Spatial and Temporal Analysis of Plasmodium knowlesi Infection in Peninsular Malaysia, 2011 to 2018
Abstract
1. Introduction
2. Materials and Methods
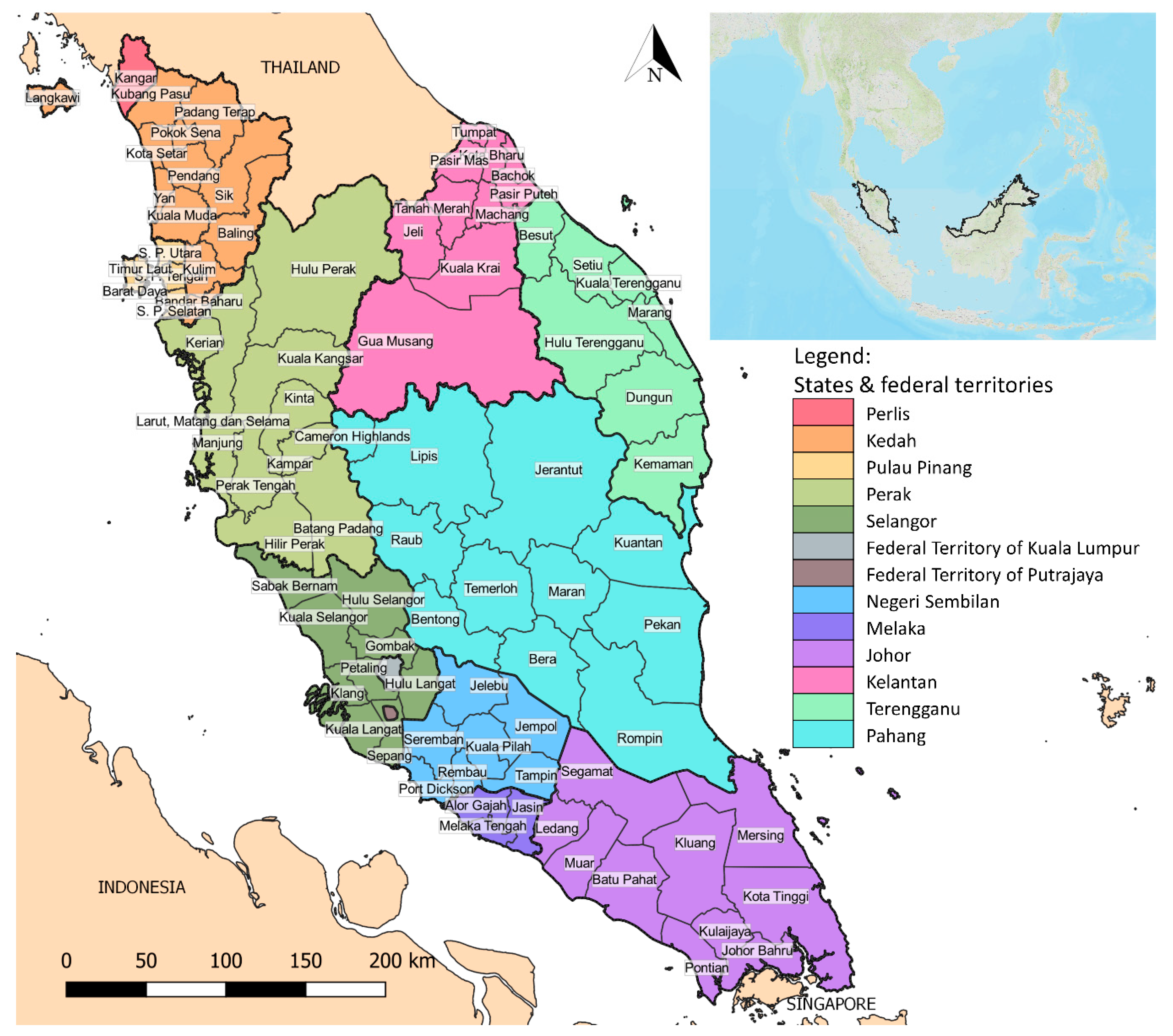
2.1. Geography and Demography of Study Areas
2.2. Data of Knowlesi Malaria in Peninsular Malaysia
2.3. Spatial Analysis of Knowlesi Malaria in Peninsular Malaysia
2.4. Time Series Analysis of P. knowlesi Incidence in the Hotspot Region
3. Results
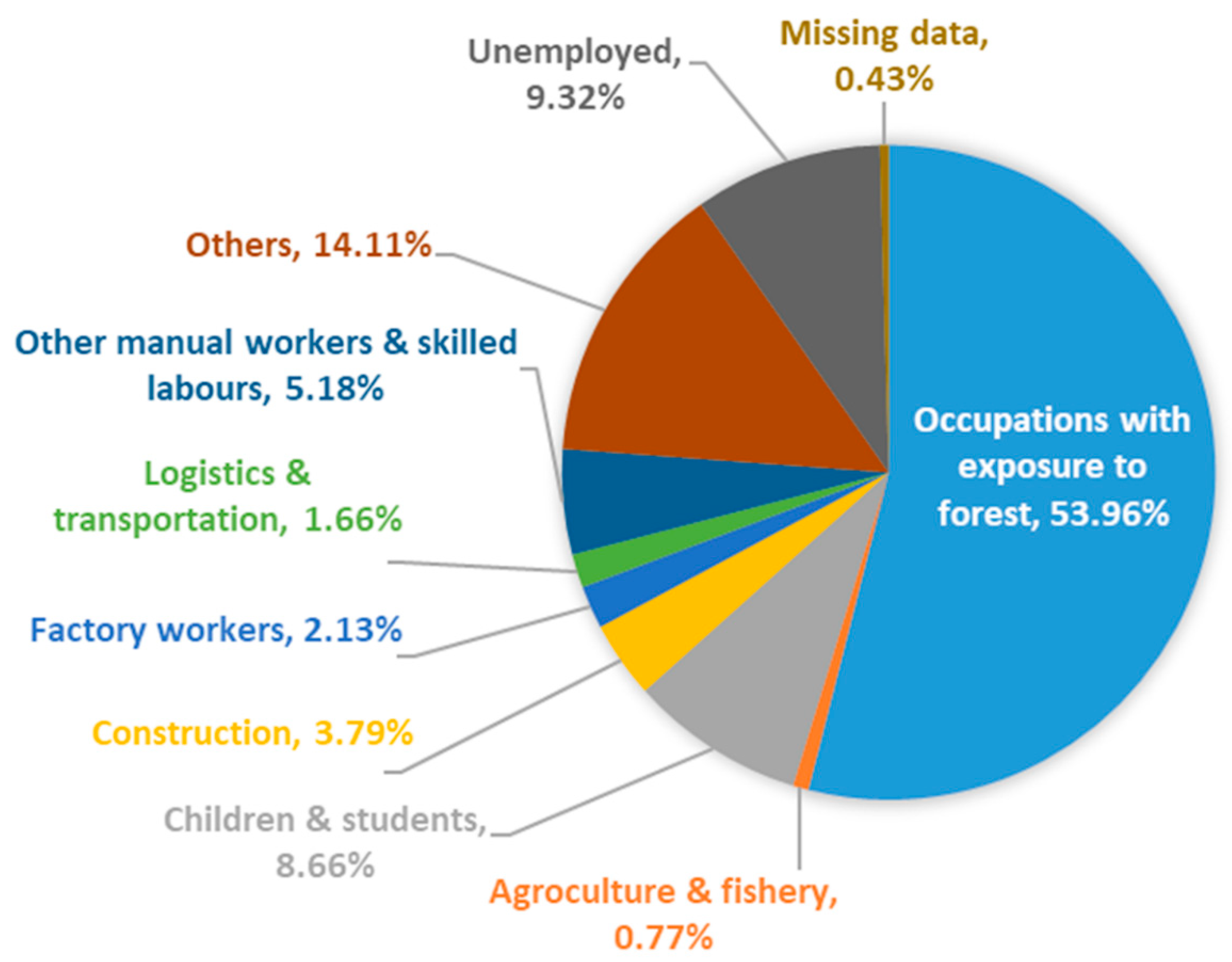
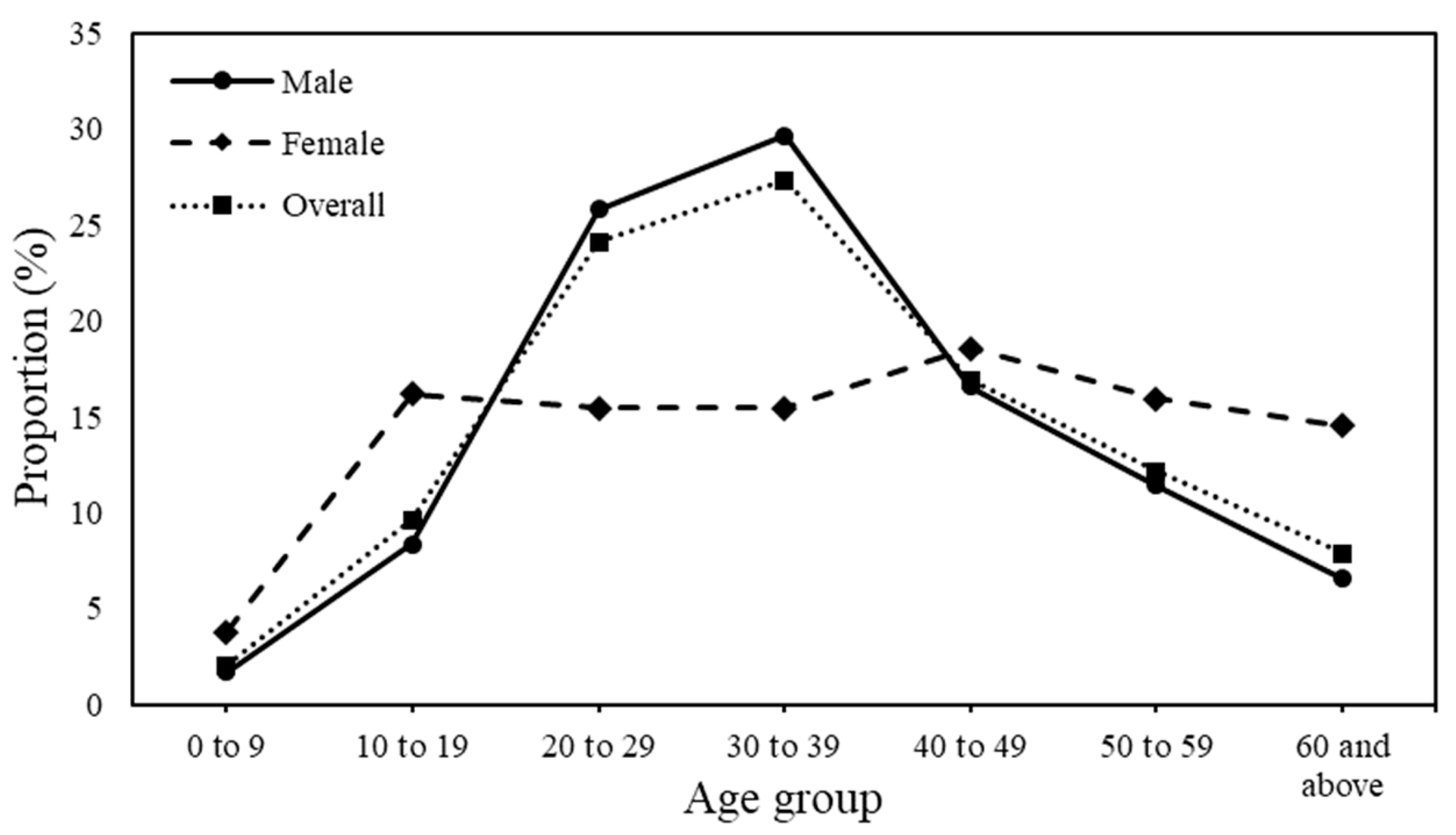
3.1. Demographic Characteristics of Knowlesi Malaria from 2011 to 2018
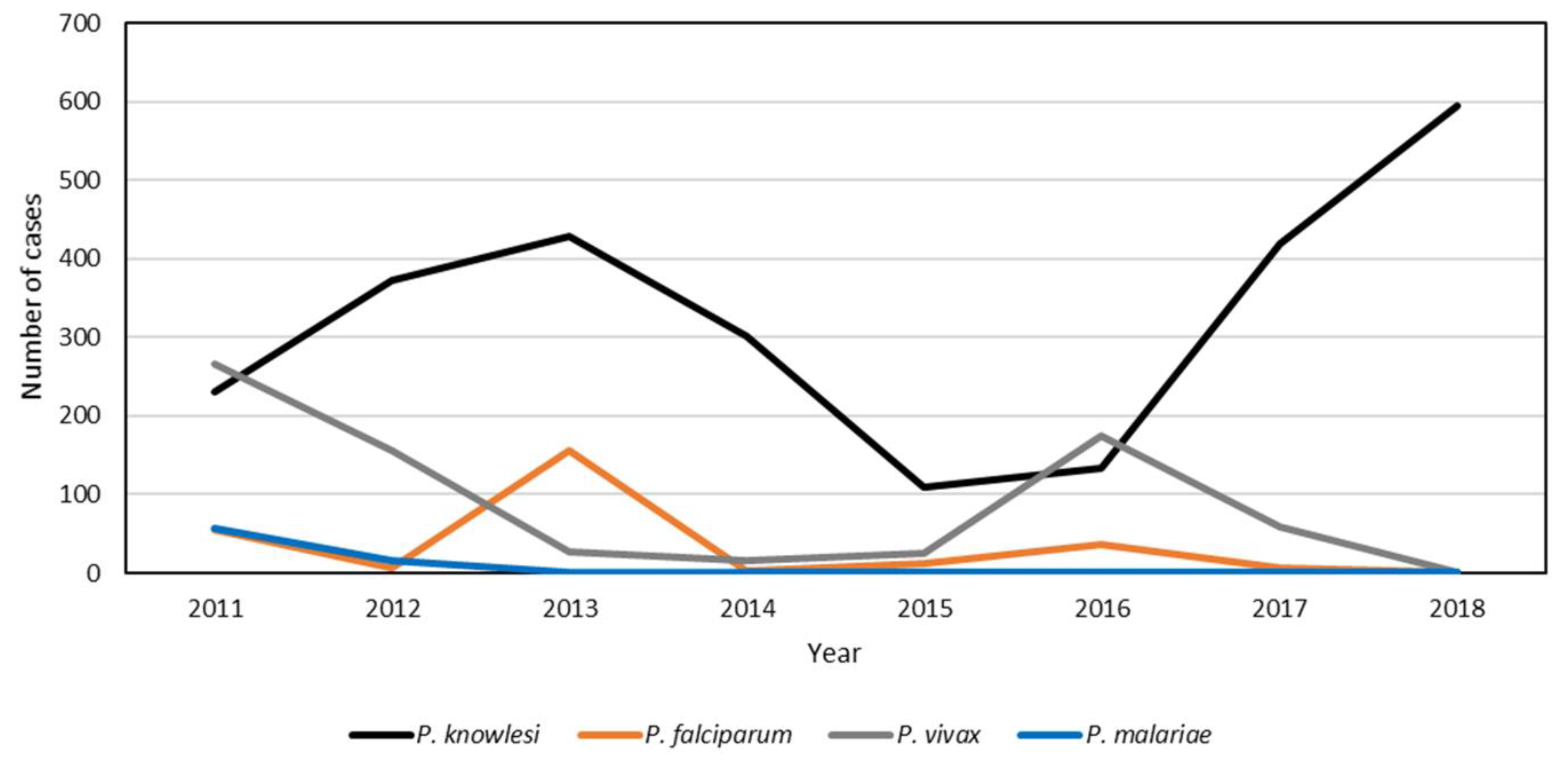
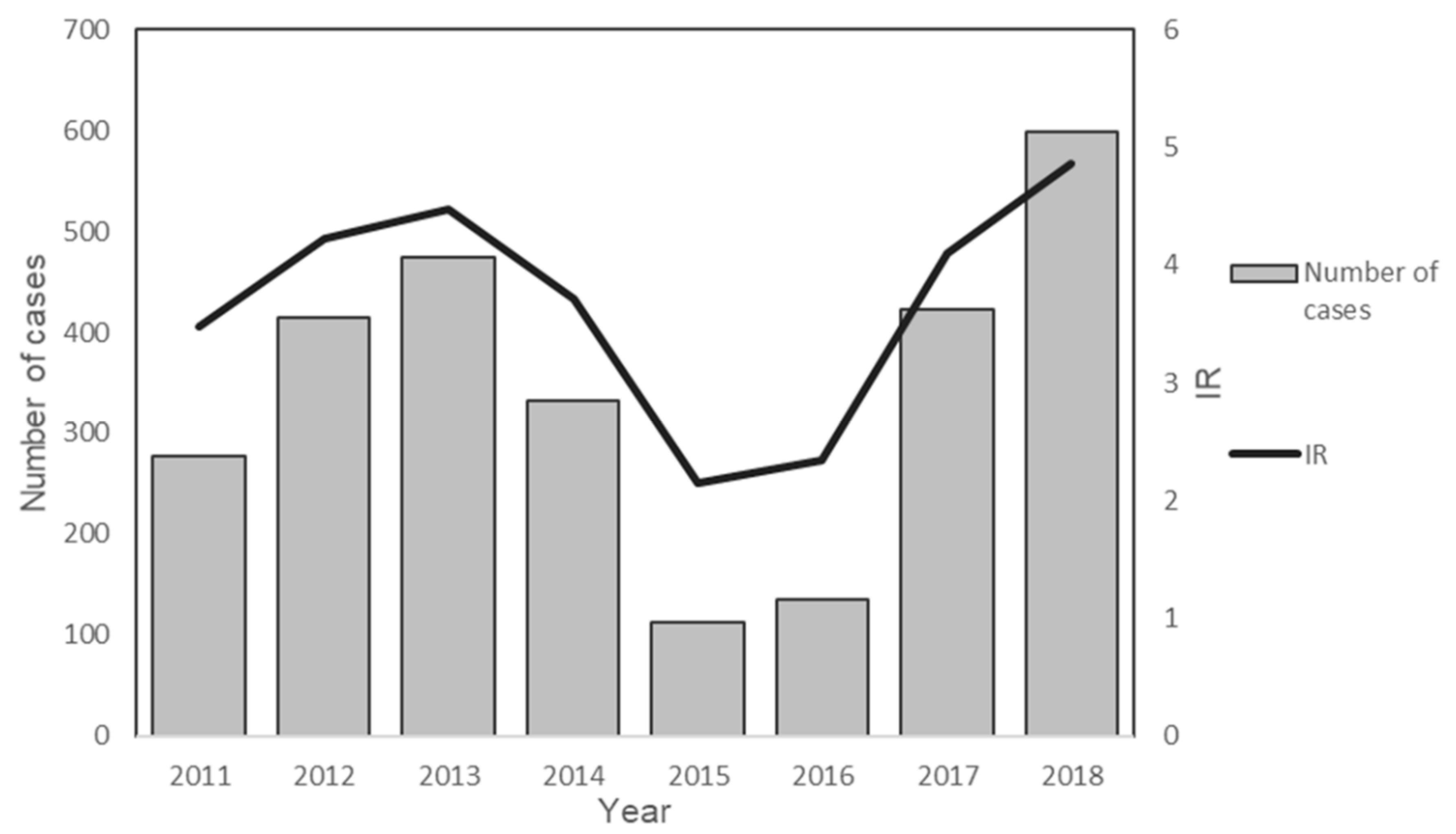
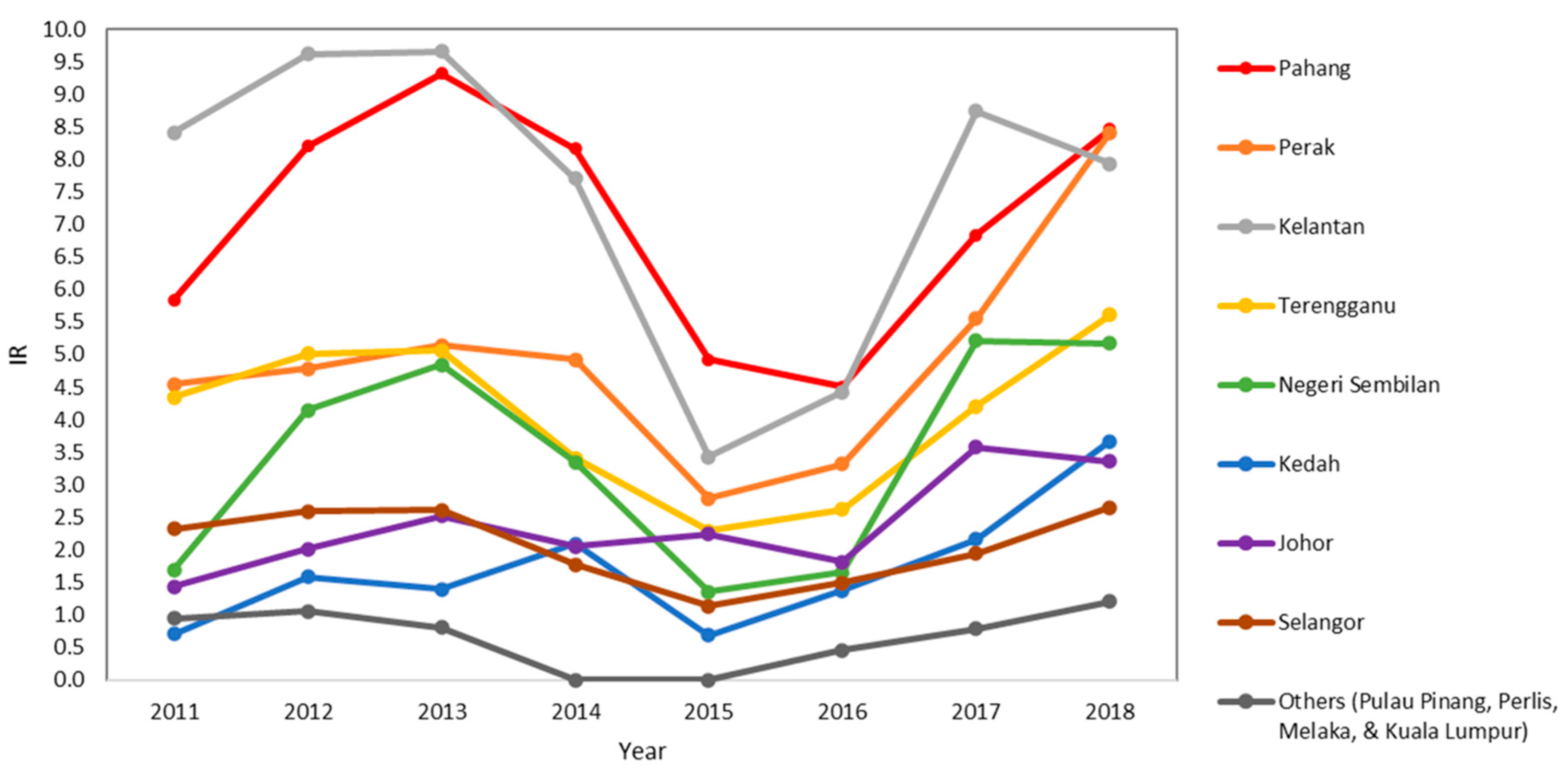
3.2. State-Level Trend of P. knowlesi Incidence in Peninsular Malaysia from 2011 to 2018
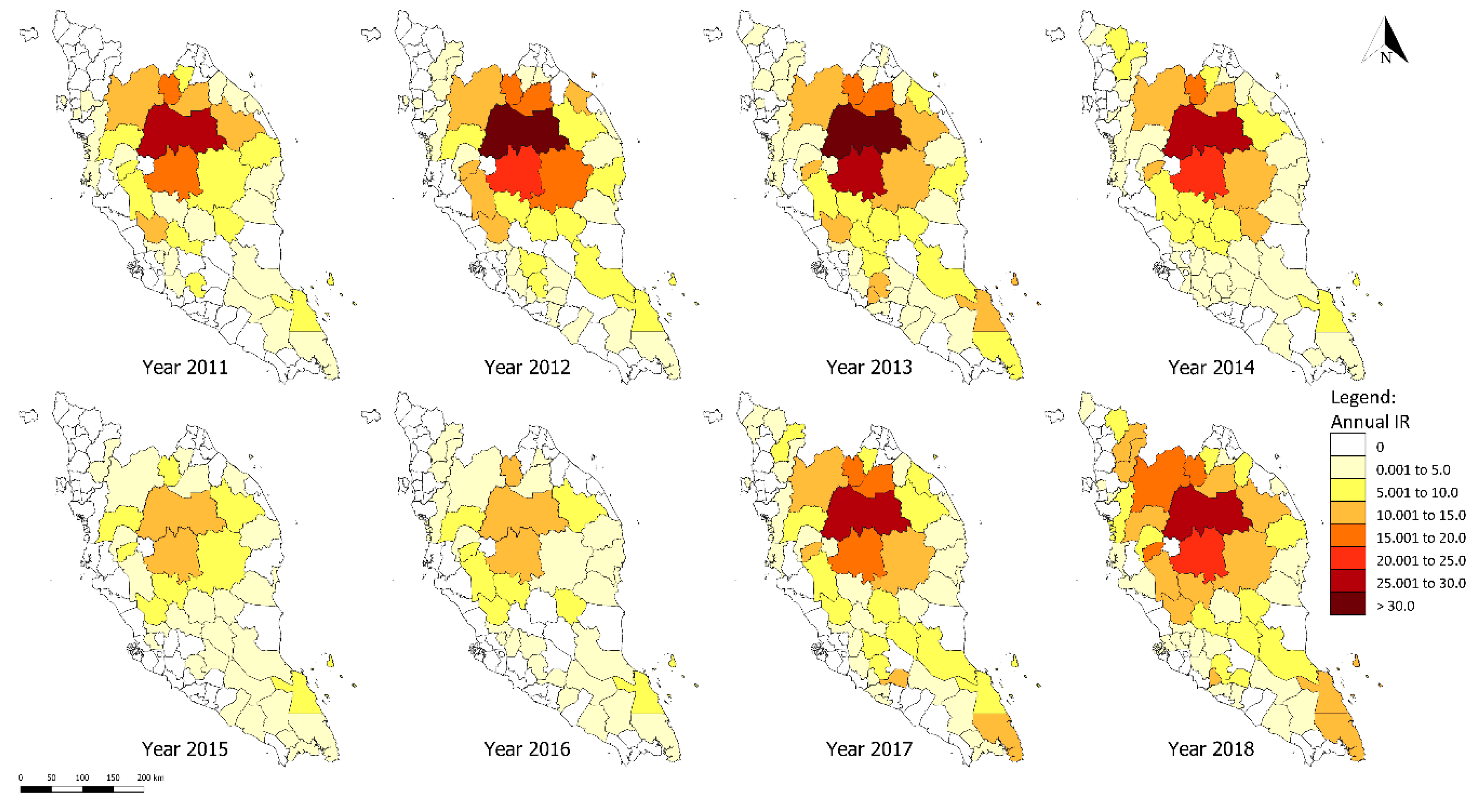
3.3. District-Level Spatial Analysis of P. knowlesi Infection
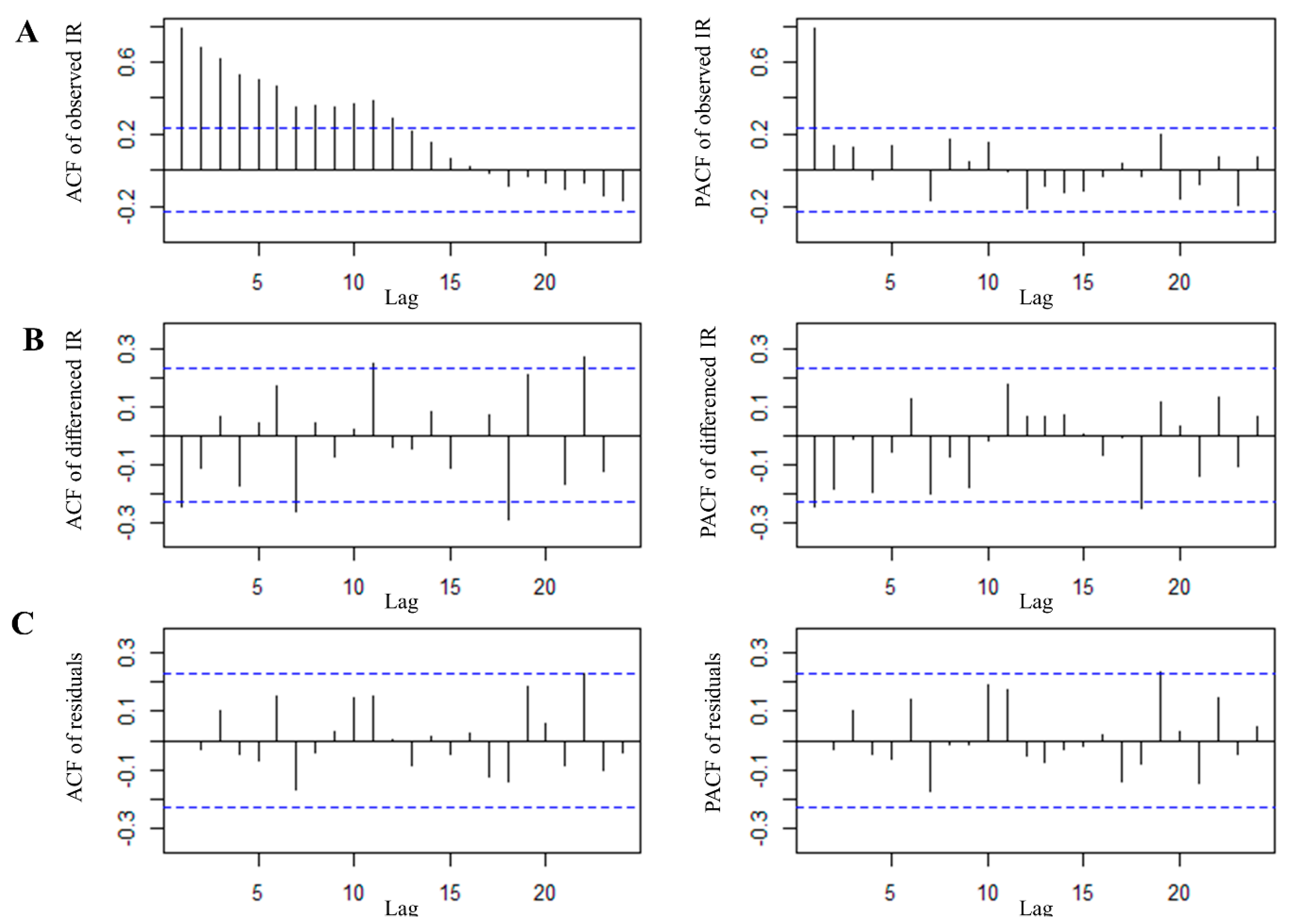
3.4. Time Series Analysis of P. knowlesi Incidence in the Hotspot Region
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- White, N.J.; Pukrittayakamee, S.; Hien, T.T.; Faiz, M.A.; Mokuolu, O.A.; Dondorp, A.M. Malaria. Lancet 2014, 383, 723–735. [Google Scholar] [CrossRef]
- World Health Organization. World Malaria Report 2019; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Lim, E.S. Current status of malaria in Malaysia. Southeast Asian J. Trop. Med. Public Health 1992, 23 (Suppl. 4), 43–49. [Google Scholar] [PubMed]
- White, N.J. Plasmodium knowlesi: The fifth human malaria parasite. Clin. Infect. Dis. 2008, 46, 172–173. [Google Scholar] [CrossRef] [PubMed]
- Daneshvar, C.; Davis, T.M.; Cox-Singh, J.; Rafa’ee, M.Z.; Zakaria, S.K.; Divis, P.C.; Singh, B. Clinical and laboratory features of human Plasmodium knowlesi infection. Clin. Infect. Dis. 2009, 49, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Hussin, N.; Lim, Y.A.; Goh, P.P.; William, T.; Jelip, J.; Mudin, R.N. Updates on malaria incidence and profile in Malaysia from 2013 to 2017. Malar. J. 2020, 19, 55. [Google Scholar] [CrossRef] [PubMed]
- Cox-Singh, J.; Davis, T.M.; Lee, K.S.; Shamsul, S.S.; Matusop, A.; Ratnam, S.; Rahman, H.A.; Conway, D.J.; Singh, B. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis. 2008, 46, 165–171. [Google Scholar] [CrossRef]
- Singh, B.; Lee, K.S.; Matusop, A.; Radhakrishnan, A.; Shamsul, S.S.; Cox-Singh, J.; Thomas, A.; Conway, D.J. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 2004, 363, 1017–1024. [Google Scholar] [CrossRef]
- Vythilingam, I.; Lim, Y.A.; Venugopalan, B.; Ngui, R.; Leong, C.S.; Wong, M.L.; Khaw, L.; Goh, X.; Yap, N.; Sulaiman, W.Y.; et al. Plasmodium knowlesi malaria an emerging public health problem in Hulu Selangor, Selangor, Malaysia (2009–2013): Epidemiologic and entomologic analysis. Parasit. Vectors 2014, 7, 436. [Google Scholar] [CrossRef]
- Marchand, R.P.; Culleton, R.; Maeno, Y.; Quang, N.T.; Nakazawa, S. Co-infections of Plasmodium knowlesi, P. falciparum, and P. vivax among humans and Anopheles dirus mosquitoes, southern Vietnam. Emerg. Infect. Dis. 2011, 17, 1232–1239. [Google Scholar] [CrossRef]
- Dhimal, M.; O’Hara, R.B.; Karki, R.; Thakur, G.D.; Kuch, U.; Ahrens, B. Spatio-temporal distribution of malaria and its association with climatic factors and vector-control interventions in two high-risk districts of Nepal. Malar. J. 2014, 13, 457. [Google Scholar] [CrossRef]
- Hundessa, S.H.; Williams, G.; Li, S.; Guo, J.; Chen, L.; Zhang, W.; Guo, Y. Spatial and space-time distribution of Plasmodium vivax and Plasmodium falciparum malaria in China, 2005–2014. Malar. J. 2016, 15, 595. [Google Scholar] [CrossRef] [PubMed]
- Mercado, C.E.G.; Lawpoolsri, S.; Sudathip, P.; Kaewkungwal, J.; Khamsiriwatchara, A.; Pan-Ngum, W.; Yimsamran, S.; Lawawirojwong, S.; Ho, K.; Ekapirat, N.; et al. Spatiotemporal epidemiology, environmental correlates, and demography of malaria in Tak Province, Thailand (2012–2015). Malar. J. 2019, 18, 240. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.J.; Rajahram, G.S.; William, T.; Jelip, J.; Mohammad, R.; Benedict, J.; Alaza, D.A.; Malacova, E.; Yeo, T.W.; Grigg, M.J.; et al. Plasmodium knowlesi malaria in Sabah, Malaysia, 2015–2017: Ongoing increase in incidence despite near-elimination of the human-only Plasmodium species. Clin. Infect. Dis. 2020, 70, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Fornace, K.M.; Brock, P.M.; Abidin, T.R.; Grignard, L.; Herman, L.S.; Chua, T.H.; Daim, S.; William, T.; Patterson, C.; Hall, T.; et al. Environmental risk factors and exposure to the zoonotic malaria parasite Plasmodium knowlesi across northern Sabah, Malaysia: A population-based cross-sectional survey. Lancet Planet Health 2019, 3, e179–e186. [Google Scholar] [CrossRef]
- William, T.; Rahman, H.A.; Jelip, J.; Ibrahim, M.Y.; Menon, J.; Grigg, M.J.; Yeo, T.W.; Anstey, N.M.; Barber, B.E. Increasing incidence of Plasmodium knowlesi malaria following control of P. falciparum and P. vivax Malaria in Sabah, Malaysia. PLoS Negl. Trop. Dis. 2013, 7, e2026. [Google Scholar] [CrossRef]
- Department of Statistics Malaysia. Population Distribution and Basic Demographic Characteristics 2010; Department of Statistics Malaysia: Putrajaya, Malaysia, 2011.
- Forestry Department Peninsular Malaysia. Forestry Statistics. Available online: https://www.forestry.gov.my/en/2016-06-07-02-53-46/2016-06-07-03-12-29 (accessed on 21 December 2019).
- Suhaila, J.; Deni, S.M.; Zin, W.W.; Jemain, A.A.J.S.M. Trends in peninsular Malaysia rainfall data during the southwest monsoon and northeast monsoon seasons: 1975–2004. Sains Malays. 2010, 39, 533–542. [Google Scholar]
- Ministry of Health Malaysia. Management Guidelines of Malaria in Malaysia; Ministry of Health Malaysia: Putrajaya, Malaysia, 2014.
- Department of Statistics Malaysia. Malaysia @ a Glance. Available online: https://www.dosm.gov.my/v1/index.php?r=column/cone&menu_id=ZmVrN2FoYnBvZE05T1AzK0RLcEtiZz09 (accessed on 30 October 2019).
- World Health Organization. WHO Malaria Terminology; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Moran, P.A.P. The interpretation of statistical maps. J. R. Stat. Soc. B 1948, 10, 243–251. [Google Scholar] [CrossRef]
- Anselin, L. Local indicators of spatial association-LISA. Geogr. Anal. 1995, 27, 93–115. [Google Scholar] [CrossRef]
- Cleveland, R.; Cleveland, W.S.; McRae, J.; Terpenning, I.J. STL: A seasonal-trend decomposition procedure based on loess. J. Off. Stat. 1990, 6, 3–33. [Google Scholar]
- Fabozzi, F.J.; Focardi, S.M.; Rachev, S.T.; Ashanapalli, B.G. The Basics of Financial Econometrics: Tools, Concepts, and Asset Management Applications; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Grigg, M.J.; Cox, J.; William, T.; Jelip, J.; Fornace, K.M.; Brock, P.M.; von Seidlein, L.; Barber, B.E.; Anstey, N.M.; Yeo, T.W.; et al. Individual-level factors associated with the risk of acquiring human Plasmodium knowlesi malaria in Malaysia: A case-control study. Lancet Planet Health 2017, 1, e97–e104. [Google Scholar] [CrossRef]
- Masron, T.; Masami, F.; Ismail, N. Orang Asli in Peninsular Malaysia: Population, spatial distribution and socio-economic condition. J. Ritsumeikan Soc. Sci. Hum. 2013, 6, 75–115. [Google Scholar]
- Norhayati, M.; Rohani, A.K.; Hayati, M.I.; Halimah, A.S.; Sharom, M.Y.; Abidin, A.H.; Fatmah, M.S. Clinical features of malaria in Orang Asli population in Pos Piah, Malaysia. Med. J. Malaysia 2001, 56, 271–274. [Google Scholar] [PubMed]
- Liew, J.W.K.; Mahpot, R.B.; Dzul, S.; Abdul Razak, H.A.B.; Ahmad Shah Azizi, N.A.B.; Kamarudin, M.B.; Russell, B.; Lim, K.L.; de Silva, J.R.; Lim, B.S.; et al. Importance of proactive malaria case surveillance and management in Malaysia. Am. J. Trop. Med. Hyg. 2018, 98, 1709–1713. [Google Scholar] [CrossRef] [PubMed]
- Mahdy, M.; Chan, B.; Noor Hayati, M.; Sa’iah, H.; Ismail, M.; Jeffery, J.J.T.B. The distribution of malaria parasites among Orang Asli populations living in the interior areas of Pahang and Kelantan, Malaysia. Trop. Biomed. 2004, 21, 101–105. [Google Scholar]
- Jiram, A.I.; Hisam, H.; Reuben, H.; Husin, S.Z.; Roslan, A.; Ismail, W.R.W. Submicroscopic evidence of the simian malaria parasite, P. knowlesi, in orang asli community. Southeast Asian J. Trop. Med. Public Health 2016, 47, 591–599. [Google Scholar]
- Kardooni, R.; Kari, F.; Yahaya, S.; Yusup, S. Traditional knowledge of orang asli on forests in Peninsular Malaysia. Indian J. Tradit. Know. 2014, 13, 283–291. [Google Scholar]
- Kamal, S.F.; Lim, V.-C. Forest reserve as an inclusive or exclusive space? Engaging orang asli as stakeholder in protected area management. J. Trop. For. Sci. 2019, 31, 278–285. [Google Scholar] [CrossRef]
- Samsurijan, M.S.; Rahman, N.N.N.A.; Ishak, M.I.S.; Masron, T.A.; Kadir, O.A. Land use change in Kelantan: Review of the environmental impact assessment (EIA) reports. Geografia Malays. J. Soc. Space 2018, 14, 322–331. [Google Scholar] [CrossRef]
- Karuppannan, K.; Saaban, S.; Mustapa, A.R.; Zainal Abidin, F.A.; Azimat, N.A.; Keliang, C. Population status of long-tailed macaque (Macaca fascicularis) in Peninsular Malaysia. J. Primatol. 2014, 3, 1000118. [Google Scholar] [CrossRef]
- Vythilingam, I.; Wong, M.L.; Wan-Yussof, W.S. Current status of Plasmodium knowlesi vectors: A public health concern? Parasitology 2018, 145, 32–40. [Google Scholar] [CrossRef]
- Amir, A.; Shahari, S.; Liew, J.W.K.; de Silva, J.R.; Khan, M.B.; Lai, M.Y.; Snounou, G.; Abdullah, M.L.; Gani, M.; Rovie-Ryan, J.J.; et al. Natural Plasmodium infection in wild macaques of three states in Peninsular Malaysia. Acta Trop. 2020, 211, 105596. [Google Scholar] [CrossRef] [PubMed]
- Akter, R.; Vythilingam, I.; Khaw, L.T.; Qvist, R.; Lim, Y.A.L.; Sitam, F.T.; Venugopalan, B.; Sekaran, S.D. Simian malaria in wild macaques: First report from Hulu Selangor district, Selangor, Malaysia. Malar. J. 2015, 14, 386. [Google Scholar] [CrossRef] [PubMed]
- Vythilingam, I.; NoorAzian, Y.M.; Tan, C.H.; Jiram, A.I.; Yusri, Y.M.; Azahari, A.H.; NorParina, I.; NoorRain, A.; LokmanHakim, S. Plasmodium knowlesi in humans, macaques and mosquitoes in Peninsular Malaysia. Parasit. Vectors 2008, 1, 26. [Google Scholar] [CrossRef] [PubMed]
- Warren, M.; Cheong, W.H.; Fredericks, H.K.; Coatney, G.R. Cycles of jungle malaria in West Malaysia. Am. J. Trop. Med. Hyg. 1970, 19, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Chaves, L.F.; Ramírez Rojas, M.; Prado, M.; Garcés, J.L.; Salas Peraza, D.; Marín Rodríguez, R. Health policy impacts on malaria transmission in Costa Rica. Parasitology 2020, 147, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T.; Loha, E.; Deressa, W.; Gari, T.; Lindtjorn, B. Spatiotemporal clustering of malaria in southern-central Ethiopia: A community-based cohort study. PLoS ONE 2019, 14, e0222986. [Google Scholar] [CrossRef]
- Samsuddin, N.A.C.; Khan, M.F.; Maulud, K.N.A.; Hamid, A.H.; Munna, F.T.; Rahim, M.A.A.; Latif, M.T.; Akhtaruzzaman, M. Local and transboundary factors’ impacts on trace gases and aerosol during haze episode in 2015 El Nino in Malaysia. Sci. Total Environ. 2018, 630, 1502–1514. [Google Scholar] [CrossRef]
- Miyamoto, M.; Mamat Mohd, P.; Zakaria Noor, A.; Michinaka, T. Proximate and underlying causes of forest cover change in Peninsular Malaysia. For. Policy Econ. 2014, 44, 18–25. [Google Scholar] [CrossRef]
- Guerra, C.A.; Snow, R.W.; Hay, S.I. A global assessment of closed forests, deforestation and malaria risk. Ann. Trop. Med. Parasitol. 2006, 100, 189–204. [Google Scholar] [CrossRef]
- Coatney, G.R.; Collins, W.E.; Warren, M.; Gontacos, P.G. The Primate Malarias; Centers for Disease Control and Prevention: Atlanta, GA, USA, 1971. [Google Scholar]
- Muh, F.; Ahmed, M.A.; Han, J.H.; Nyunt, M.H.; Lee, S.K.; Lau, Y.L.; Kaneko, O.; Han, E.T. Cross-species analysis of apical asparagine-rich protein of Plasmodium vivax and Plasmodium knowlesi. Sci. Rep. 2018, 8, 5781. [Google Scholar] [CrossRef]
- Van Rooyen, C.E.; Pile, G.R. Observations on infection by Plasmodium knowlesi (ape malaria) in the treatment of general paralysis of the insane. Br. Med. J. 1935, 2, 662–666. [Google Scholar] [CrossRef] [PubMed]
- William, T.; Jelip, J.; Menon, J.; Anderios, F.; Mohammad, R.; Awang Mohammad, T.A.; Grigg, M.J.; Yeo, T.W.; Anstey, N.M.; Barber, B.E. Changing epidemiology of malaria in Sabah, Malaysia: Increasing incidence of Plasmodium knowlesi. Malar. J. 2014, 13, 390. [Google Scholar] [CrossRef]
- Fornace, K.M.; Herman, L.S.; Abidin, T.R.; Chua, T.H.; Daim, S.; Lorenzo, P.J.; Grignard, L.; Nuin, N.A.; Ying, L.T.; Grigg, M.J.; et al. Exposure and infection to Plasmodium knowlesi in case study communities in Northern Sabah, Malaysia and Palawan, The Philippines. PLoS Negl. Trop. Dis. 2018, 12, e0006432. [Google Scholar] [CrossRef]
- Huestis, D.L.; Dao, A.; Diallo, M.; Sanogo, Z.L.; Samake, D.; Yaro, A.S.; Ousman, Y.; Linton, Y.M.; Krishna, A.; Veru, L.; et al. Windborne long-distance migration of malaria mosquitoes in the Sahel. Nature 2019, 574, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Dao, A.; Yaro, A.S.; Diallo, M.; Timbine, S.; Huestis, D.L.; Kassogue, Y.; Traore, A.I.; Sanogo, Z.L.; Samake, D.; Lehmann, T. Signatures of aestivation and migration in Sahelian malaria mosquito populations. Nature 2014, 516, 387–390. [Google Scholar] [CrossRef]
- Anwar, M.Y.; Lewnard, J.A.; Parikh, S.; Pitzer, V.E. Time series analysis of malaria in Afghanistan: Using ARIMA models to predict future trends in incidence. Malar. J. 2016, 15, 566. [Google Scholar] [CrossRef] [PubMed]
- Ebhuoma, O.; Gebreslasie, M.; Magubane, L. A seasonal autoregressive integrated moving average (SARIMA) forecasting model to predict monthly malaria cases in KwaZulu-Natal, South Africa. S. Afr. Med. J. 2018, 108, 573–578. [Google Scholar] [CrossRef]
- Permanasari, A.E.; Hidayah, I.; Bustoni, I.A. SARIMA (seasonal ARIMA) implementation on time series to forecast the number of malaria incidence. In Proceedings of the 2013 International Conference on Information Technology and Electrical Engineering (ICITEE), Yogyakarta, Indonesia, 7–8 October 2013; pp. 203–207. [Google Scholar]
- Martinez, E.Z.; Silva, E.A.; Fabbro, A.L. A SARIMA forecasting model to predict the number of cases of dengue in Campinas, State of Sao Paulo, Brazil. Rev. Soc. Bras. Med. Trop. 2011, 44, 436–440. [Google Scholar] [CrossRef]
- Cong, J.; Ren, M.; Xie, S.; Wang, P. Predicting seasonal influenza based on SARIMA model, in Mainland China from 2005 to 2018. Int. J. Environ. Res. Public. Health 2019, 16, 4760. [Google Scholar] [CrossRef]
- He, Z.; Tao, H. Epidemiology and ARIMA model of positive-rate of influenza viruses among children in Wuhan, China: A nine-year retrospective study. Int. J. Infect. Dis. 2018, 74, 61–70. [Google Scholar] [CrossRef]
- Xu, Q.; Li, R.; Liu, Y.; Luo, C.; Xu, A.; Xue, F.; Xu, Q.; Li, X. Forecasting the incidence of mumps in Zibo city based on a SARIMA model. Int. J. Environ. Res. Public Health 2017, 14, 925. [Google Scholar] [CrossRef] [PubMed]
- Ooi, C.H.; Bujang, M.A.; Tg Abu Bakar Sidik, T.M.I.; Ngui, R.; Lim, Y.A. Over two decades of Plasmodium knowlesi infections in Sarawak: Trend and forecast. Acta Trop. 2017, 176, 83–90. [Google Scholar] [CrossRef] [PubMed]


| Variable | N (%) |
|---|---|
| Age group | |
| 0 to 9 | 53 (2.0) |
| 10 to 19 | 249 (9.6) |
| 20 to 29 | 624 (24.1) |
| 30 to 39 | 707 (27.3) |
| 40 to 49 | 437 (16.9) |
| 50 to 59 | 314 (12.1) |
| 60 and above | 203 (7.8) |
| Gender | |
| Male | 2167 (83.8) |
| Female | 420 (16.2) |
| Ethnicity | |
| Chinese | 145 (5.6) |
| Indian | 44 (1.7) |
| Malay | 1518 (58.7) |
| Orang Asli (aborigine community) | 372 (14.4) |
| Sabahan | 27 (1.0) |
| Sarawakian | 34 (1.3) |
| Others | 429 (16.6) |
| No data | 18 (0.7) |
| Occupation class | |
| Agroculture and fishery | 20 (0.8) |
| Army and police * | 138 (5.3) |
| Children and students | 224 (8.7) |
| Construction | 98 (3.8) |
| Ecotourism, forest, and wildlife management * | 39 (1.5) |
| Estate, farm, and plantation workers * | 897 (34.7) |
| Factory workers | 55 (2.1) |
| Forest resource gatherers * | 82 (3.2) |
| Logging * | 121 (4.7) |
| Logistics and transportation | 43 (1.7) |
| Mining * | 51 (2.0) |
| Other manual workers and skilled labors | 134 (5.2) |
| Village labors * | 68 (2.6) |
| Others | 365 (14.1) |
| Unemployed | 241 (9.3) |
| No data | 11 (0.4) |
| Citizenship | |
| Locals | 2161 (83.5) |
| Foreigners | 423 (16.4) |
| No data | 3 (0.1) |
| Age Group | Number of Indigenous Knowlesi Malaria Monoinfection Cases According to States (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Johor | Kedah | Kelantan | Melaka | Negeri Sembilan | Pahang | Perak | Perlis | Pulau Pinang | Selangor | Terengganu | Kuala Lumpur | |
| 0 to 9 | 0 (0.0) | 1 (1.7) | 18 (2.7) | 0 (0.0) | 3 (2.5) | 16 (2.4) | 14 (2.7) | 0 (0.0) | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0 (0.0) |
| 10 to 19 | 4 (2.4) | 0 (0.0) | 83 (12.2) | 1 (14.3) | 6 (5.1) | 59 (8.9) | 60 (11.6) | 0 (0.0) | 1 (16.7) | 24 (11.4) | 11 (6.9) | 0 (0.0) |
| 20 to 29 | 47 (28.5) | 8 (13.3) | 165 (24.3) | 2 (28.6) | 39 (33.1) | 167 (25.2) | 107 (20.7) | 0 (0.0) | 0 (0.0) | 55 (26.2) | 34 (21.3) | 0 (0.0) |
| 30 to 39 | 58 (35.2) | 24 (40.0) | 176 (26.0) | 1 (14.3) | 24 (20.3) | 163 (24.6) | 137 (26.4) | 1 (100.0) | 2 (33.3) | 66 (31.4) | 55 (34.4) | 0 (0.0) |
| 40 to 49 | 33 (20.00) | 12 (20.0) | 114 (16.8) | 1 (14.3) | 19 (16.1) | 108 (16.3) | 93 (18.0) | 0 (0.0) | 0 (0.0) | 29 (13.8) | 27 (16.9) | 1 (100.0) |
| 50 to 59 | 17 (10.3) | 12 (20.0) | 75 (11.1) | 1 (14.3) | 17 (14.4) | 86 (13.0) | 66 (12.7) | 0 (0.0) | 1 (16.7) | 20 (9.5) | 19 (11.9) | 0 (0.0) |
| 60 and above | 6 (3.6) | 3 (5.0) | 47 (6.9) | 1 (14.3) | 10 (8.5) | 64 (9.7) | 41 (7.9) | 0 (0.0) | 2 (33.3) | 15 (7.1) | 14 (8.8) | 0 (0.0) |
| Total | 165 (100.0) | 60 (100.0) | 678 (100.0) | 7 (100.0) | 118 (100.0) | 663 (100.0) | 518 (100.0) | 1 (100.0) | 6 (100.0) | 210 (100.0) | 160 (100.0) | 1 (100.0) |
| State | Number of Indigenous Knowlesi Malaria Cases According to Gender (%) | Total (%) | |
|---|---|---|---|
| Male | Female | ||
| Johor | 157 (93.15) | 8 (4.85) | 165 (100.00) |
| Kedah | 56 (93.33) | 4 (6.67) | 60 (100.00) |
| Kelantan | 576 (84.96) | 102 (15.04) | 678 (100.00) |
| Melaka | 6 (85.71) | 1 (14.29) | 7 (100.00) |
| Negeri Sembilan | 98 (83.05) | 20 (16.95) | 118 (100.00) |
| Pahang | 546 (82.35) | 117 (17.65) | 663 (100.00) |
| Perak | 414 (79.92) | 104 (20.08) | 518 (100.00) |
| Perlis | 1 (100.00) | 0 (0.00) | 1 (100.00) |
| Pulau Pinang | 4 (66.66) | 2 (33.33) | 6 (100.00) |
| Selangor | 181 (86.19) | 29 (13.81) | 210 (100.00) |
| Terengganu | 128 (80.00) | 32 (20.00) | 160 (100.00) |
| Kuala Lumpur | 0 (0.00) | 1 (100.00) | 1 (100.00) |
| States | Annual Number of Knowlesi Malaria Cases | Total | % | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | |||
| Total | 277 | 414 | 474 | 332 | 113 | 136 | 423 | 598 | 2767 | 100 |
| Johor | 7 | 14 | 22 | 15 | 18 | 12 | 47 | 42 | 177 | 6.40 |
| Kedah | 1 | 5 | 4 | 9 | 1 | 4 | 10 | 29 | 63 | 2.28 |
| Kelantan | 111 | 148 | 152 | 99 | 20 | 34 | 135 | 113 | 812 | 29.35 |
| Melaka | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 4 | 8 | 0.29 |
| Negeri Sembilan | 3 | 18 | 25 | 12 | 2 | 3 | 30 | 30 | 123 | 4.45 |
| Pahang | 52 | 104 | 136 | 106 | 39 | 33 | 77 | 119 | 666 | 24.07 |
| Perak | 49 | 55 | 64 | 59 | 19 | 27 | 76 | 175 | 524 | 18.94 |
| Perlis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0.04 |
| Pulau Pinang | 3 | 4 | 2 | 0 | 0 | 0 | 1 | 1 | 11 | 0.40 |
| Selangor | 30 | 38 | 40 | 19 | 8 | 14 | 24 | 45 | 218 | 7.88 |
| Terengganu | 20 | 27 | 28 | 13 | 6 | 8 | 21 | 38 | 161 | 5.82 |
| Federal Territory of Kuala Lumpur | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 3 | 0.11 |
| Federal Territory of Putrajaya | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 |
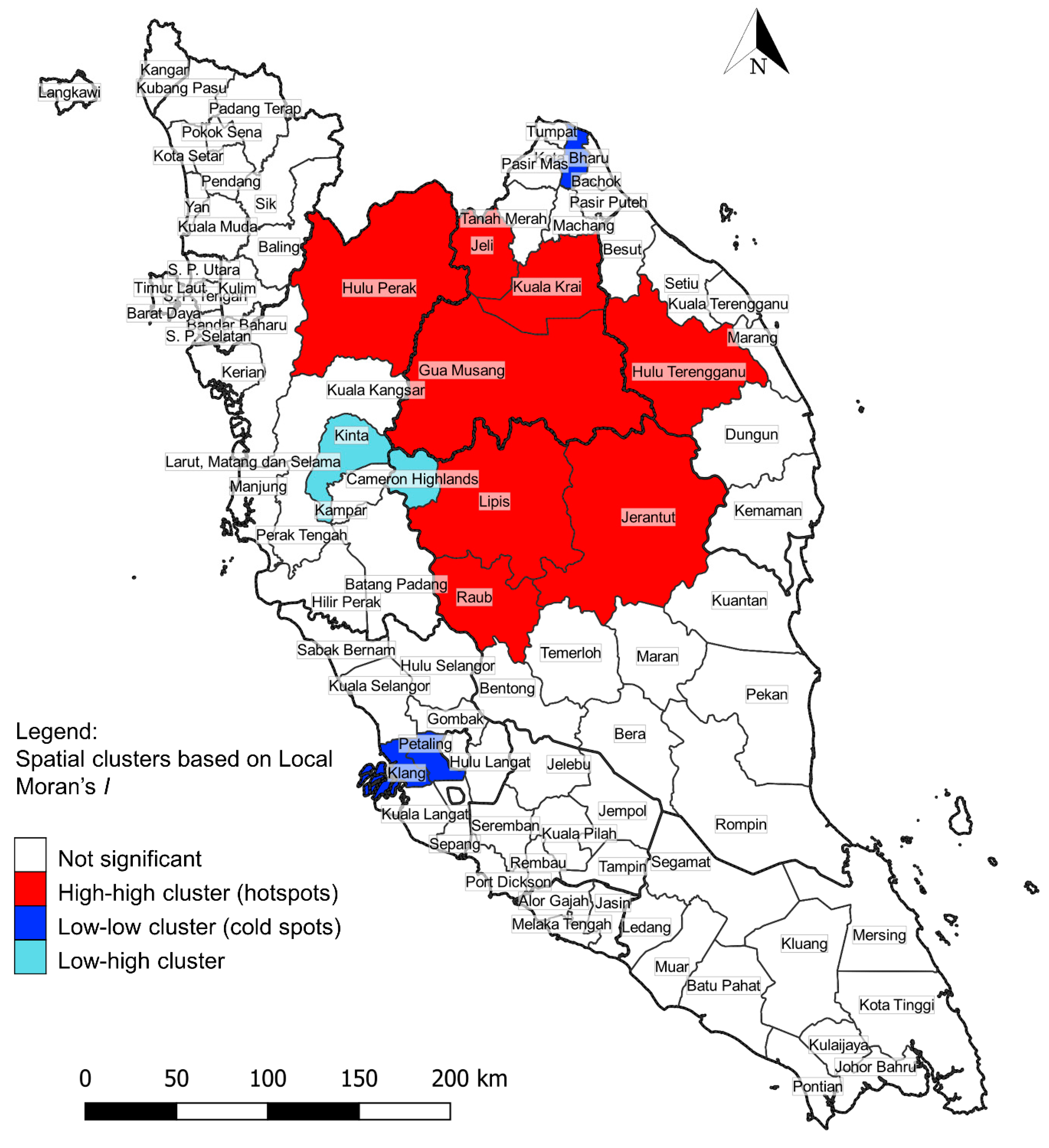
| Cluster Type | District | Number of Cases | IR | LISA Index | p-Value |
|---|---|---|---|---|---|
| High-high | Jeli | 95 | 46.24 | 5.88 | <0.001 |
| Kuala Krai | 167 | 37.35 | 2.92 | 0.001 | |
| Jerantut | 101 | 32.28 | 2.16 | <0.001 | |
| Lipis | 329 | 58.70 | 5.97 | <0.001 | |
| Raub | 66 | 25.49 | 1.51 | 0.003 | |
| Hulu Perak | 98 | 32.23 | 2.14 | 0.003 | |
| Hulu Terengganu | 68 | 29.42 | 1.36 | 0.008 | |
| Gua Musang | 506 | 71.68 | 6.88 | <0.001 | |
| Low-low | Kota Bharu | 1 | 1.36 | 0.48 | 0.006 |
| Klang | 1 | 1.03 | 0.54 | 0.008 | |
| Petaling | 3 | 1.22 | 0.44 | 0.008 | |
| Low-high | Cameron Highlands | 2 | 6.99 | −0.58 | <0.001 |
| Kinta | 48 | 7.78 | −0.25 | 0.006 |
| Models | AIC | BIC | RMSE | MAPE | Ljung–Box Test # |
|---|---|---|---|---|---|
| SARIMA (0,1,0)(2,1,0)12 | 190.37 | 196.60 | 0.93 | 18.92 | 0.08 |
| SARIMA (0,1,1)(2,1,0)12 | 184.62 | 192.93 | 0.90 | 18.51 | 0.50 |
| SARIMA (0,1,1)(2,1,1)12 | 186.65 | 197.03 | 0.87 | 18.93 | 0.19 |
| SARIMA (1,1,0)(2,1,0)12 | 186.14 | 194.45 | 0.89 | 18.52 | 0.36 |
| SARIMA (2,1,0)(2,1,0)12 | 185.91 | 196.30 | 0.89 | 18.17 | 0.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phang, W.K.; Hamid, M.H.A.; Jelip, J.; Mudin, R.N.; Chuang, T.-W.; Lau, Y.L.; Fong, M.Y. Spatial and Temporal Analysis of Plasmodium knowlesi Infection in Peninsular Malaysia, 2011 to 2018. Int. J. Environ. Res. Public Health 2020, 17, 9271. https://doi.org/10.3390/ijerph17249271
Phang WK, Hamid MHA, Jelip J, Mudin RN, Chuang T-W, Lau YL, Fong MY. Spatial and Temporal Analysis of Plasmodium knowlesi Infection in Peninsular Malaysia, 2011 to 2018. International Journal of Environmental Research and Public Health. 2020; 17(24):9271. https://doi.org/10.3390/ijerph17249271
Chicago/Turabian StylePhang, Wei Kit, Mohd Hafizi Abdul Hamid, Jenarun Jelip, Rose Nani Mudin, Ting-Wu Chuang, Yee Ling Lau, and Mun Yik Fong. 2020. "Spatial and Temporal Analysis of Plasmodium knowlesi Infection in Peninsular Malaysia, 2011 to 2018" International Journal of Environmental Research and Public Health 17, no. 24: 9271. https://doi.org/10.3390/ijerph17249271
APA StylePhang, W. K., Hamid, M. H. A., Jelip, J., Mudin, R. N., Chuang, T.-W., Lau, Y. L., & Fong, M. Y. (2020). Spatial and Temporal Analysis of Plasmodium knowlesi Infection in Peninsular Malaysia, 2011 to 2018. International Journal of Environmental Research and Public Health, 17(24), 9271. https://doi.org/10.3390/ijerph17249271





