Abstract
Copy number variants (CNVs) play an important role in the genetic underpinnings of neuropsychiatric/neurodevelopmental disorders. The chromosomal region 16p11.2 (BP4–BP5) harbours both deletions and duplications that are associated in carriers with neurodevelopmental and neuropsychiatric conditions as well as several rare disorders including congenital malformation syndromes. The aim of this article is to provide a review of the current knowledge of the diverse neurodevelopmental disorders (NDD) associated with 16p11.2 deletions and duplications reported in published cohorts. A literature review was conducted using the PubMed/MEDLINE electronic database limited to papers published in English between 1 January 2010 and 31 July 2020, describing 16p11.2 deletions and duplications carriers’ cohorts. Twelve articles meeting inclusion criteria were reviewed from the 75 articles identified by the search. Of these twelve papers, eight described both deletions and duplications, three described deletions only and one described duplications only. This study highlights the heterogeneity of NDD descriptions of the selected cohorts and inconsistencies concerning accuracy of data reporting.
1. Introduction
Copy-number variation (CNVs) is a type of structural variant involving alterations in the number of copies of specific DNA sequences, which can be deleted and/or duplicated. CNVs vary considerably in size, gene content and prevalence [1]. Although often benign, a major advance in our understanding is that CNVs contribute significantly to risk for several neurodevelopmental and neuropsychiatric conditions, rare diseases and congenital malformation syndromes. However, variable penetrance and expression, syndromes, as well as pleiotropy, is reported [2]. Improved understanding of these risks has implications for more accurate and timely communication of the associated clinical risks.
The 16p11.2 region encompasses several distinct CNVs, responsible for five rare disorders identified as ORPHA entities in the Orphanet portal for rare diseases and orphan drugs [3] (Table 1). CNVs in the 16p11.2 region are associated with exchange of chromosomal material in regions of repetitive DNA sequence [4]. The most common CNV is a recurrent interstitial deletion of ∼600 kb, defined by breakpoints 4 and 5 (BP4–BP5) containing 26 known genes, four of which are OMIM morbid genes; most of the genes within this region are expressed in different regions of the brain (ORPHA:261197).

Table 1.
16p11.2 CNV syndromes reported in Orphanet.
These CNVs (deletion and/or duplication) arise with a frequency of about 3/10.000 (0.03%) [5]. This specific BP4–BP5 deletion has a population prevalence of approximately 1/2.000 (0.05%) and 0.5% among those with autism spectrum disorders (ASD) [5,6,7]. While about 71% of 16p11.2 deletions are de novo, ~70% of 16p11.2 duplications are inherited [8]; 16p11.2 duplications have been estimated to occur in about 3 in 10.000 people and are present in about 4 in 10.000 people who have mental health problems or difficulties with speech and language [9]. It is one of the most frequent single causes of neurodevelopmental disorders (NDDs) and autism spectrum disorders [10].
Several studies have described carriers of 16p11.2 rearrangements associated with increased risk of developmental/psychomotor delay, intellectual disability, ASD, obsessive and repetitive behaviours, other behavioural problems, and schizophrenia [8,9,11,12,13,14]. These clinical findings are also linked to a large number of patients with rare diseases [15]. Dysmorphic facial features and major malformations have also been observed, particularly in those 16p11.2 microduplications syndromal rare diseases [16]. 16p11.2 rearrangements may be overlooked by clinicians as they have variable penetrance and pleiotropic effects [2]. Their detection, however, may identify important genetic causes of neurodevelopmental and neuropsychiatric presentation that are useful in a clinical genetics context to inform genetic counselling and more broadly to inform clinical care for deletion carriers. Therefore, we need a better understanding of the neurodevelopmental and neuropsychiatric outcomes for 16p11.2 carriers to support evidence-based clinical care.
The BP4–BP5 region contains 26 UCSC genes (a set of gene predictions based on data from RefSeq, GenBank, CCDS, Rfam, and the tRNA Genes track, and includes both protein-coding genes and non-coding RNA genes) all of which are protein coding (Figure 1). Four genes are OMIM morbid genes [17]: KIF22 associated with autosomal dominant spondyloepimetaphyseal dysplasia with joint laxity, type 2 (OMIM 603546); ALDOA associated with autosomal recessive glycogen storage disease XII (OMIM 611881), and TBX6 associated with autosomal dominant and recessive Spondylocostal dysostosis 5 (OMIM 122600). The last gene PRRT2 is associated with three distinct autosomal dominant disorders, familial infantile convulsions with paroxysmal choreoathetosis (OMIM 602066), episodic kinesigenic dyskinesia 1 (OMIM 128200) and benign familial infantile seizures 2 (OMIM 605751). According to GTEx expression data, most of the genes, except for a few such as SPN, C16orf54 and ZG16, are expressed (though in different levels) in different regions of the brain [18]. Notably, CDIPT, SEZ6L2, ASPHD1, DOC2A, FAM57B and the OMIM morbid gene PRRT2, which is associated with seizures and dyskinesia, are highly expressed in different brain regions and some of them are highly brain specific. Genes MAZ and TAOK2, which are also brain expressed genes, are classified as “extremely loss-of-function intolerant genes” according to the computational pLI score [19].
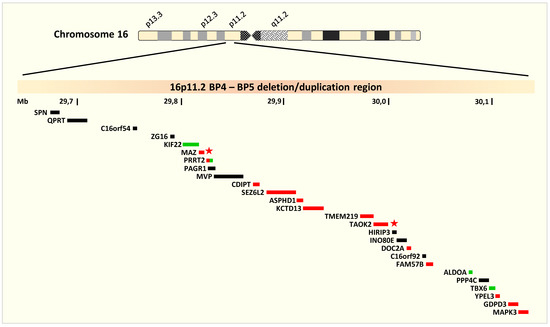
Figure 1.
16p11.2 deletion/duplication region (GRC37/hg19 chr16:29,641,983-30,180,793). The ideogram highlighting the deletion/duplication region of interest on the short arm of chromosome 16 and the genes encompassed by BP4 and BP5. The green bars represent the OMIM morbid genes and the red bars represent the genes expressed in the brain according to GTEx data. PRRT2 (red-green bar) is an OMIM morbid gene, expressed in the brain. MAZ and TAOK2 (marked with an asterisk) are two extremely loss-of-function intolerant genes.
This article provides a review of the current understanding of neurodevelopmental and neuropsychiatric phenotypes described in the cohorts with 16p11.2 (BP4–BP5) CNVs, involving deletions and/or duplications. The aim is to clarify the reported frequency of neurodevelopmental or neuropsychiatric phenotypes in the literature to date, to identify inconsistencies in phenotype descriptions that may improve or standardize the approach in the future.
2. Materials and Methods
2.1. Searching Strategy
The literature review was conducted using the PubMed/MEDLINE electronic database. The search was limited to English language papers published between 1 January 2010 and 31 July 2020. Descriptors used for the search were “16p11.2”, “Schizophrenia”, “Intellectual disabilities”, “Mental retardation”, “Intellectual disabilities”, “Autism spectrum disorders”, “Attention deficit hyperactivity disorder”, “Communication disorders”, “Specific learning disorders” and “Motor disorders” in the title and/or abstract of all papers. The Boolean operators used were “AND” and “OR”.
2.2. Study Selection: Inclusion and Exclusion Criteria
Regarding the inclusion criteria, we selected only studies based on patients’ cohorts and referring to cross-sectional, prospective or retrospective studies (minimum of five carriers/patients described); subjects in the cohort had molecular confirmation of 16p11.2 BP4–BP5 microdeletions and microduplications (e.g., through FISH, MLPA, or microarray/aCGH analysis); neurodevelopmental and/or neuropsychiatric phenotypes/outcomes as described by search descriptors were reported either in the paper or, alternatively, in the paper’s tables.
Exclusion criteria were: papers regarding non neurodevelopmental and/or neuropsychiatric phenotypes/outcomes (e.g., head size, obesity); papers reporting neuroimaging findings and systematic reviews or meta-analyses papers.
All articles were screened by title and abstracts to identify those that fitted the inclusion criteria. Next, the full text of the paper was evaluated. The papers were assessed independently for inclusion by two or three authors and imported to Mendeley, a free web-based manager for organizing references; any duplicates were excluded (Table 2 and Table 3).

Table 2.
Neurodevelopmental features in probands reported in association with proximal 16p11.2 (BP4–BP5) microdeletions.

Table 3.
Neurodevelopmental features in proband reported in association with proximal 16p11.2 (BP4–BP5) microduplications.
2.3. Data Extraction
The following information was extracted from the selected articles: (1) study identification (author, year); (2) ascertainment (16p11.2); (3) age range of subjects (years); (4) sample size; (5) the proportion or percentage of 16p11.2 deletions and/or duplications with neurodevelopmental or neuropsychiatric phenotypes according to the following domains: cognition, speech and language problems, NDDs, mood disorders, seizures and behaviour problems. The neurodevelopmental or neuropsychiatric phenotypes phenotypic data for 16p11.2 (BP4–BP5) microdeletions and microduplications are presented separately (Table 2 and Table 3).
3. Results
3.1. Review of the Literature
A total of 75 articles were identified and screened for inclusion. Sixty-three papers were excluded because they used population cohorts, the genetic diagnosis was not clinically confirmed, the studies did not include NDD outcomes, they did not report on 16p11.2 (BP4–BP5) or they were review or meta-analysis articles about 16p11.2.
The 12 selected articles included eight papers describing phenotypes for both deletions and duplications, three reporting phenotypes for deletions only and one for duplications only. Two tables were constructed to support this review of phenotypic features (noted frequency, rate) included in the articles. Table 2 and Table 3 describe frequency of phenotypic outcomes for the 16p11.2 microdeletions (nine articles) 16p11.2 microduplications (seven articles), respectively.
3.2. Proximal Deletions
Nine papers reported neurodevelopmental or neuropsychiatric outcomes for microdeletions (Table 2). The selected papers described phenotypes in children, adolescents and adults of both male and female genders ranging in age from five months to fifty-nine years. Males were overrepresented among all carriers with the exception of one study where the CNV was preferentially transmitted from mothers [4]. This was the only study noting transmission data.
Probands were determined using a variety of methods, which differed between studies. These included identification through clinical genetic testing, on the basis of having a neurodevelopmental disorder or as part of cascade screening in families where a prior carrier was identified. Clinical phenotypes were reported based on either direct assessment or clinical reports.
Cognitive functioning was described to be impaired in carriers in all studies but the proportion of those affected varied. Three smaller studies reported intellectual disability in almost all of the probands [14,20,21]. Other studies noted ID in 10–26% of probands [4,13,22,23]. One study did not report cognitive ability [8], and one other only stated the range of IQ for probands [24].
Impairments in speech and language functioning have previously been highlighted in 16p11.2 carriers. Of the nine studies reviewed here speech and language delay, non-verbal language status and articulation difficulties were reported variably between studies. The proportion affected was addressed in six studies, indicating speech and language difficulties in 54–100% of probands [4,13,14,20,21,22].
Reported neurodevelopmental disorders included autism diagnosis or autism features, ADHD/ADHD features and psychosis (including schizophrenia). Autism and autism features were described in all studies and the range varied between 5–100% of cases [4,8,13,14,20,21,22,23,24]. ADHD or ADHD features were reported in six studies ranging from 19–60% [14,20,21,22,23]. Schizophrenia was not found in any of the probands in five studies [8,20,21,23,24] and was not noted in four studies [4,13,14,22].
Four studies investigated the presence of seizures in probands. One study found no evidence of seizures [20]. D’Angelo et al. 2016 [8] noted seizures in 22% of cases, while Zufferey et al. 2012 [4] and Shinawi et al. 2010 [16] reported seizures in 24% and 30% of cases respectively.
Other psychiatric and behavioural disorders were also communicated. Oppositional defiant disorder (ODD) and aggression were reported in six studies with rates ranging from 0% to 13%. Depression, anxiety disorders and phobias were also described in a small number of studies; however, the proportions of cases with these diagnoses were low and appeared comparable with rates in the general population.
3.3. Proximal Duplications
Seven studies described phenotypes in subjects with 16p11.2 proximal duplications [8,14,20,21,22,23,25]. Only one of the studies did not overlap with others reporting phenotypes in proximal deletions [25]. The cohorts in these studies included children, adolescents (ranging from one to 17 years) and adults; one study reported only the participants mean age; it included adults with a mean age of 24.2 years [8]. Male carriers were overrepresented in all cohorts with one exception [20].
Cognitive functioning and/or evidence of developmental delay in carriers was described in five studies; the rates varied widely. Developmental regression was reported in three studies; it was detected in a small number of individuals. Speech and language delay ranged from 26–86% in four studies. Two further studies reported articulation difficulties in 22% [25] and 35% of cases [22].
The presence or absence of seizures was noted in four studies and rates ranged from none to 19.4% of cases [8,14,20,21].
The most common described neurodevelopmental disorder was autism spectrum disorder, which was found in 20–27% of cases in four larger studies [8,22,23,25]. Fernandez et al. 2010 reported ASD or autism traits as an outcome in three observed cases [20]. Rosenfeld et al. 2010 found one case of ASD among seven carriers [21] and Shinawi et al. 2010 found evidence of autism traits in one of seven carriers [16]. Schizophrenia was not noted as an outcome in any study. ADHD or ADHD traits were described in five studies. ADHD was reported in 26–44% of cases. One study also highlighted Developmental Coordination Disorder in 54% of cases.
The prevalence of behavioural problems ranged from 13% to 30%, and the proportion found to have mood and/or anxiety disorders also varied substantially between studies. Anxiety disorders were more common (4–17.4%) [25], while only a small number of individuals were reported to have depression or OCD.
4. Discussion
The objective of this review was to examine recently reported neurodevelopmental and neuropsychiatric outcomes associated with a 16p11.2 proximal (BP4–BP5) microdeletion or microduplication. These two CNV syndromes are associated with intellectual disability (ID) and developmental delay (DD) or neuropsychiatric presentations, particularly ASD [23], [26], other neurological, behaviour and mental disorders. In brief, our review of these recent studies is consistent with the emerging view that a substantial proportion of carriers of both the 16p11.2 proximal deletions and the proximal duplications are affected by ID/DD and neurodevelopmental disorders (NDD), particularly ASD and ADHD.
Intellectual disability or developmental delay (ID/DD) affected carriers of both deletions and duplications, and may occur more frequently in carriers of the proximal deletion; ID/DD was reported in approximately 70% of deletion cases compared to approximately 35% in those with duplications. Many carriers of both deletions and duplications had a history of speech and language delay, but there was greater variability among those with proximal duplications.
Neurodevelopmental disorders were observed to affect between 20% and 30% of carriers with deletions and duplications. In the deletion carriers there was significant variability in the proportion with ASD or autism traits between studies. Larger studies tended to find higher rates of ASD or autism traits, but used patient collections that had systematically screened for ASD such as the Simons VIP consortium/Searchlight [26], the ECHO study, the 16p11.2 European Consortium and the IMAGINE-ID study [8,27]. The was a relatively high prevalence of ADHD in both the deletion and duplication carriers, with a higher rate in those with duplications. Duplication carriers have previously been shown to have more than double the risk of ADHD compared with deletion carriers [23].
The prevalence of major psychiatric disorders (psychosis and schizophrenia) was not elevated in any of the cohorts reviewed here [23], contrasting with previous reports of psychotic symptoms in association with 16p11.2 duplications [28,29]. A recent meta-analysis of 36.676 schizophrenia patients and 48.331 healthy controls from 24 independent samples showed a significantly increased odds of developing schizophrenia in carriers of the 16p11.2 microduplication [30]. In contrast, a study of 217 deletion carriers, 114 duplication carriers, and family-based controls did not identify cases with schizophrenia beyond four who had been ascertained on the basis of schizophrenia diagnosis [23]. The absence of psychosis or schizophrenia among the studies reviewed here may reflect the relatively young mean age of the cohorts, who were yet to enter the age range usually associated with highest risk of emerging symptoms. Although many had an Autism Spectrum Disorder, the risks of psychosis are independent of autism in 16p11.2 duplication carriers [2].
We found evidence of an increased risk of recurrent seizures in association with 16p11.2 CNV. Shinawi et al. reported a series of 16p11.2 deletion and duplication patients; 3/10 duplication patients had seizures, one of which was associated with a de novo rearrangement [16]. Bijlsma et al. reported three patients with 16p11.2 deletions and history of developmental delay and seizures, of which one was a de novo rearrangement [31]. Kumar et al. also reported one ASD patient with a 16p11.2 deletion and history of seizures [11]. Ghebranious et al. described a 16p11.2 microdeletion in monozygotic twins with complex phenotypes that included seizure disorder with onset at 11.5 and 13 years of age, along with mental retardation and heart defects [32]. It appears from this review that vulnerability to seizures affects approximately 20% of both deletion and duplication CNV carriers, a proportion that is similar to that associated with ID/DD and ASD of diverse genetic aetiology, suggesting that the association with 16p11.2 is non-specific.
Behavioural phenotypes are characteristically found among individuals with pathogenic CNV. These include problems, as defined by the Human Phenotype Ontology (HPO) [33], that encompass abnormalities of mental functioning including various affective, behavioural, cognitive and perceptual abnormalities. In other words, a behavioural phenotype is a characteristic pattern of social, linguistic, cognitive and motor observations consistently associated with a genetic disorder. In general, these problems were infrequent, mostly around 10% of cases in both CNV types, and vary between reports. Their presentation is not static and typically varies according to the level of learning disability and a host of environmental, developmental and therapeutic influences; it changes with increasing age.
Finally, 16p11.2 deletion and duplication carriers have also been considered to confer an increased risk of other mental health problems including anxiety and depression. In our reviewed studies, a small number of individuals with microdeletions or duplications exhibited other mental health problems, such as anxiety, depressions, specific phobia or Obsessive-Compulsive Disorder (OCD). We found that frequency of anxiety, depression and phobias are almost the same (8-12%) in all the samples and both CNV types, as reported in Zufferey et al. 2012 [4], Hanson et al. 2015 [14] and Niarchou et al. 2019 [23].
In summary, these reports show the strongest association between the presence of a 16p11.2 CNV and ID/DD and ASD. For the 16p11.2 microdeletion, this appears to present in ID/DD in approximately 70% of carriers, dropping to approximately 30% for ID/DD in duplications and for NDD for both CNV types. Further studies will be required to confirm whether the relatively high frequency of ID/DD in association with the microdeletion is a consistent feature. Although other psychiatric, behavioural and mood disorders have also been reported, neither their frequency nor association with the CNV carriers is strong. An increased risk of recurrent seizures occurs; however, it is no greater than expected within a sample of young people with ID/DD and ASD of genetic aetiology.
Future Perspectives
Our review of these studies emphasises that there is a wide range of phenotypic variation and differences in severity of the clinical outcomes associated with both deletions and duplications of 16p11.2. This aligns with the previously reported variable penetrance of 31% and 34% for 16p11.2 deletions and duplications, respectively [34]. There may, however, be several factors leading to variation in clinical presentation, some of which may be reduced or eliminated, through experimental design and standardisation; others are inherent to these syndromes and will need to be accommodated by our analytical approaches.
A significant concern will always be variation and bias in patient ascertainment and clinical assessment. Most reports reviewed here used subjects that had been recruited from the Simons VIP Consortium, thus ensuring that patient data had been collected through a common protocol and in accordance with recognized data standards [26]. In addition, this recruitment was genetics-led; participants were recruited primarily because they possessed a 16p11.2 CNV potentially preceding a standardised clinical assessment.
Conversely, in a few other reports the phenotyping methodology that followed clinical referral is not detailed and some cases considered “not seriously affected” by ID or behavioural characteristics may have been excluded from further assessment. This exclusion would have lead to ascertainment bias, which is more likely in smaller studies and should be considered when comparisons are made between reports or prevalence. Furthermore, prior selection of patient groups based upon particular diagnostic criteria such as ASD, could lead to the overrepresentation of diagnostic sub-groups of the CNV cohorts.
We conclude that it is possible to draw high-level inferences by pooling data, bearing in mind that differences in ascertainment, recruitment and phenotyping methodologies will impact on the depth of phenotypic analysis that can be achieved with pooled data.
We recommend the future development of innovative and standard tools to detect, annotate and interpret CNVs and to improve and standardize the phenotype descriptions of cohorts carrying the CNVs. A step forward would be to use standard ontologies for phenotypic description such as HPO [33] and NCIt [35]. Using standard ways of describing syndromal rare diseases will help to clarify diagnostic enquiries and permit the identification of similar patients in different cohorts. Standardisation is essential for clinical diagnosis, and for identifying disease-causing genes. All data from cohort studies will need to adhere to the FAIR (Findable, Accessible, Interoperable and Reusable) principles and should promote the prospective collection of standardised phenotypic data for epidemiological and clinical research. Future studies need to ensure that collected data and bioresources are compatible with large-scale objectives to compile and share data for rare NDD patients, in line with Elixir recommendations. Rare diseases cohorts, such as 16p11.2 carriers can strongly contribute to these large-scale objectives.
Beyond the goals of reducing ascertainment bias, establishing standard clinical assessments and the creation of interoperable datasets, there is a need better to understand the origins of variability within sample populations due to genetic and environmental factors. Family background factors have been shown to influence cognitive ability, social behaviour and neuromotor performance in 16p11.2 deletion carriers, suggesting that environmental factors and polygenic genetic influences affect outcomes [36].
The most immediately addressable factor with current technology and practice is that introduced by genetic variation. Several evidence-based data reported in the literature are consistent with the presence of additional rare (<0.1% frequency in control individuals) or larger CNVs as modifiers of disorder severity [37,38,39]. In fact, genetic variation and dosage imbalance at other loci could contribute to the observed phenotypic heterogeneity, resulting in an additive or synergistic effect on neurodevelopmental pathways and disease outcomes. A potential explanation for variable expressivity associated with microdeletion/microduplication 16p11.2 syndromes hass been proposed. This is the “two-hit” model, in which the compound effect of a relatively small number of rare variants of large effect contribute to the phenotypic heterogeneity [38,39]. The consideration of secondary mutational hits modifying the phenotypic expression of 16p11.2 CNV would entail further exome and sequencing analysis of all patients. We aknowledge that this will increase the complexity of future patient studies and accentuate the need for larger-scale investigations with the appropriate resources.
A parallel approach is to integrate clinical data with the analysis of animal and cell models of disease, although such studies are not without limitations. Both animal and cell studies provide information on the underlying biological mechanisms that may lead to patient pathology, but no one model can capture all aspects of the patient condition. A 0.44 Mb syntenic region of mouse chromosome 7 has been used to generate animal models that simulate 16p11.2 deletions and duplications. They are associated with reciprocal differences in brain structure and behaviour [40]. Mice are maintained in standardized conditions and bred to have a similar genetic background, so the phenotypic consequences of these manipulations are not readily generalizable. Use of patient-induced pluripotent stem cells (iPSC) can capture genetic diversity [41] but as yet, neither the numbers of lines that are available for study nor the depth of phenotypic analysis can explain the variation in clinical phenotypes seen in patient populations [42]. CrispR-cas9 technique can be used to generate both deletions and duplications, equivalent to those of patients [43] in human iPSC. In future, it may be possible to introduce 16p11.2 CNVs into human iPSCs that have different genetic backgrounds chosen to reflect different levels of genetic risk for the specific symptoms described above. Such an approach could allow a systematic analysis of the genetic interactions that lead to some of the phenotypic variability.
5. Conclusions
We reviewed research into neurodevelopmental and neuropsychiatric phenotypes reported in patient cohorts with 16p11.2 deletions and/or duplications. We have highlighted the heterogeneity of neurodevelopment disorders in clinical descriptions of the published cohorts, drawing attention to inconsistencies concerning the accuracy of data reporting. Our conclusions support the emerging view that carriers of 16p11.2 proximal deletions and duplications have high rates of intellectual disabilities and developmental delays, particularly autism spectrum disorders. The wide range of phenotypic variation and differences in severity of the clinical outcomes that were reported could be explained by inherent characteristics of the syndromes or related to the variability of research designs.
There is a need for good standardization of assessments and encoding of data. This can only be achieved through cooperation and collaboration within the global research community. International consortia and researcher networks, such as MINDDS [COST Action CA16210: Maximizing Impact of Research in Neurodevelopmental Disorders], are emerging to promote cooperation. Such groups are facilitating agreement and adoption of standardised methodologies, protocols and assessments, enhancing our ability for data pooling and interoperability. Working at the international level will enable researchers to build larger cohorts with good geographical coverage, and overcome the problem that no single country can provide adequate sample sizes for appropriate statistical power.
Author Contributions
Conceptualization, N.O.-T., Y.K. and D.S.; methodology, N.O.-T., Y.K., M.C.d.S. and D.S.; data collection/literature search, N.O.-T., Y.K., M.C.d.S. and P.J.; formal data analysis, N.O.-T., Y.K., M.C.d.S., P.J., S.R., G.C., L.G., I.B., S.M.-S., J.W. and D.S.; writing, N.O.-T., Y.K., M.C.d.S., P.J., G.C., L.G., S.M.-S., I.B., Z.T., A.J.H. and D.S.; writing—review and editing, N.O.-T., S.R., A.J.H. and D.S. All authors have read and agreed to the published version of the manuscript.
Funding
If this article is accepted for publication, Open Access publication will be funded by COST ACTION 16210 (MINDDS, https://mindds.eu/).
Acknowledgments
The article is a product of collaboration between basic and clinical scientists within MINDDS (Maximizing Impact of Research in Neurodevelopmental Disorders) initiative. MINDDS is a pan-European partnership and part of COST Action 16210 (European Cooperation in Science and technology), aimed to facilitate identification of individuals with pathogenic CNVs, standardization of assessment and research protocols, as well as data sharing and knowledge exchange (https://mindds.eu/). UMIB is supported by National Funds through the FCT—Fundação para a Ciência e a Tecnologia in the frameworks of the UIDP/00215/2020 and UIDB/00215/2020 projects—Unit for Multidisciplinary Research in Biomedicine—UMIB/ICBAS/UP.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Thapar, A.; Cooper, M. Copy Number Variation: What Is It and What. J. Am. Acad. Child Adolesc. Psychiatry 2013, 52, 772–774. [Google Scholar] [CrossRef]
- Jutla, A.; Turner, J.B.; Snyder, L.G.; Chung, W.K.; Veenstra-VanderWeele, J. Psychotic symptoms in 16p11.2 copy-number variant carriers. Autism Res. 2019, 13, 187–198. [Google Scholar] [CrossRef]
- Orphanet. Available online: http://www.orpha.net/consor/cgi-bin/Disease_Search.php?lng=EN&data_id=12136&Disease_Disease_Search_diseaseGroup=hae&Disease_Disease_Search_diseaseType=Pat&Disease(s)/group of diseases=Hereditary-angioedema--HAE-&title=Hereditary-angioedema--HAE-&search=Di (accessed on 22 July 2020).
- Zufferey, F.; Sherr, E.H.; Beckmann, N.D.; Hanson, E.; Maillard, A.M.; Hippolyte, L.; Macé, A.; Ferrari, C.; Kutalik, Z.; Andrieux, J.; et al. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med Genet. 2012, 49, 660–668. [Google Scholar] [CrossRef]
- Weiss, L.; Shen, Y.; Korn, J.M.; Arking, D.E.; Miller, D.T.; Fossdal, R.; Saemundsen, E.; Stefansson, H.; Ferreira, M.A.; Green, T.; et al. Association between Microdeletion and Microduplication at 16p11.2 and Autism. New Engl. J. Med. 2008, 358, 667–675. [Google Scholar] [CrossRef]
- Jacquemont, S.; Reymond, A.; Zufferey, F.; Harewood, L.; Walters, R.G.; Kutalik, Z.; Martinet, D.; Shen, Y.; Valsesia, A.; Beckmann, N.D.; et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nat. Cell Biol. 2011, 478, 97–102. [Google Scholar] [CrossRef]
- Hudac, C.M.; Bove, J.; Barber, S.; Duyzend, M.; Wallace, A.; Martin, C.L.; Ledbetter, D.H.; Hanson, E.; Goin-Kochel, R.P.; Green-Snyder, L.; et al. Evaluating heterogeneity in ASD symptomatology, cognitive ability, and adaptive functioning among 16p11.2 CNV carriers. Autism Res. 2020. [Google Scholar] [CrossRef]
- D’Angelo, D.; Lebon, S.; Chen, Q.; Martin-Brevet, S.; Snyder, L.G.; Hippolyte, L.; Hanson, E.; Maillard, A.M.; Faucett, W.A.; Macé, A.; et al. Defining the Effect of the 16p11.2 Duplication on Cognition, Behavior, and Medical Comorbidities. JAMA Psychiatry 2016, 73, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Genetics Home Reference. Available online: https://ghr.nlm.nih.gov/condition/16p112-duplication#statistics (accessed on 20 August 2020).
- Sanders, K.B.S.J.; Ercan-Sencicek, A.G.; Hus, V.; Luo, R.; Murtha, M.T.; Moreno-De-Luca, D.; Chu, S.H.; Moreau, M.P.; Gupta, A.R.; Thomson, S.A.; et al. Multiple recurrent de novo copy number variations (CNVs), including duplications of the 7q11.23 Williams-Beuren syndrome region, are strongly associated with autism. Neuron 2011, 70, 863–885. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.A.; Karamohamed, S.; Sudi, J.; Conrad, D.F.; Brune, C.; Badner, J.A.; Gilliam, T.C.; Nowak, N.J.; Cook, E.H.; Dobyns, W.B.; et al. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2007, 17, 628–638. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.R.; Noor, A.; Vincent, J.B.; Lionel, A.C.; Feuk, L.; Skaug, J.; Shago, M.; Moessner, R.; Pinto, D.; Ren, Y.; et al. Structural Variation of Chromosomes in Autism Spectrum Disorder. Am. J. Hum. Genet. 2008, 82, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Rees, E.; Walters, J.T.R.; Georgieva, L.; Isles, A.R.; Chambert, K.D.; Richards, A.L.; Mahoney-Davies, G.; Legge, S.E.; Moran, J.L.; McCarroll, S.A.; et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br. J. Psychiatry 2014, 204, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Hanson, E.; Bernier, R.; Porche, K.; Jackson, F.I.; Goin-Kochel, R.P.; Snyder, L.G.; Snow, A.V.; Wallace, A.S.; Campe, K.L.; Zhang, Y.; et al. The Cognitive and Behavioral Phenotype of the 16p11.2 Deletion in a Clinically Ascertained Population. Biol. Psychiatry 2015, 77, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Kvarnung, M.; Nordgren, A. Intellectual Disability & Rare Disorders: A Diagnostic Challenge. Atherosclerosis 2017, 1031, 39–54. [Google Scholar] [CrossRef]
- Shinawi, M.; Liu, P.; Kang, S.-H.L.; Shen, J.; Belmont, J.W.; Scott, D.; Probst, F.J.; Craigen, W.J.; Graham, B.H.; Pursley, A.; et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J. Med Genet. 2010, 47, 332–341. [Google Scholar] [CrossRef]
- OMIN. Available online: https://omim.org/ (accessed on 20 August 2020).
- GTEx Expression Data. Available online: https://www.gtexportal.org/home/ (accessed on 20 August 2020).
- The Genome Aggregation Database. Available online: https://gnomad.broadinstitute.org/ (accessed on 20 August 2020).
- Fernandez, B.; Roberts, W.S.; Chung, B.; Weksberg, R.; Meyn, S.; Szatmari, P.; Joseph-George, A.M.; Mackay, S.; Whitten, K.; Noble, B.; et al. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J. Med Genet. 2009, 47, 195–203. [Google Scholar] [CrossRef]
- Rosenfeld, J.A.; Coppinger, J.; Bejjani, B.A.; Girirajan, S.; Eichler, E.E.; Shaffer, L.G.; Ballif, B.C. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. J. Neurodev. Disord. 2009, 2, 26–38. [Google Scholar] [CrossRef]
- Goin-Kochel, R.P.; Hudac, C.M.; Chen, Q.; Zeng, C.; Wallace, A.S.; Gerdts, J.; Earl, R.; Peterson, J.; Wolken, A.; Peters, A.; et al. Developmental trajectories for young children with 16p11.2 copy number variation. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2017, 174, 367–380. [Google Scholar] [CrossRef]
- Niarchou, M.; Chawner, S.J.; Doherty, J.L.; Maillard, A.M.; Jacquemont, S.; Chung, W.K.; Green-Snyder, L.; Bernier, R.A.; Goin-Kochel, R.P.; Hanson, E.; et al. Psychiatric disorders in children with 16p11.2 deletion and duplication. Transl. Psychiatry 2019, 9, 1–8. [Google Scholar]
- Hippolyte, L.; Maillard, A.M.; Rodriguez-Herreros, B.; Pain, A.; Martin-Brevet, S.; Ferrari, C.; Conus, P.; Macé, A.; Hadjikhani, N.; Metspalu, A.; et al. The Number of Genomic Copies at the 16p11.2 Locus Modulates Language, Verbal Memory, and Inhibition. Biol. Psychiatry 2016, 80, 129–139. [Google Scholar] [CrossRef]
- Snyder, L.G.; D’Angelo, D.; Chen, Q.; Bernier, R.; Goin-Kochel, R.P.; Wallace, A.S.; Gerdts, J.; Kanne, S.; Berry, L.; Blaskey, L.; et al. Autism Spectrum Disorder, Developmental and Psychiatric Features in 16p11.2 Duplication. J. Autism Dev. Disord. 2016, 46, 2734–2748. [Google Scholar] [CrossRef]
- The Simons VIP Consortium. Simons Variation in Individuals Project (Simons VIP): A Genetics-First Approach to Studying Autism Spectrum and Related Neurodevelopmental Disorders. Neuron 2012, 73, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Shadan, K.; Rodriguez, E.A. Available online. Met. Powder Rep. 2008, 63, 11. [Google Scholar] [CrossRef]
- McCarthy, S.; Makarov, V.; Kirov, G.; Addington, A.M.; McClellan, J.; Yoon, S.; Perkins, D.; Dickel, D.E.; Kusenda, M.; Krause, V. Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 2009, 41, 1223–1227. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.R.; Howrigan, D.P.; Merico, D.; Thiruvahindrapuram, B.; Wu, W.; Greer, D.S.; Antaki, D.; Shetty, A.; Holmans, P.A.; Pinto, D.; et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017, 49, 27–35. [Google Scholar] [CrossRef]
- Chang, Y.S.; Owen, J.P.; Pojman, N.J.; Thieu, T.; Bukshpun, P.; Wakahiro, M.L.; Marco, E.J.; Berman, J.I.; Spiro, J.E.; Chung, W.K.; et al. Reciprocal white matter alterations due to 16p11.2 chromosomal deletions versus duplications. Hum. Brain Mapp. 2016, 37, 2833–2848. [Google Scholar] [CrossRef]
- Bijlsma, E.; Gijsbers, A.C.J.; Schuurs-Hoeijmakers, J.H.M.; Van Haeringen, A.; Van De Putte, D.E.F.; Anderlid, B.-M.; Lundin, J.; Lapunzina, P.; Jurado, L.A.P.; Chiaie, B.D.; et al. Extending the phenotype of recurrent rearrangements of 16p11.2: Deletions in mentally retarded patients without autism and in normal individuals. Eur. J. Med. Genet. 2009, 52, 77–87. [Google Scholar] [CrossRef]
- Ghebranious, N.; Giampietro, P.F.; Wesbrook, F.P.; Rezkalla, S.H. A novel microdeletion at 16p11.2 harbors candidate genes for aortic valve development, seizure disorder, and mild mental retardation. Am. J. Med. Genet. Part A 2007, 221, 1462–1471. [Google Scholar] [CrossRef]
- Human Phenotype Ontology. Available online: https://hpo.jax.org/app (accessed on 20 August 2020).
- Kirov, G.; Rees, E.; Walters, J.T.; Escott-Price, V.; Georgieva, L.; Richards, A.L.; Chambert, K.D.; Davies, G.; Legge, S.E.; Moran, J.L.; et al. The Penetrance of Copy Number Variations for Schizophrenia and Developmental Delay. Biol. Psychiatry 2014, 75, 378–385. [Google Scholar] [CrossRef]
- NCIt. Available online: https://www.ebi.ac.uk/ols/ontologies/ncit (accessed on 20 August 2020).
- Moreno-De-Luca, A.; Evans, D.W.; Boomer, K.B.; Hanson, E.; Bernier, R.; Goin-Kochel, R.P.; Myers, S.M.; Challman, T.D.; Moreno-De-Luca, D.; Slane, M.M.; et al. The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatry 2015, 72, 119–126. [Google Scholar] [CrossRef]
- Duyzend, M.H.; Nuttle, X.; Coe, B.P.; Baker, C.; Nickerson, D.A.; Bernier, R.; Eichler, E.E. Maternal Modifiers and Parent-of-Origin Bias of the Autism-Associated 16p11.2 CNV. Am. J. Hum. Genet. 2016, 98, 45–57. [Google Scholar] [CrossRef]
- Girirajan, P.D.S.; Rosenfeld, J.A.; Coe, B.P.; Parikh, S.; Friedman, N.; Goldstein, A.; Filipink, R.A.; McConnell, J.S.; Angle, B.; Meschino, W.S.; et al. Phenotypic Heterogeneity of Genomic Disorders and Rare Copy- Number Variants. N. Engl. J. Med. 2013, 367, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- Redaelli, S.; Maitz, S.; Crosti, F.; Sala, E.; Villa, N.; Spaccini, L.; Selicorni, A.; Rigoldi, M.; Conconi, D.; Dalpra, L.; et al. Refining the Phenotype of Recurrent Rearrangements of Chromosome 16. Int. J. Mol. Sci. 2019, 20, 1095. [Google Scholar] [CrossRef] [PubMed]
- Horev, G.; Ellegood, J.; Lerch, J.P.; Son, Y.-E.E.; Muthuswamy, L.; Vogel, H.; Krieger, A.M.; Buja, A.; Henkelman, R.M.; Wigler, M.; et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc. Natl. Acad. Sci. USA 2011, 108, 17076–17081. [Google Scholar] [CrossRef] [PubMed]
- Drakulic, D.; Djurovic, S.; Syed, Y.A.; Trattaro, S.; Caporale, N.; Falk, A.; Ofir, R.; Heine, V.M.; Chawner, S.J.R.A.; Rodriguez-Moreno, A.; et al. Copy number variants (CNVs): A powerful tool for iPSC-based modelling of ASD. Mol. Autism 2020, 11. [Google Scholar] [CrossRef]
- Deshpande, A.; Yadav, S.; Dao, D.Q.; Wu, Z.Y.; Hokanson, K.C.; Cahill, M.K.; Wiita, A.P.; Jan, Y.-N.; Ullian, E.M.; Weiss, L.A.; et al. Cellular phenotypes in human iPSC-derived neurons from a genetic model of autism spectrum disorder. Cell Rep. 2017, 21, 2678–2687. [Google Scholar] [CrossRef]
- Tai, D.J.C.; Ragavendran, A.; Manavalan, P.; Stortchevoi, A.; Seabra, C.M.; Erdin, S.; Collins, R.L.; Blumenthal, I.; Chen, X.; Shen, Y.; et al. Engineering microdeletions and microduplications by targeting segmental duplications with CRISPR. Nat. Neurosci. 2016, 19, 517–522. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).