Impact of Sediment Concentration on the Survival of Wastewater-Derived blaCTX-M-15-Producing E. coli, and the Implications for Dispersal into Estuarine Waters
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Sample Collection and Characterisation
2.3. Microcosm Study
2.3.1. Preparation and Inoculation of Turbid Water
2.3.2. Artificial Sunlight Exposure
2.3.3. Isolation and Confirmation of E. coli
2.3.4. Detection of the uidA Gene through Single-Plex PCR
2.3.5. Detection of the blaCTX-M-15 Gene through Single-Plex PCR
2.3.6. Statistical Analysis
2.4. Model Development
3. Results
3.1. Characterization of Samples
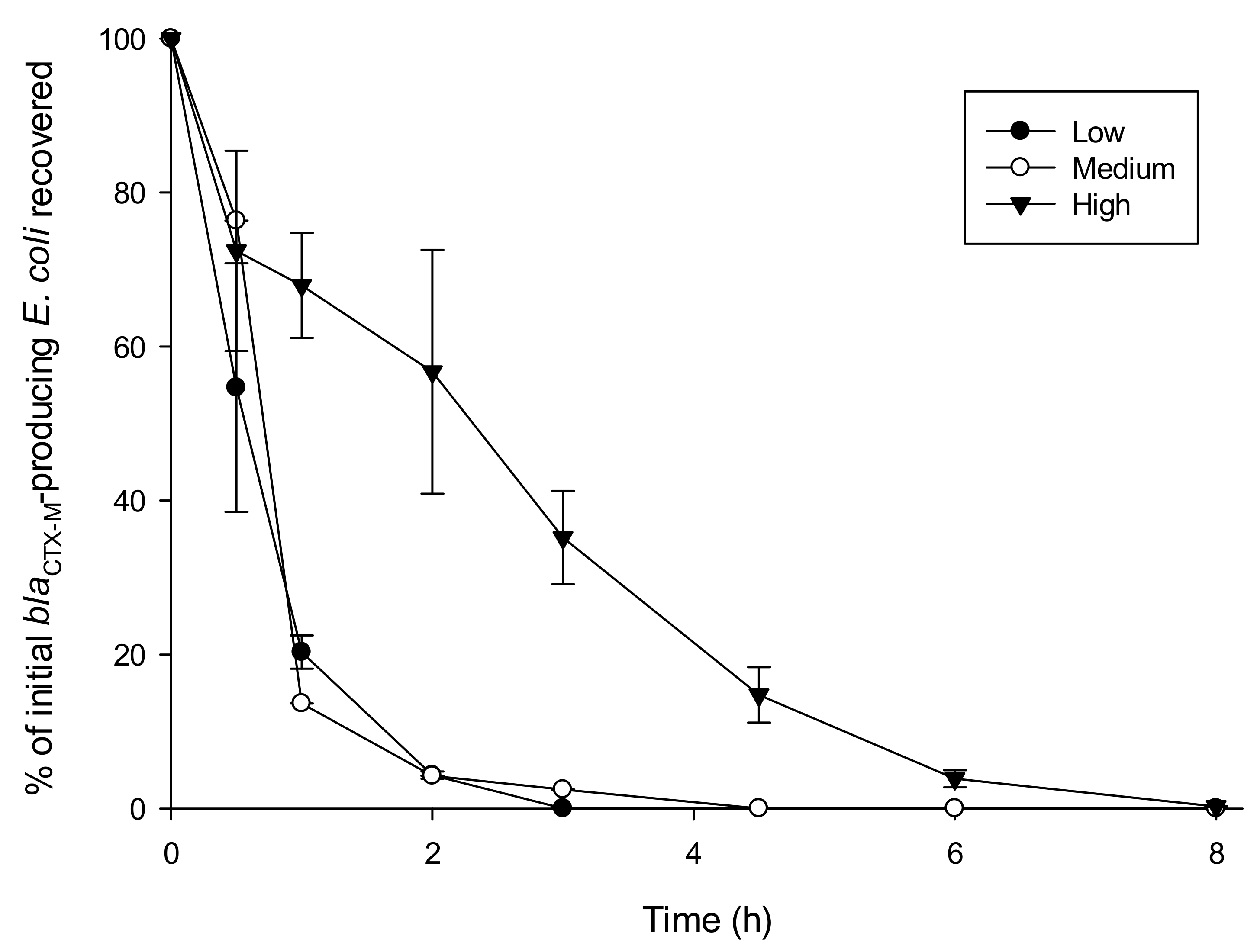
3.2. Influence of Turbidity on Survival of blaCTX-M-15-Producing E. coli
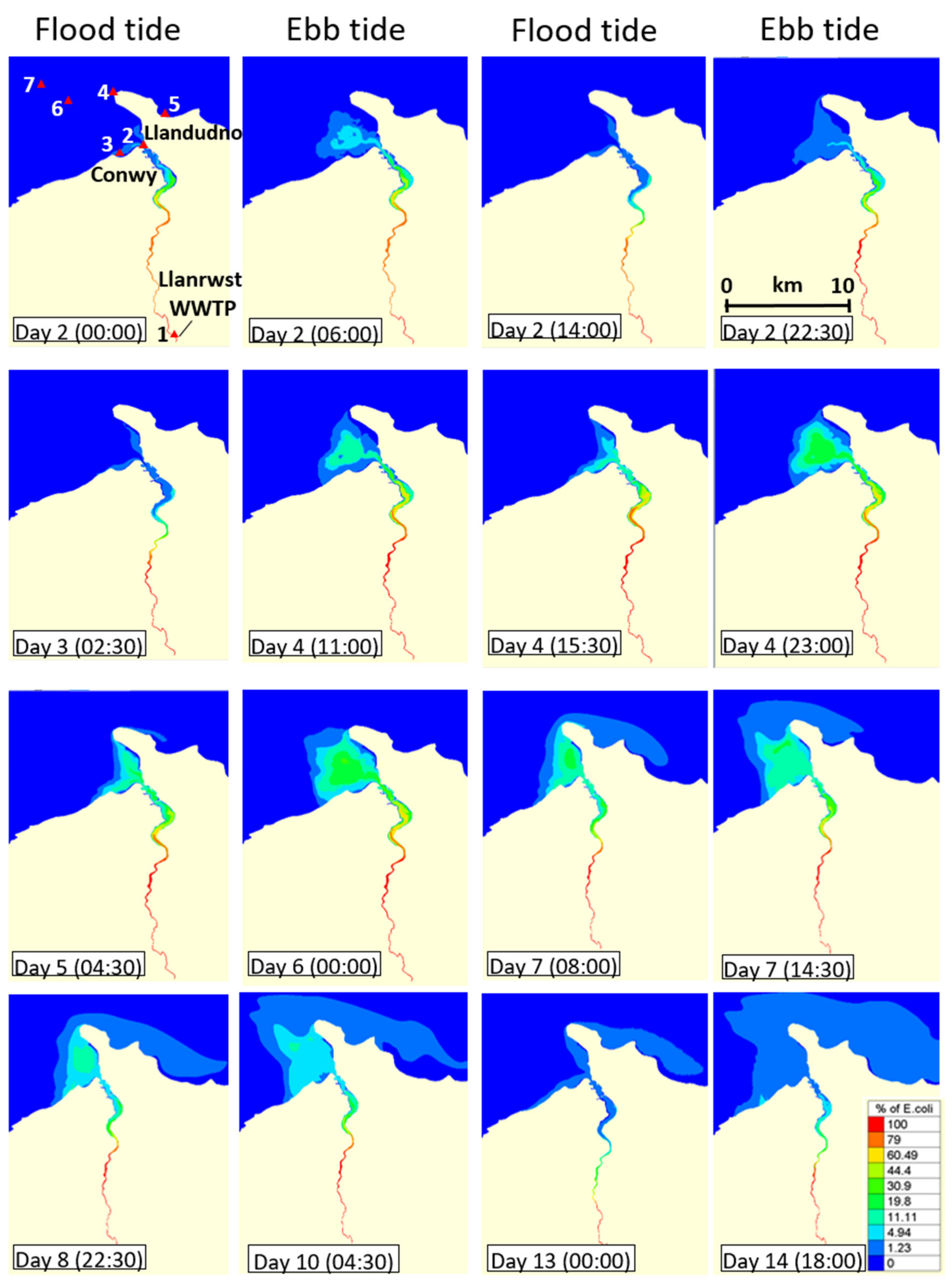
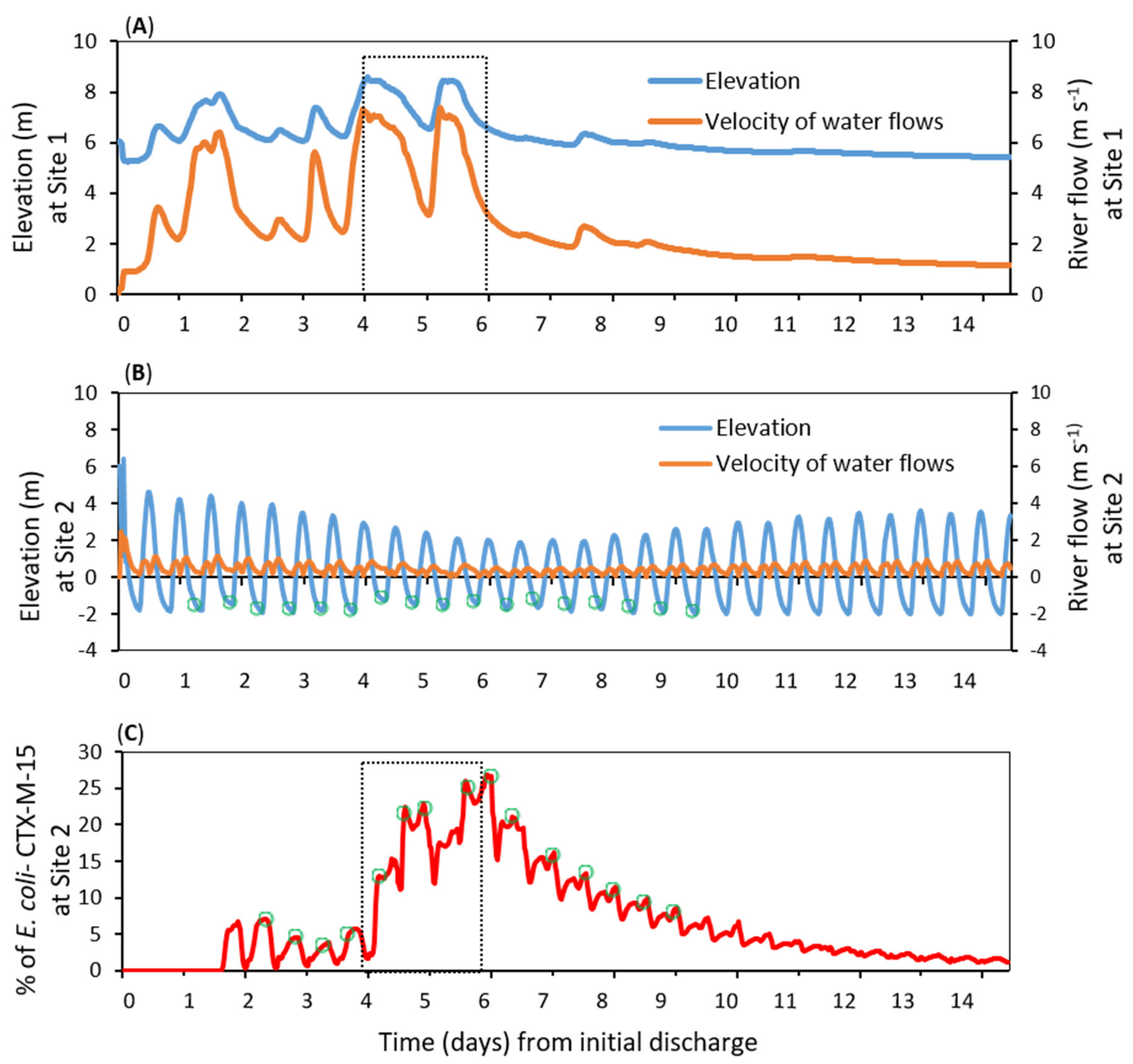
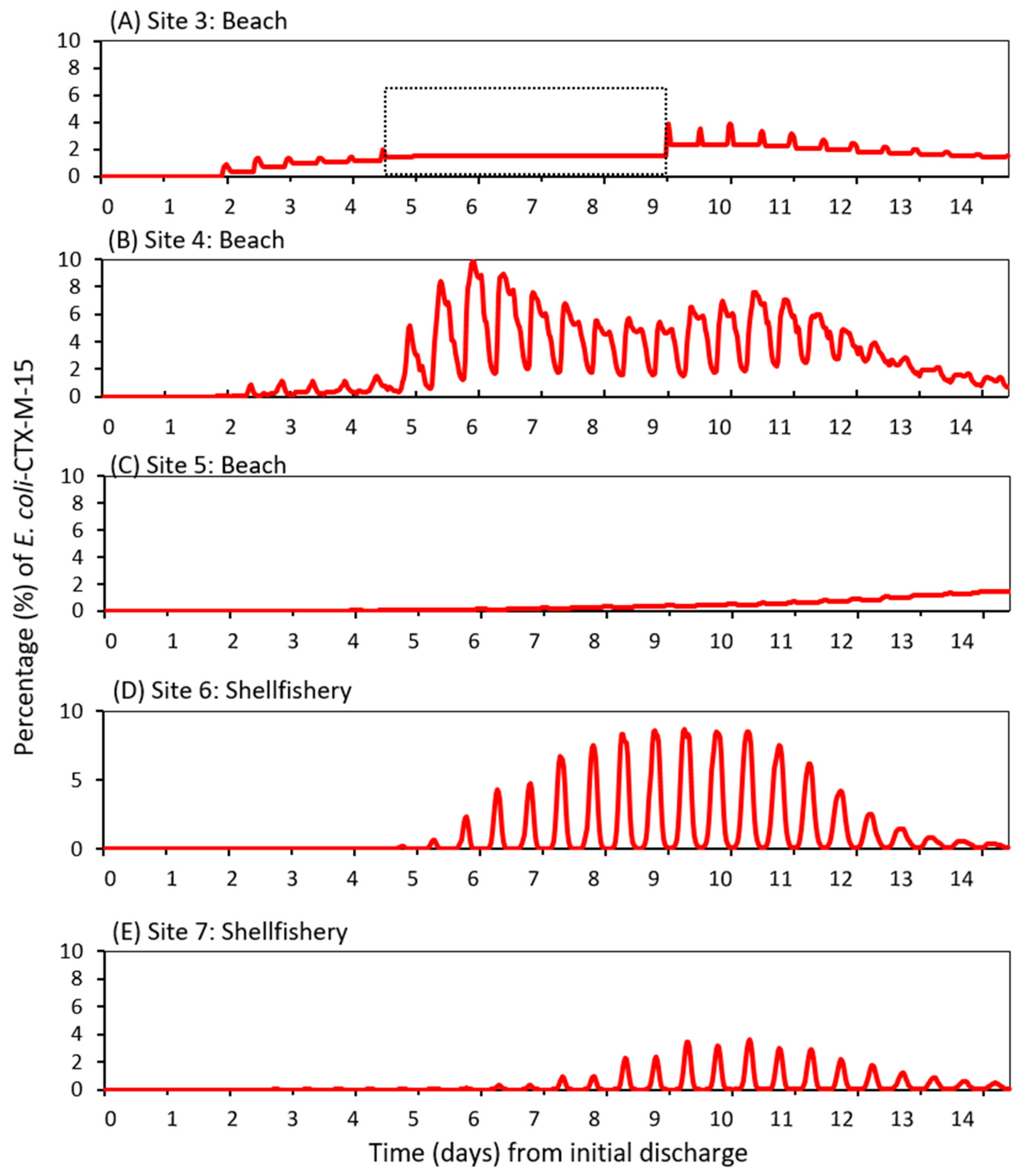
3.3. Dispersal of blaCTX-M-15-Producing E. coli within the Receiving Estuary
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pitout, J.D.D.; Nordmann, P.; Laupland, K.B.; Poirel, L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 2005, 56, 52–59. [Google Scholar] [CrossRef]
- Samaha-kfoury, J.N.; Araj, G.F. Recent developments in β lactamases and extended spectrum β lactamases. Br. Med. J. 2003, 327, 1209–1213. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Munir, M.; Xagoraraki, I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 2012, 421–422, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Novo, A.; André, S.; Viana, P.; Nunes, O.C.; Manaia, C.M. Antibiotic resistance, Antimicrobial residues and bacterial community composition in urban wastewater. Water Res. 2013, 47, 1875–1887. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Fiorentino, A.; Anselmo, A. Advanced treatment of urban wastewater by UV radiation: Effect on antibiotics and antibiotic-resistant E. coli strains. Chemosphere 2013, 92, 171–176. [Google Scholar] [CrossRef]
- Wellington, E.M.H.; Boxall, A.B.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Bonnet, R. Growing Group of Extended-Spectrum β-Lactamases: The CTX-M Enzymes. Antimicrob. Agents Chemother. Agents Chemother. 2004, 48, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bollache, L.; Bardet, E.; Depret, G.; Motreuil, S.; Neuwirth, C.; Moreau, J.; Hartmann, A. Dissemination of CTX-M-Producing Escherichia coli in Freshwater Fishes From a French Watershed (Burgundy). Front. Microbiol. 2019, 9, 3239. [Google Scholar] [CrossRef]
- Sellera, F.P.; Fernandes, M.R.; Moura, Q.; Souza, T.A.; Cerdeira, L.; Lincopan, N. Draft genome sequence of Enterobacter cloacae ST520 harbouring blaKPC-2, blaCTX-M-15 and blaOXA-17 isolated from coastal waters of the South Atlantic Ocean. J. Glob. Antimicrob. Resist. 2017, 10, 279–280. [Google Scholar] [CrossRef]
- Bréchet, C.; Plantin, J.; Sauget, M.; Thouverez, M.; Talon, D.; Cholley, P.; Guyeux, C.; Hocquet, D.; Bertrand, X. Wastewater treatment plants release large amounts of extended-spectrum β-lactamase-producing Escherichia coli into the environment. Clin. Infect. Dis. 2014, 58, 1658–1665. [Google Scholar] [CrossRef]
- Williams, A.P.; Avery, L.M.; Killham, K.; Jones, D.L. Persistence, dissipation, and activity of Escherichia coli O157:H7 within sand and seawater environments. FEMS Microbiol. Ecol. 2007, 60, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Avery, L.M.; Williams, P.; Killham, K.; Jones, D.L. Survival of Escherichia coli O157: H7 in waters from lakes, rivers, puddles and animal-drinking troughs. Sci. Total Environ. 2008, 389, 378–385. [Google Scholar] [CrossRef]
- Marti, E.; Variatza, E.; Balcazar, J.L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Hassard, F.; Gwyther, C.L.; Farkas, K.; Andrews, A.; Jones, V.; Cox, B.; Brett, H.; Jones, D.L.; McDonald, J.E.; Malham, S.K. Abundance and distribution of enteric bacteria and viruses in coastal and estuarine sediments-a review. Front. Microbiol. 2016, 7, 1692. [Google Scholar] [CrossRef] [PubMed]
- Malham, S.K.; Rajko-Nenow, P.; Howlett, E.; Tuson, K.E.; Perkins, T.L.; Pallett, D.W.; Wang, H.; Jago, C.F.; Jones, D.L.; McDonald, J.E. The interaction of human microbial pathogens, particulate material and nutrients in estuarine environments and their impacts on recreational and shellfish waters. Environ. Sci. Process. Impacts 2014, 16, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.Y.; Zhang, Y.L.; Geng, S.N.; Li, T.Y.; Ye, Z.M.; Zhang, D.S.; Zou, F.; Zhou, H.W. High diversity of extended-spectrum beta-lactamase-producing bacteria in an urban river sediment habitat. Appl. Environ. Microbiol. 2010, 76, 5972–5976. [Google Scholar] [CrossRef]
- Robins, P.E.; Lewis, M.J.; Simpson, J.H.; Howlett, E.R.; Malham, S.K. Future variability of solute transport in a macrotidal estuary. Estuar. Coast. Shelf Sci. 2014, 151, 88–99. [Google Scholar] [CrossRef]
- Robins, P.E.; Lewis, M.J.; Freer, J.; Cooper, D.M.; Skinner, C.J.; Coulthard, T.J. Improving estuary models by reducing uncertainties associated with river flows. Estuar. Coast. Shelf Sci. 2018, 207, 63–73. [Google Scholar] [CrossRef]
- Howlett, E.R.; Bowers, D.G.; Malarkey, J.; Jago, C.F. Stratification in the presence of an axial convergent front: Causes and implications. Estuar. Coast. Shelf Sci. 2015, 161, 1–10. [Google Scholar] [CrossRef]
- Bussi, G.; Whitehead, P.G.; Thomas, A.R.C.; Masante, D.; Jones, L.; Jack Cosby, B.; Emmett, B.A.; Malham, S.K.; Prudhomme, C.; Prosser, H. Climate and land-use change impact on faecal indicator bacteria in a temperate maritime catchment (the River Conwy, Wales). J. Hydrol. 2017, 553, 248–261. [Google Scholar] [CrossRef]
- Hassard, F.; Andrews, A.; Jones, D.L.; Parsons, L.; Jones, V.; Cox, B.A.; Daldorph, P.; Brett, H.; McDonald, J.E.; Malham, S.K. Physicochemical factors influence the abundance and culturability of human enteric pathogens and fecal indicator organisms in estuarine water and sediment. Front. Microbiol. 2017, 8, 1996. [Google Scholar] [CrossRef]
- Farkas, K.; Cooper, D.M.; McDonald, J.E.; Malham, S.K.; de Rougemont, A.; Jones, D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018, 634, 1174–1183. [Google Scholar] [CrossRef] [PubMed]
- Emmett, B.A.; Cooper, D.; Smart, S.; Jackson, B.; Thomas, A.; Cosby, B.; Evans, C.; Glanville, H.; McDonald, J.E.; Malham, S.K.; et al. Spatial patterns and environmental constraints on ecosystem services at a catchment scale. Sci. Total Environ. 2016, 572, 1586–1600. [Google Scholar] [CrossRef]
- Murphy, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Miranda, K.M.; Espey, M.G.; Wink, D.A. A rapid, simple, spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 2001, 5, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.D.; Bogaerts, P.; Berhin, C.; Guisset, A.; Glupczynski, Y. Evaluation of Brilliance ESBL Agar, a novel chromogenic medium for detection of extended-spectrum-beta- lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 2010, 48, 2091–2096. [Google Scholar] [CrossRef]
- Lawler, D.M.; Foster, I.D.L.; Petts, G.E.; Harper, S.; Morrissey, I.P. Suspended sediment dynamics for June storm events in the urbanized River Tame, UK. In Proceedings of the Sediment Dynamics and the Hydromorphology of Fluvial Systems (IAHS Symposium), Dundee, UK, 2–7 July 2006; International Association of Hydrological Sciences Publ.: Wallingford, UK, 2006; pp. 96–103. Available online: https://iahs.info/uploads/dms/13538.16-96-103-45-306-Lawler-et-al-edited.pdf (accessed on 16 January 2020).
- Mallin, M.A.; Mciver, M.R.; Spivey, M.I.H.; Song, B. 2009 Environmental Quality of Wilmington and New Hanover County Watersheds. 2008. Available online: www.uncw.edu/cmsr/aquaticecology/tidalcreeks (accessed on 20 March 2018).
- Chandra, V.; Goswami, P.N. Detection of TEM & SHV genes in Extended Spectrum Beta Lactamase (ESBL) producing E. coli & Klebsiella pneumoniae isolated from a tertiary care cancer hospital. Natl. J. Med. Res. 2014, 4, 201–204. [Google Scholar]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Bej, A.K.; Mccarty, S.C.; Atlas, R.M. Detection of coliform bacteria and Escherichia coli by multiplex polymerase chain reaction: Comparison with defined substrate and plating methods for water quality monitoring. Appl. Environ. Microbiol. 1991, 57, 2429–2432. [Google Scholar] [CrossRef]
- Hervouet, J.-M. Hydrodynamics of Free Surface Flows: Modelling with the Finite Element Method; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2007; ISBN 978-0-470-03558-0. [Google Scholar]
- Robins, P.E.; Farkas, K.; Jones, D.L.; Cooper, D.; Malham, S.K. Viral dispersal in the coastal zone: A method to quantify water quality risk. Environ. Int. 2019, 126, 430–442. [Google Scholar] [CrossRef]
- Egbert, G.D.; Bennett, A.F.; Foreman, M.G. TOPEX/POSEIDON tides estimated using a global inverse model. J. Geophys. Res. 1994, 99, 24821–24852. [Google Scholar] [CrossRef]
- Bueno, I.; Williams-Nguyen, J.; Hwang, H.; Sargeant, J.M.; Nault, A.J.; Singer, R.S. Systematic Review: Impact of point sources on antibiotic-resistant bacteria in the natural environment. Zoonoses Public Health 2018, 65, e162–e184. [Google Scholar] [CrossRef] [PubMed]
- Amos, G.C.A.; Hawkey, P.M.; Gaze, W.H.; Wellington, E.M. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob. Chemother. 2014, 69, 1785–1791. [Google Scholar] [CrossRef] [PubMed]
- Connor, D.W.; Gilliand, P.; Golding, N.; Robinson, P.; Todd, D.; Verling, E. UKSeaMap: The Mapping of Seabed and Water Column Features of UK Seas; Joint Nature Conservation Committee: Peterborough, UK, 2006; ISBN 9781861075901. [Google Scholar]
- Perkins, T.L.; Perrow, K.; Rajko-Nenow, P.; Jago, C.F.; Jones, D.L.; Malham, S.K.; McDonald, J.E. Decay rates of faecal indicator bacteria from sewage and ovine faeces in brackish and freshwater microcosms with contrasting suspended particulate matter concentrations. Sci. Total Environ. 2016, 572, 1645–1652. [Google Scholar] [CrossRef] [PubMed]
- Berney, M.; Weilenmann, H.U.; Egli, T. Flow-cytometric study of vital cellular functions in Escherichia coli during solar disinfection (SODIS). Microbiology 2006, 152, 1719–1729. [Google Scholar] [CrossRef]
- Bosshard, F.; Bucheli, M.; Meur, Y.; Egli, T. The respiratory chain is the cell’s Achilles’ heel during UVA inactivation in Escherichia coli. Microbiology 2010, 156, 2006–2015. [Google Scholar] [CrossRef]
- Pachepsky, Y.A.; Shelton, D.R. Escherichia coli and Fecal Coliforms in Freshwater and Estuarine Sediments. Crit. Rev. Environ. Sci. Technol. 2011, 41, 1067–1110. [Google Scholar] [CrossRef]
- Wait, D.A.; Sobsey, M.D. Comparative survival of enteric viruses and bacteria in Atlantic Ocean seawater. Water Sci. Technol. 2001, 43, 139–142. [Google Scholar] [CrossRef]
- Silva, I.; Tacão, M.; Tavares, R.D.S.; Miranda, R.; Araújo, S. Fate of cefotaxime-resistant Enterobacteriaceae and ESBL-producers over a full-scale wastewater treatment process with UV disinfection. Sci. Total Environ. 2018, 639, 1028–1037. [Google Scholar] [CrossRef]
- Muela, A.; García-Bringas, J.M.; Arana, I.; Barcina, I. The effect of simulated solar radiation on Escherichia coli: The relative roles of UV-B, UV-A, and photosynthetically active radiation. Microb. Ecol. 2000, 39, 65–71. [Google Scholar] [CrossRef]
- Sinton, L.; Hall, C.; Braithwaite, R. Sunlight inactivation of Campylobacter jejuni and Salmonella enterica, compared with Escherichia coli, in seawater and river water. J. Water Health 2007, 5, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Locatelli, A.; Amoureux, L.; Depret, G.; Jolivet, C.; Gueneau, E.; Neuwirth, C. Occurrence of CTX-M producing Escherichia coli in soils, cattle, and farm environment in France (Burgundy region). Front. Microbiol. 2012, 3, 83. [Google Scholar] [CrossRef] [PubMed]
- Starlander, G.; Yin, H.; Edquist, P.; Melhus, Å. Survival in the environment is a possible key factor for the expansion of Escherichia coli strains producing extended-spectrum β-lactamases. J. Pathol. Microbiol. Immunol. 2014, 122, 59–67. [Google Scholar]
| Parameter | Mean ± Standard Error of Mean (SEM) |
|---|---|
| pH | 7.41 ± 0.09 |
| Electrical conductivity (mS/cm) | 0.10 ± 0.01 |
| Total organic carbon (mg/L) | 5.92 ± 1.10 |
| Total organic nitrogen (mg/L) | 1.42 ± 0.19 |
| Nitrate (mg/L) | 0.89 ± 0.01 |
| Phosphate (mg/L) | 0.02 ± 0.01 |
| ESBL *-producing E. coli | None detected |
| Turbidity Level | k-Value | SEM | R2 |
|---|---|---|---|
| Low | 2.80 | 0.09 | 0.88 |
| Medium | 1.95 | 0.06 | 0.96 |
| High | 0.62 | 0.03 | 0.96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bashawri, Y.M.; Robins, P.; Cooper, D.M.; McDonald, J.E.; Jones, D.L.; Williams, A.P. Impact of Sediment Concentration on the Survival of Wastewater-Derived blaCTX-M-15-Producing E. coli, and the Implications for Dispersal into Estuarine Waters. Int. J. Environ. Res. Public Health 2020, 17, 7608. https://doi.org/10.3390/ijerph17207608
Bashawri YM, Robins P, Cooper DM, McDonald JE, Jones DL, Williams AP. Impact of Sediment Concentration on the Survival of Wastewater-Derived blaCTX-M-15-Producing E. coli, and the Implications for Dispersal into Estuarine Waters. International Journal of Environmental Research and Public Health. 2020; 17(20):7608. https://doi.org/10.3390/ijerph17207608
Chicago/Turabian StyleBashawri, Yasir M., Peter Robins, David M. Cooper, James E. McDonald, Davey L. Jones, and A. Prysor Williams. 2020. "Impact of Sediment Concentration on the Survival of Wastewater-Derived blaCTX-M-15-Producing E. coli, and the Implications for Dispersal into Estuarine Waters" International Journal of Environmental Research and Public Health 17, no. 20: 7608. https://doi.org/10.3390/ijerph17207608
APA StyleBashawri, Y. M., Robins, P., Cooper, D. M., McDonald, J. E., Jones, D. L., & Williams, A. P. (2020). Impact of Sediment Concentration on the Survival of Wastewater-Derived blaCTX-M-15-Producing E. coli, and the Implications for Dispersal into Estuarine Waters. International Journal of Environmental Research and Public Health, 17(20), 7608. https://doi.org/10.3390/ijerph17207608





