pneumoniae | hospital A/raw sewage | R | R | S | S | R | R | R | R | R | R | R | 3 | ||
| FA9C | Escherichia coli | hospital A/raw sewage | R | R | S | S | S | S | S | S | S | R | S | 3 | ||
| FA11C | Klebsiella pneumoniae | hospital A/raw sewage | R | R | S | S | I | I | S | S | R | R | R | 15 | ||
| FA1C | Escherichia coli | hospital A/raw sewage | R | R | R | S | I | S | R | R | R | R | R | 123 | ||
| FA3C | Klebsiella pneumoniae | hospital A/raw sewage | R | R | S | S | I | R | R | R | R | R | R | 15 | ||
| FA4C | Escherichia coli | hospital A/raw sewage | R | R | R | S | R | R | I | R | R | R | R | 15 | ||
| FA6C | Enterobacter sp. | hospital A/raw sewage | R | R | R | S | R | S | S | S | S | S | S | 65 | ||
| FA7C | Escherichia coli | hospital A/raw sewage | R | R | S | S | S | S | R | R | R | S | R | 55 | ||
| FA3M | Klebsiella pneumoniae | hospital A/raw sewage | R | R | R | R | R | S | I | R | R | S | R | 5 | ||
| FA5M | Klebsiella pneumoniae | hospital A/raw sewage | R | R | R | R | R | S | I | R | R | S | R | 5 | ||
| FA7M | Raoultella planticola | hospital A/raw sewage | R | R | R | R | R | R | S | S | R | R | R | 1 | 2 | 14 |
| FA9M | Escherichia coli | hospital A/raw sewage | R | R | R | R | R | R | R | R | R | R | R | 5 | 55 | |
| FA4M | Citrobacter braakii | hospital A/raw sewage | R | R | R | R | R | R | S | S | R | R | R | 2 | ||
| ZA4M | Escherichia coli | hospital B/raw sewage | R | R | R | R | R | S | S | I | R | R | R | 5 | 14 | |
| ZA3M | Escherichia coli | hospital B/raw sewage | R | R | R | R | I | S | S | S | S | R | R | 5 | 15 | |
| ZA1M | Escherichia coli | hospital B/raw sewage | R | R | R | R | R | R | R | R | R | R | R | 5 | 199 | |
| ZA10M | Escherichia coli | hospital B/raw sewage | R | R | R | R | R | R | R | R | R | R | R | 5 | 199 | |
| ZA9M | Klebsiella pneumoniae | hospital B/raw sewage | R | R | R | R | R | S | R | R | R | R | R | 27 | ||
| ZA7M | Citrobacter sp. | hospital B/raw sewage | R | R | R | R | R | R | S | S | R | R | R | 1 | 2 | 14 |
| ZA5M | Escherichia coli | hospital B/raw sewage | R | R | R | R | R | R | S | S | S | R | R | 5 | 123 | |
| ZA7C | Klebsiella pneumoniae | hospital B/raw sewage | R | R | R | S | R | S | S | S | R | S | R | |||
| ZA1C | Escherichia coli | hospital B/raw sewage | R | R | S | S | S | R | R | S | R | S | R | 55 | ||
| ZA2C | Escherichia coli | hospital B/raw sewage | R | R | R | S | I | S | I | S | S | R | R | 55 | ||
| ZA4C | Escherichia coli | hospital B/raw sewage | R | R | R | S | R | R | R | R | R | I | S | 55 | ||
| ZA5C | Escherichia coli | hospital B/raw sewage | R | R | S | S | S | R | R | S | R | S | R | 55 | ||
| ZA6C | Escherichia coli | hospital B/raw sewage | R | R | S | S | I | R | S | S | R | R | R | 55 | ||
| RA2M | Escherichia coli | hospital C/raw sewage | R | R | R | R | R | R | R | R | R | R | R | 5 | 55 | |
| RA1M | Enterobacter kobei | hospital C/raw sewage | R | R | R | R | R | S | S | S | S | S | S | 2 | 9 | |
| RA13C | Raoultella planticola | hospital C/raw sewage | R | R | S | R | R | R | S | S | S | S | R | 2 | ||
| RA14M | Klebsiella pneumoniae | hospital C/raw sewage | R | R | R | R | R | S | I | R | R | R | S | 2 | ||
| RA11M | Klebsiella pneumoniae | hospital C/raw sewage | R | R | R | R | R | R | S | R | S | R | R | 2 | 65 | |
| RA6C | Klebsiella pneumoniae | hospital C/raw sewage | R | R | R | R | R | R | S | S | S | S | R | 2 | ||
| RA5C | Escherichia coli | hospital C/raw sewage | R | R | S | S | I | S | R | R | R | S | R | 55 | ||
| RA4C | Escherichia coli | hospital C/raw sewage | R | R | S | R | I | R | I | S | R | R | R | 1 | 14 | |
| RA2C | Escherichia coli | hospital C/raw sewage | R | R | S | S | R | S | R | I | R | R | R | 55 | ||
| RA1C | Escherichia coli | hospital C/raw sewage | R | R | S | S | R | S | I | R | R | S | S | 55 | ||
| RA10M | Enterobacter sp. | hospital C/raw sewage | R | R | R | R | R | S | S | S | S | S | S | 2 | 55 | |
| RA7M | Escherichia coli | hospital C/raw sewage | R | R | R | R | R | R | R | R | R | R | R | 5 | + | |
| RA12C | Escherichia coli | hospital C/raw sewage | R | R | S | R | R | S | S | S | S | R | S | 1 | 55 | |
| RA11C | Escherichia coli | hospital C/raw sewage | R | R | S | R | S | S | S | S | S | R | S | 1 | ||
| RA8C | Klebsiella pneumoniae | hospital C/raw sewage | R | R | R | S | R | R | S | R | R | R | R | 27 | ||
| RA7C | Escherichia coli | hospital C/raw sewage | R | R | S | S | R | R | S | S | R | R | R | 15 | ||
| RA8M | Escherichia coli | hospital C/raw sewage | R | R | R | R | R | R | R | R | R | S | R | 55 | ||
| RB1C | Escherichia coli | hospital C/effluent | R | R | S | S | I | R | R | R | R | R | R | 55 | ||
| RB7C | Escherichia coli | hospital C/effluent | R | R | I | R | S | S | I | S | S | R | R | 2 | 14 | |
| RB6C | Escherichia coli | hospital C/effluent | R | R | S | S | I | R | R | R | R | R | R | 55 | ||
| RB4C | Escherichia coli | hospital C/effluent | R | R | S | S | R | S | S | R | R | S | S | 55 | ||
| RB3M | Klebsiella pneumoniae | hospital C/effluent | R | R | R | R | R | R | R | R | R | R | R | 5 | 55 | |
| RB6M | Klebsiella variicola | hospital C/effluent | R | R | R | R | R | R | R | R | R | S | R | 5 | 55 | |
| CS3C | Escherichia coli | river water | R | R | S | S | S | S | S | S | S | R | R | 14 | ||
| CS4C | Enterobacter tabaci | river water | R | R | R | S | R | S | S | S | S | S | S | + | ||
| CS8C | Escherichia coli | river water | R | R | S | S | R | S | R | S | R | R | R | 55 | ||
| TX1C | Klebsiella variicola | river water | R | R | R | S | R | S | S | S | R | S | R | + | ||
| TX2C | Klebsiella pneumoniae | river water | R | R | S | S | S | S | S | R | R | S | R | 27 | ||
| TX5C | Klebsiella pneumoniae | river water | R | R | R | S | R | S | R | S | R | S | R | + | ||
| TS1C | Klebsiella pneumoniae | river water | R | R | R | S | S | S | S | S | S | S | S | + | ||
| TS11C | Escherichia coli | river water | R | R | S | S | S | S | S | S | R | S | S | |||
| TS3M | Escherichia coli | river water | R | R | R | R | R | R | R | R | R | R | R | 5 | ||
| TS4M | Klebsiella pneumoniae | river water | R | R | R | R | R | R | R | R | R | R | R | 5 | ||
| CX1M | Escherichia coli | river water | R | R | R | R | R | R | R | S | R | R | S | 1 | ||
| TX5M | Citrobacter freundii | river water | R | R | R | R | R | R | R | R | R | R | R | 5 | ||
| TX6M | Escherichia coli | river water | R | R | R | R | R | R | I | R | R | S | R |
| Isolation No. a | Isolation Source | CTX b | FOX | MEM | CN | TET | CIP | SXT | NDM | OXA | CTX-M |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FA2C | hospital A/raw sewage | R | R | S | S | S | S | S | 65 | ||
| FA2M | hospital A/raw sewage | R | R | R | R | R | R | R | 1 | ||
| ZA2M | hospital B/raw sewage | R | R | R | R | R | I | R | 1 | 58 | |
| ZA6M | hospital B/raw sewage | R | R | R | R | R | S | R | 1 | ||
| ZA8M | hospital B/raw sewage | R | S | S | R | S | S | I | |||
| ZA8C | hospital B/raw sewage | R | R | S | S | S | S | S | 55 | ||
| RA5M | hospital C/raw sewage | R | R | R | S | R | S | R | 1 | ||
| RA9M | hospital C/raw sewage | R | R | R | R | R | R | R | 1 | 58 | |
| RA12M | hospital C/raw sewage | R | R | R | R | R | I | R | 5 | 58 | 3 |
| RA3M | hospital C/raw sewage | R | R | R | S | R | I | R | 1 | 58 | |
| RA4M | hospital C/raw sewage | R | R | R | R | R | I | R | 1 | 58 | |
| RB4M | hospital C/effluent | R | R | R | R | R | S | R | 1 | ||
| RB5C | hospital C/effluent | R | R | R | R | R | S | R | 1 | 55 | |
| RB1M | hospital C/effluent | R | R | R | R | R | I | R | 55 | ||
| RB2M | hospital C/effluent | R | R | R | R | R | S | R | 1 | 58 | 55 |
| RB5M | hospital C/effluent | R | R | R | S | R | I | I | 1 | 58 | 3 |
| CX9C | river water | R | I | S | S | S | S | S | |||
| CX11C | river water | R | S | S | S | R | S | S | |||
| TX3C | river water | R | R | S | S | R | S | R | |||
| CS1M | river water | R | R | R | S | S | S | S | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Ma, X.; Luo, L.; Hu, N.; Duan, J.; Tang, Z.; Zhong, R.; Li, Y. The Prevalence and Characterization of Extended-Spectrum β-Lactamase- and Carbapenemase-Producing Bacteria from Hospital Sewage, Treated Effluents and Receiving Rivers. Int. J. Environ. Res. Public Health 2020, 17, 1183. https://doi.org/10.3390/ijerph17041183
Zhang L, Ma X, Luo L, Hu N, Duan J, Tang Z, Zhong R, Li Y. The Prevalence and Characterization of Extended-Spectrum β-Lactamase- and Carbapenemase-Producing Bacteria from Hospital Sewage, Treated Effluents and Receiving Rivers. International Journal of Environmental Research and Public Health. 2020; 17(4):1183. https://doi.org/10.3390/ijerph17041183
Chicago/Turabian StyleZhang, Luhua, Xinyue Ma, Li Luo, Nan Hu, Jiayao Duan, Zhongjian Tang, Rujie Zhong, and Ying Li. 2020. "The Prevalence and Characterization of Extended-Spectrum β-Lactamase- and Carbapenemase-Producing Bacteria from Hospital Sewage, Treated Effluents and Receiving Rivers" International Journal of Environmental Research and Public Health 17, no. 4: 1183. https://doi.org/10.3390/ijerph17041183
APA StyleZhang, L., Ma, X., Luo, L., Hu, N., Duan, J., Tang, Z., Zhong, R., & Li, Y. (2020). The Prevalence and Characterization of Extended-Spectrum β-Lactamase- and Carbapenemase-Producing Bacteria from Hospital Sewage, Treated Effluents and Receiving Rivers. International Journal of Environmental Research and Public Health, 17(4), 1183. https://doi.org/10.3390/ijerph17041183





