The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae
Abstract
:1. Introduction
2. Virulence
3. Biofilm
4. Antibiotic Resistance
4.1. Aminoglycoside Resistance Gene
4.2. Quinolone Resistance Gene
4.3. β-lactam Resistance Gene
4.4. Polymyxin Resistance Gene
4.5. Tigecycline Resistance Gene
5. Application of Whole Genome Sequencing
6. Application of Global Proteomics
7. Clinical Study of K. pneumoniae
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chew, K.L.; Lin, R.T.P.; Teo, J.W.P. Klebsiella pneumoniae in Singapore: Hypervirulent Infections and the Carbapenemase Threat. Front. Cell. Infect. Microbiol. 2017, 7, 515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yao, Z.; Zhan, S.; Yang, Z.; Wei, D.; Zhang, J.; Li, J.; Kyaw, M.H. Disease burden of intensive care unit-acquired pneumonia in China: A systematic review and meta-analysis. Int. J. Infect. Dis. 2014, 29, 84–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, Q.; Yin, Y.; Chen, H.; Jin, L.; Gu, B.; Xie, L.; Yang, C.; Ma, X.; Li, H.; et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae Infections: Report from the China CRE Network. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Martin, R.M.; Bachman, M.A. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Cubero, M.; Grau, I.; Tubau, F.; Pallares, R.; Dominguez, M.A.; Linares, J.; Ardanuy, C. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin. Microbiol. Infect. 2016, 22, 154–160. [Google Scholar] [CrossRef] [Green Version]
- Kishibe, S.; Okubo, Y.; Morino, S.; Hirotaki, S.; Tame, T.; Aoki, K.; Ishii, Y.; Ota, N.; Shimomura, S.; Sakakibara, H.; et al. Pediatric hypervirulent Klebsiella pneumoniae septic arthritis. Pediatr. Int. 2016, 58, 382–385. [Google Scholar] [CrossRef]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [Green Version]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Piperaki, E.T.; Syrogiannopoulos, G.A.; Tzouvelekis, L.S.; Daikos, G.L. Klebsiella pneumoniae: Virulence, Biofilm and Antimicrobial Resistance. Pediatr. Infect. Dis. J. 2017, 36, 1002–1005. [Google Scholar] [CrossRef] [PubMed]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Happel, K.I.; Dubin, P.J.; Zheng, M.; Ghilardi, N.; Lockhart, C.; Quinton, L.J.; Odden, A.R.; Shellito, J.E.; Bagby, G.J.; Nelson, S.; et al. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J. Exp. Med. 2005, 202, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.F.; Yang, F.L.; Chiu, H.W.; Chou, J.C.; Dong, W.C.; Lin, C.N.; Lin, C.Y.; Wang, J.T.; Li, L.H.; Chiu, H.W.; et al. Capsular Polysaccharide Is Involved in NLRP3 Inflammasome Activation by Klebsiella pneumoniae Serotype K1. Infect. Immun. 2015, 83, 3396–3409. [Google Scholar] [CrossRef] [Green Version]
- Shon, A.S.; Bajwa, R.P.; Russo, T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Sun, S.; Zhao, Z.Y.; Sun, Y. The pathogenicity of rmpA or aerobactin-positive Klebsiella pneumoniae in infected mice. J. Int. Med. Res. 2019, 47, 4344–4352. [Google Scholar] [CrossRef] [Green Version]
- Merino, S.; Camprubi, S.; Alberti, S.; Benedi, V.J.; Tomas, J.M. Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect. Immun. 1992, 60, 2529–2535. [Google Scholar] [CrossRef] [Green Version]
- Llobet, E.; Martinez-Moliner, V.; Moranta, D.; Dahlstrom, K.M.; Regueiro, V.; Tomas, A.; Cano, V.; Perez-Gutierrez, C.; Frank, C.G.; Fernandez-Carrasco, H.; et al. Deciphering tissue-induced Klebsiella pneumoniae lipid A structure. Proc. Natl. Acad. Sci. USA 2015, 112, E6369–E6378. [Google Scholar] [CrossRef] [Green Version]
- Lam, M.M.C.; Wyres, K.L.; Judd, L.M.; Wick, R.R.; Jenney, A.; Brisse, S.; Holt, K.E. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018, 10, 77. [Google Scholar] [CrossRef] [Green Version]
- Russo, T.A.; Shon, A.S.; Beanan, J.M.; Olson, R.; MacDonald, U.; Pomakov, A.O.; Visitacion, M.P. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS ONE 2011, 6, e26734. [Google Scholar] [CrossRef]
- Lan, Y.; Zhou, M.; Jian, Z.; Yan, Q.; Wang, S.; Liu, W. Prevalence of pks gene cluster and characteristics of Klebsiella pneumoniae-induced bloodstream infections. J. Clin. Lab. Anal. 2019, 33, e22838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.; Fu, Y.; Fang, Y.; Xu, H.; Kong, H.; Liu, Y.; Chen, Y.; Li, L. High prevalence of KPC-2-producing hypervirulent Klebsiella pneumoniae causing meningitis in Eastern China. Infect. Drug Resist. 2019, 12, 641–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turton, J.; Davies, F.; Turton, J.; Perry, C.; Payne, Z.; Pike, R. Hybrid Resistance and Virulence Plasmids in “High-Risk” Clones of Klebsiella pneumoniae, Including Those Carrying blaNDM-5. Microorganisms 2019, 7, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostafavi, M.; Wang, L.; Xie, L.; Takeoka, K.T.; Richie, D.L.; Casey, F.; Ruzin, A.; Sawyer, W.S.; Rath, C.M.; Wei, J.R.; et al. Interplay of Klebsiella pneumoniae fabZ and lpxC Mutations Leads to LpxC Inhibitor-Dependent Growth Resulting from Loss of Membrane Homeostasis. mSphere 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, P.F.; Hsu, C.R.; Chen, C.T.; Lin, T.L.; Wang, J.T. The Klebsiella pneumoniae YfgL (BamB) lipoprotein contributes to outer membrane protein biogenesis, type-1 fimbriae expression, anti-phagocytosis, and in vivo virulence. Virulence 2016, 7, 587–601. [Google Scholar] [CrossRef] [Green Version]
- Saurel, O.; Iordanov, I.; Nars, G.; Demange, P.; Le Marchand, T.; Andreas, L.B.; Pintacuda, G.; Milon, A. Local and Global Dynamics in Klebsiella pneumoniae Outer Membrane Protein a in Lipid Bilayers Probed at Atomic Resolution. J. Am. Chem. Soc. 2017, 139, 1590–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doi, Y.; Wachino, J.I.; Arakawa, Y. Aminoglycoside Resistance: The Emergence of Acquired 16S Ribosomal RNA Methyltransferases. Infect. Dis. Clin. N. Am. 2016, 30, 523–537. [Google Scholar] [CrossRef]
- Peirano, G.; Ahmed-Bentley, J.; Fuller, J.; Rubin, J.E.; Pitout, J.D. Travel-related carbapenemase-producing Gram-negative bacteria in Alberta, Canada: The first 3 years. J. Clin. Microbiol. 2014, 52, 1575–1581. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, V.B.; Rajamohan, G. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob. Agents Chemother. 2013, 57, 4449–4462. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, V.B.; Venkataramaiah, M.; Mondal, A.; Vaidyanathan, V.; Govil, T.; Rajamohan, G. Functional characterization of a novel outer membrane porin KpnO, regulated by PhoBR two-component system in Klebsiella pneumoniae NTUH-K2044. PLoS ONE 2012, 7, e41505. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.K.; Liou, C.H.; Lin, J.C.; Ma, L.; Fung, C.P.; Chang, F.Y.; Siu, L.K. A suitable streptomycin-resistant mutant for constructing unmarked in-frame gene deletions using rpsL as a counter-selection marker. PLoS ONE 2014, 9, e109258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, Y.S.; Cho, S.Y.; Yang, H.Y.; Park, K.S.; Jang, J.H.; Kim, Y.T.; Jeong, J.W.; Suh, J.T.; Lee, H.J. Investigation of mutation distribution in DNA gyrase and topoisomerase IV genes in ciprofloxacin-non-susceptible Enterobacteriaceae isolated from blood cultures in a tertiary care university hospital in South Korea, 2005–2010. Int. J. Antimicrob. Agents 2013, 41, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Martinez, L.; Hernandez-Alles, S.; Alberti, S.; Tomas, J.M.; Benedi, V.J.; Jacoby, G.A. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum-cephalosporins. Antimicrob. Agents Chemother. 1996, 40, 342–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzariol, A.; Zuliani, J.; Cornaglia, G.; Rossolini, G.M.; Fontana, R. AcrAB Efflux System: Expression and Contribution to Fluoroquinolone Resistance in Klebsiella spp. Antimicrob. Agents Chemother. 2002, 46, 3984–3986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ping, Y.; Ogawa, W.; Kuroda, T.; Tsuchiya, T. Gene cloning and characterization of KdeA, a multidrug efflux pump from Klebsiella pneumoniae. Biol. Pharm. Bull. 2007, 30, 1962–1964. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.H.; Chan, E.W.; Chen, S. Evolution and dissemination of OqxAB-like efflux pumps, an emerging quinolone resistance determinant among members of Enterobacteriaceae. Antimicrob. Agents Chemother. 2015, 59, 3290–3297. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, E.; Saenz, Y.; Zarazaga, M.; Rocha-Gracia, R.; Martinez-Martinez, L.; Arlet, G.; Torres, C. qnr, aac(6’)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp.: Genetic environments and plasmid and chromosomal location. J. Antimicrob. Chemother. 2012, 67, 886–897. [Google Scholar] [CrossRef]
- Sirot, D.; Sirot, J.; Labia, R.; Morand, A.; Courvalin, P.; Darfeuille-Michaud, A.; Perroux, R.; Cluzel, R. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: Identification of CTX-1, a novel beta-lactamase. J. Antimicrob. Chemother. 1987, 20, 323–334. [Google Scholar] [CrossRef]
- Calbo, E.; Garau, J. The changing epidemiology of hospital outbreaks due to ESBL-producing Klebsiella pneumoniae: The CTX-M-15 type consolidation. Future Microbiol. 2015, 10, 1063–1075. [Google Scholar] [CrossRef]
- Jimenez-Castellanos, J.C.; Wan Nur Ismah, W.A.K.; Takebayashi, Y.; Findlay, J.; Schneiders, T.; Heesom, K.J.; Avison, M.B. Envelope proteome changes driven by RamA overproduction in Klebsiella pneumoniae that enhance acquired beta-lactam resistance. J. Antimicrob. Chemother. 2018, 73, 88–94. [Google Scholar] [CrossRef]
- Evans, B.A.; Amyes, S.G. OXA beta-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, P.A. Extended-spectrum beta-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippon, A.; Slama, P.; Deny, P.; Labia, R. A Structure-Based Classification of Class A beta-Lactamases, a Broadly Diverse Family of Enzymes. Clin. Microbiol. Rev. 2016, 29, 29–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Shen, K.; Zhang, Y.; Ying, J.; Zhu, T.; Liu, Y.; Xu, L.; Lin, C.; Zhang, K.; Li, P.; et al. Characterization of a Novel blaKLUC Variant with Reduced beta-Lactam Resistance From an IncA/C Group Plasmid in a Clinical Klebsiella pneumoniae Isolate. Front. Microbiol. 2018, 9, 1908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clements, A.; Tull, D.; Jenney, A.W.; Farn, J.L.; Kim, S.H.; Bishop, R.E.; McPhee, J.B.; Hancock, R.E.; Hartland, E.L.; Pearse, M.J.; et al. Secondary acylation of Klebsiella pneumoniae lipopolysaccharide contributes to sensitivity to antibacterial peptides. J. Biol. Chem. 2007, 282, 15569–15577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Majumdar, S.; Veleba, M.; Finn, S.; Fanning, S.; Schneiders, T. Elucidating the regulon of multidrug resistance regulator RarA in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2013, 57, 1603–1609. [Google Scholar] [CrossRef] [Green Version]
- Mitrophanov, A.Y.; Jewett, M.W.; Hadley, T.J.; Groisman, E.A. Evolution and dynamics of regulatory architectures controlling polymyxin B resistance in enteric bacteria. PLoS Genet. 2008, 4, e1000233. [Google Scholar] [CrossRef] [Green Version]
- Llobet, E.; Campos, M.A.; Gimenez, P.; Moranta, D.; Bengoechea, J.A. Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect. Immun. 2011, 79, 3718–3732. [Google Scholar] [CrossRef] [Green Version]
- Jayol, A.; Poirel, L.; Brink, A.; Villegas, M.V.; Yilmaz, M.; Nordmann, P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 2014, 58, 4762–4766. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Jayol, A.; Bontron, S.; Villegas, M.V.; Ozdamar, M.; Turkoglu, S.; Nordmann, P. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2015, 70, 75–80. [Google Scholar] [CrossRef]
- Pal, S.; Verma, J.; Mallick, S.; Rastogi, S.K.; Kumar, A.; Ghosh, A.S. Absence of the glycosyltransferase WcaJ in Klebsiella pneumoniae ATCC13883 affects biofilm formation, increases polymyxin resistance and reduces murine macrophage activation. Microbiology 2019, 165, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Osei Sekyere, J.; Govinden, U.; Bester, L.A.; Essack, S.Y. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: Emerging resistance mechanisms and detection methods. J. Appl. Microbiol. 2016, 121, 601–617. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Chen, Q.; Shi, K.; Li, X.; Shi, Q.; He, F.; Zhou, J.; Yu, Y.; Hua, X. Step-Wise Increase in Tigecycline Resistance in Klebsiella pneumoniae Associated with Mutations in ramR, lon and rpsJ. PLoS ONE 2016, 11, e0165019. [Google Scholar] [CrossRef]
- Kallman, O.; Motakefi, A.; Wretlind, B.; Kalin, M.; Olsson-Liljequist, B.; Giske, C.G. Cefuroxime non-susceptibility in multidrug-resistant Klebsiella pneumoniae overexpressing ramA and acrA and expressing ompK35 at reduced levels. J. Antimicrob. Chemother. 2008, 62, 986–990. [Google Scholar] [CrossRef]
- Ahn, C.; Yoon, S.S.; Yong, T.S.; Jeong, S.H.; Lee, K. The Resistance Mechanism and Clonal Distribution of Tigecycline-Nonsusceptible Klebsiella pneumoniae Isolates in Korea. Yonsei Med. J. 2016, 57, 641–646. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Clegg, S.; Murphy, C.N. Epidemiology and Virulence of Klebsiella pneumoniae. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Fux, C.A.; Costerton, J.W.; Stewart, P.S.; Stoodley, P. Survival strategies of infectious biofilms. Trends Microbiol. 2005, 13, 34–40. [Google Scholar] [CrossRef]
- Yang, S.K.; Yusoff, K.; Ajat, M.; Thomas, W.; Abushelaibi, A.; Akseer, R.; Lim, S.E.; Lai, K.S. Disruption of KPC-producing Klebsiella pneumoniae membrane via induction of oxidative stress by cinnamon bark (Cinnamomum verum J. Presl) essential oil. PLoS ONE 2019, 14, e0214326. [Google Scholar] [CrossRef] [Green Version]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poulikakos, P.; Falagas, M.E. Aminoglycoside therapy in infectious diseases. Expert Opin. Pharmacother. 2013, 14, 1585–1597. [Google Scholar] [CrossRef] [PubMed]
- Cirit, O.S.; Fernandez-Martinez, M.; Yayla, B.; Martinez-Martinez, L. Aminoglycoside resistance determinants in multiresistant Escherichia coli and Klebsiella pneumoniae clinical isolates from Turkish and Syrian patients. Acta Microbiol. Immunol. Hung. 2019, 66, 327–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Wang, L.; Pan, J.; Yao, D.; Chen, C.; Zhu, T.; Lou, Q.; Hu, J.; Wu, Y.; Zhang, X.; et al. Prevalence of 16S rRNA methylase genes in Klebsiella pneumoniae isolates from a Chinese teaching hospital: Coexistence of rmtB and armA genes in the same isolate. Diagn. Microbiol. Infect. Dis. 2009, 64, 57–63. [Google Scholar] [CrossRef] [PubMed]
- El-Badawy, M.F.; Tawakol, W.M.; El-Far, S.W.; Maghrabi, I.A.; Al-Ghamdi, S.A.; Mansy, M.S.; Ashour, M.S.; Shohayeb, M.M. Molecular Identification of Aminoglycoside-Modifying Enzymes and Plasmid-Mediated Quinolone Resistance Genes among Klebsiella pneumoniae Clinical Isolates Recovered from Egyptian Patients. Int. J. Microbiol. 2017, 2017, 8050432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guven Gokmen, T.; Nagiyev, T.; Meral, M.; Onlen, C.; Heydari, F.; Koksal, F. NDM-1 and rmtC-Producing Klebsiella pneumoniae Isolates in Turkey. Jundishapur. J. Microbiol. 2016, 9, e33990. [Google Scholar] [CrossRef] [Green Version]
- Naeem, A.; Badshah, S.L.; Muska, M.; Ahmad, N.; Khan, K. The Current Case of Quinolones: Synthetic Approaches and Antibacterial Activity. Molecules 2016, 21, 268. [Google Scholar] [CrossRef] [Green Version]
- Redgrave, L.S.; Sutton, S.B.; Webber, M.A.; Piddock, L.J. Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef]
- Guillard, T.; de Jong, A.; Limelette, A.; Lebreil, A.L.; Madoux, J.; de Champs, C.; ComPath Study, G. Characterization of quinolone resistance mechanisms in Enterobacteriaceae recovered from diseased companion animals in Europe. Vet. Microbiol. 2016, 194, 23–29. [Google Scholar] [CrossRef]
- Zheng, J.X.; Lin, Z.W.; Sun, X.; Lin, W.H.; Chen, Z.; Wu, Y.; Qi, G.B.; Deng, Q.W.; Qu, D.; Yu, Z.J. Overexpression of OqxAB and MacAB efflux pumps contributes to eravacycline resistance and heteroresistance in clinical isolates of Klebsiella pneumoniae. Emerg. Microbes Infect. 2018, 7, 139. [Google Scholar] [CrossRef] [Green Version]
- Jacoby, G.A.; Strahilevitz, J.; Hooper, D.C. Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirzaii, M.; Jamshidi, S.; Zamanzadeh, M.; Marashifard, M.; Malek Hosseini, S.A.A.; Haeili, M.; Jahanbin, F.; Mansouri, F.; Darban-Sarokhalil, D.; Khoramrooz, S.S. Determination of gyrA and parC mutations and prevalence of plasmid-mediated quinolone resistance genes in Escherichia coli and Klebsiella pneumoniae isolated from patients with urinary tract infection in Iran. J. Glob. Antimicrob. Resist. 2018, 13, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Surleac, M.; Czobor Barbu, I.; Paraschiv, S.; Popa, L.I.; Gheorghe, I.; Marutescu, L.; Popa, M.; Sarbu, I.; Talapan, D.; Nita, M.; et al. Whole genome sequencing snapshot of multi-drug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in Southern Romania. PLoS ONE 2020, 15, e0228079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schultsz, C.; Geerlings, S. Plasmid-mediated resistance in Enterobacteriaceae: Changing landscape and implications for therapy. Drugs 2012, 72, 1–16. [Google Scholar] [CrossRef]
- Bush, K. Bench-to-bedside review: The role of beta-lactamases in antibiotic-resistant Gram-negative infections. Crit. Care 2010, 14, 224. [Google Scholar] [CrossRef] [Green Version]
- Canton, R.; Akova, M.; Carmeli, Y.; Giske, C.G.; Glupczynski, Y.; Gniadkowski, M.; Livermore, D.M.; Miriagou, V.; Naas, T.; Rossolini, G.M.; et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2012, 18, 413–431. [Google Scholar] [CrossRef] [Green Version]
- Livermore, D.M. Current epidemiology and growing resistance of gram-negative pathogens. Korean J. Intern. Med. 2012, 27, 128–142. [Google Scholar] [CrossRef]
- Falagas, M.E.; Kasiakou, S.K. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [Green Version]
- Davis, B.; Lilly, H.A.; Lowbury, E.J. Gram-negative bacilli in burns. J. Clin. Pathol. 1969, 22, 634–641. [Google Scholar] [CrossRef] [Green Version]
- Antoniadou, A.; Kontopidou, F.; Poulakou, G.; Koratzanis, E.; Galani, I.; Papadomichelakis, E.; Kopterides, P.; Souli, M.; Armaganidis, A.; Giamarellou, H. Colistin-resistant isolates of Klebsiella pneumoniae emerging in intensive care unit patients: First report of a multiclonal cluster. J. Antimicrob. Chemother. 2007, 59, 786–790. [Google Scholar] [CrossRef] [Green Version]
- Wright, M.S.; Suzuki, Y.; Jones, M.B.; Marshall, S.H.; Rudin, S.D.; van Duin, D.; Kaye, K.; Jacobs, M.R.; Bonomo, R.A.; Adams, M.D. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 2015, 59, 536–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macesic, N.; Nelson, B.; McConville, T.H.; Giddins, M.J.; Green, D.A.; Stump, S.; Gomez-Simmonds, A.; Annavajhala, M.K.; Uhlemann, A.C. Emergence of Polymyxin Resistance in Clinical Klebsiella pneumoniae Through Diverse Genetic Adaptations: A Genomic, Retrospective Cohort Study. Clin. Infect. Dis. 2020, 70, 2084–2091. [Google Scholar] [CrossRef] [PubMed]
- Wink, P.L.; Caierao, J.; Nunes, A.G.A.; Collar, G.D.S.; Martins, J.B.; Dalmolin, T.V.; Pilonetto, M.; Barth, A.L. Evaluation of EDTA and Dipicolinic Acid in Broth Microdilution with Polymyxin B as a Phenotypic Test to Detect the mcr-1 Gene. Microb. Drug Resist. 2020, 26, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Dortet, L.; Broda, A.; Bernabeu, S.; Glupczynski, Y.; Bogaerts, P.; Bonnin, R.; Naas, T.; Filloux, A.; Larrouy-Maumus, G. Optimization of the MALDIxin test for the rapid identification of colistin resistance in Klebsiella pneumoniae using MALDI-TOF MS. J. Antimicrob. Chemother. 2020, 75, 110–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livermore, D.M. Tigecycline: What is it, and where should it be used? J. Antimicrob. Chemother. 2005, 56, 611–614. [Google Scholar] [CrossRef] [Green Version]
- Golan, Y. Empiric therapy for hospital-acquired, Gram-negative complicated intra-abdominal infection and complicated urinary tract infections: A systematic literature review of current and emerging treatment options. BMC Infect. Dis. 2015, 15, 313. [Google Scholar] [CrossRef] [Green Version]
- Ruzin, A.; Visalli, M.A.; Keeney, D.; Bradford, P.A. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2005, 49, 1017–1022. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Han, Y.; Zhou, Y.; Du, Z.; Wu, H.; Wang, J.; Chen, Y. Tigecycline Susceptibility and Molecular Resistance Mechanisms among Clinical Klebsiella pneumoniae Strains Isolated during Non-Tigecycline Treatment. Microb. Drug Resist. 2017, 23, 139–146. [Google Scholar] [CrossRef]
- Villa, L.; Feudi, C.; Fortini, D.; Garcia-Fernandez, A.; Carattoli, A. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob. Agents chemother. 2014, 58, 1707–1712. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Xie, Y.; Li, G.; Liu, J.; Li, X.; Tian, L.; Sun, J.; Ou, H.Y.; Qu, H. Whole-Genome-Sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence 2018, 9, 510–521. [Google Scholar] [CrossRef] [Green Version]
- Lepuschitz, S.; Schill, S.; Stoeger, A.; Pekard-Amenitsch, S.; Huhulescu, S.; Inreiter, N.; Hartl, R.; Kerschner, H.; Sorschag, S.; Springer, B.; et al. Whole genome sequencing reveals resemblance between ESBL-producing and carbapenem resistant Klebsiella pneumoniae isolates from Austrian rivers and clinical isolates from hospitals. Sci. Total Environ. 2019, 662, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Bialek-Davenet, S.; Criscuolo, A.; Ailloud, F.; Passet, V.; Jones, L.; Delannoy-Vieillard, A.S.; Garin, B.; Le Hello, S.; Arlet, G.; Nicolas-Chanoine, M.H.; et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 2014, 20, 1812–1820. [Google Scholar] [CrossRef] [PubMed]
- Struve, C.; Roe, C.C.; Stegger, M.; Stahlhut, S.G.; Hansen, D.S.; Engelthaler, D.M.; Andersen, P.S.; Driebe, E.M.; Keim, P.; Krogfelt, K.A. Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. mBio 2015, 6, e00630. [Google Scholar] [CrossRef] [Green Version]
- Rimoldi, S.G.; Gentile, B.; Pagani, C.; Di Gregorio, A.; Anselmo, A.; Palozzi, A.M.; Fortunato, A.; Pittiglio, V.; Ridolfo, A.L.; Gismondo, M.R.; et al. Whole genome sequencing for the molecular characterization of carbapenem-resistant Klebsiella pneumoniae strains isolated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012-2014. BMC Infect. Dis. 2017, 17, 666. [Google Scholar] [CrossRef] [PubMed]
- Meletis, G.; Chatzopoulou, F.; Chatzidimitriou, D.; Tsingerlioti, F.; Botziori, C.; Tzimagiorgis, G.; Skoura, L. Whole Genome Sequencing of NDM-1-Producing ST11 Klebsiella pneumoniae Isolated in a Private Laboratory in Greece. Microb. Drug Resist. 2019, 25, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Founou, R.C.; Founou, L.L.; Allam, M.; Ismail, A.; Essack, S.Y. Whole Genome Sequencing of Extended Spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae Isolated from Hospitalized Patients in KwaZulu-Natal, South Africa. Sci. Rep. 2019, 9, 6266. [Google Scholar] [CrossRef]
- Boulund, F.; Karlsson, R.; Gonzales-Siles, L.; Johnning, A.; Karami, N.; Al-Bayati, O.; Ahren, C.; Moore, E.R.B.; Kristiansson, E. Typing and Characterization of Bacteria Using Bottom-up Tandem Mass Spectrometry Proteomics. Mol. Cell. Proteom. 2017, 16, 1052–1063. [Google Scholar] [CrossRef] [Green Version]
- Bittaye, M.; Cash, P. Streptococcus pneumoniae proteomics: Determinants of pathogenesis and vaccine development. Expert Rev. Proteom. 2015, 12, 607–621. [Google Scholar] [CrossRef]
- Saleh, S.; Staes, A.; Deborggraeve, S.; Gevaert, K. Targeted Proteomics for Studying Pathogenic Bacteria. Proteomics 2019, 19, e1800435. [Google Scholar] [CrossRef]
- Kamaladevi, A.; Balamurugan, K. Global Proteomics Revealed Klebsiella pneumoniae Induced Autophagy and Oxidative Stress in Caenorhabditis elegans by Inhibiting PI3K/AKT/mTOR Pathway during Infection. Front. Cell. Infect. Microbiol. 2017, 7, 393. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Garg, A.; Kumar, M.; Khan, A.U. Proteome profiling of carbapenem-resistant K. pneumoniae clinical isolate (NDM-4): Exploring the mechanism of resistance and potential drug targets. J. Proteom. 2019, 200, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Anand, T.; Virmani, N.; Kumar, S.; Mohanty, A.K.; Pavulraj, S.; Bera, B.C.; Vaid, R.K.; Ahlawat, U.; Tripathi, B.N. Phage therapy for treatment of virulent Klebsiella pneumoniae infection in a mouse model. J. Glob. Antimicrob. Resist. 2019, 21, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.R.; Lee, J.H.; Park, K.S.; Jeon, J.H.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell. Infect. Microbiol. 2017, 7, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Guo, L.Y.; Song, W.Q.; Wang, Y.; Dong, F.; Liu, G. Risk factors for carbapenem-resistant K. pneumoniae bloodstream infection and predictors of mortality in Chinese paediatric patients. BMC Infect. Dis. 2018, 18, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demir, S.; Soysal, A.; Bakir, M.; Kaufmann, M.E.; Yagci, A. Extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in paediatric wards: A nested case-control study. J. Paediatr. Child Health 2008, 44, 548–553. [Google Scholar] [CrossRef]
- Oğuz Mızrakçı, S.; Arda, B.; Erdem, H.A.; Uyar, M.; Tünger, A.; Sipahi, O.R.; Ulusoy, S. [Risk factors for gastrointestinal colonization by ESBL-producing Klebsiella pneumoniae and Escherichia coli in anaesthesiology and reanimation intensive care unit]. Mikrobiyol. Bul. 2013, 47, 223–229. [Google Scholar] [CrossRef] [Green Version]
- Gorrie, C.L.; Mirceta, M.; Wick, R.R.; Edwards, D.J.; Thomson, N.R.; Strugnell, R.A.; Pratt, N.F.; Garlick, J.S.; Watson, K.M.; Pilcher, D.V.; et al. Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients. Clin. Infect. Dis. 2017, 65, 208–215. [Google Scholar] [CrossRef] [Green Version]
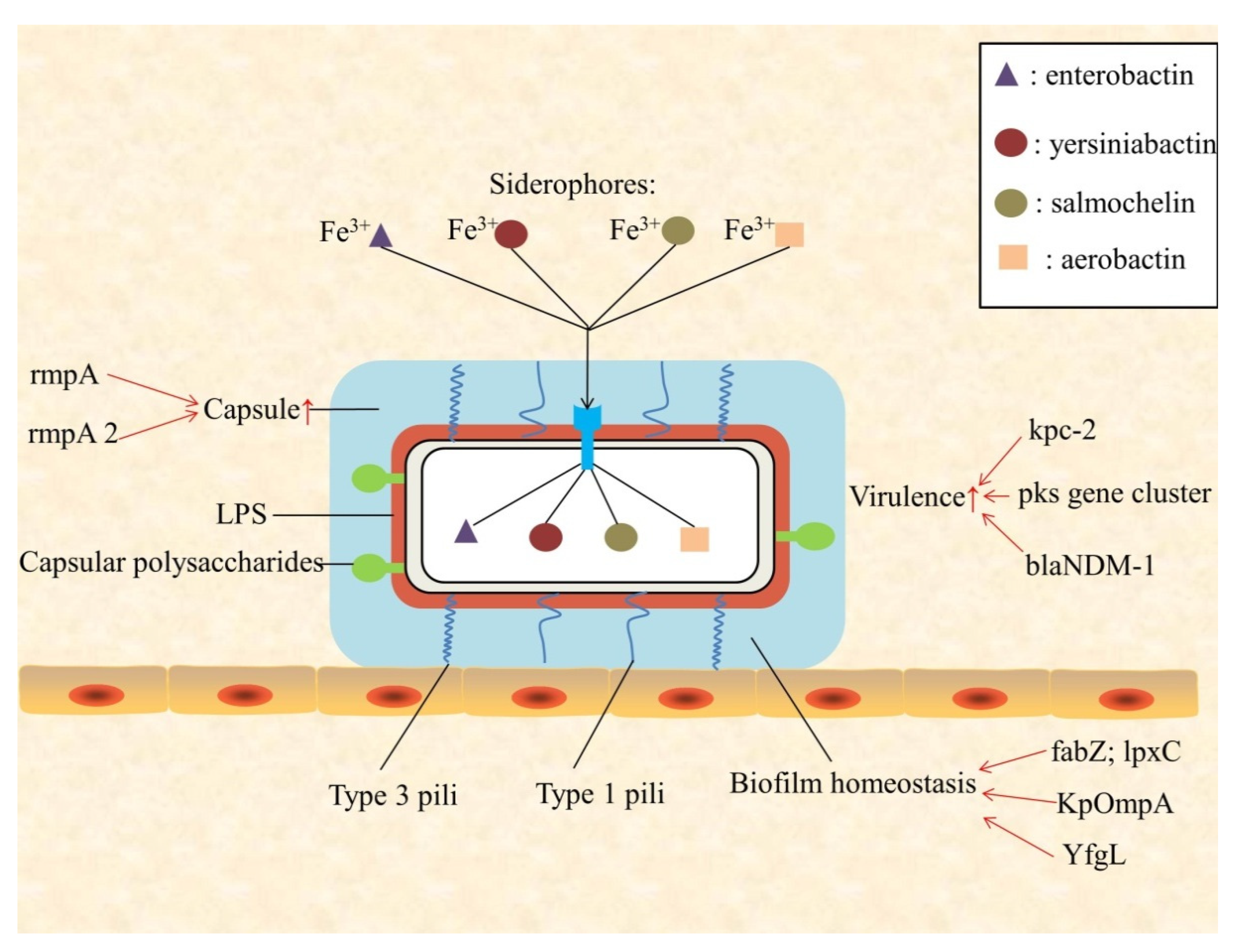
| The Characteristic of K. Pneumoniae | Gene Name | Function | References |
|---|---|---|---|
| Virulence | rmpA | Synthesis of capsular polysaccharides; high-mucus phenotype of hvKP | [16] |
| rmpA2 | Capsule up-regulation | [23] | |
| enterobactin | Iron carriers; growth and replication of bacteria | [12,13] | |
| yersiniabactin | Iron carriers; growth and replication of bacteria | [12,13] | |
| salmochelin | Iron carriers; growth and replication of bacteria; highly virulent K. pneumoniae | [12,13,19] | |
| aerobactin | Iron carriers; growth and replication of bacteria; detection rate of hvKP; highly virulent K. pneumoniae | [12,13,16] | |
| pks gene cluster | Host DNA damage; strains virulence enhancement | [21] | |
| kpc-2 | High prevalence and mortality of hvKP | [22] | |
| blaNDM-1 | Large virulent plasmid | [23] | |
| Biofilm | fabZ; lpxC | Biofilm homeostasis | [24] |
| YfgL (BamB) | biofilm formation; transcriptional expression of type 1 pili | [25] | |
| KpOmpA | Cell-cell recognition, adhesion, and immune response; pathogenicity | [26] | |
| Aminoglycoside resistance | 16S rRNA methylase | Encoding an enzyme that blocks the binding of aminoglycoside antibiotics to the 16S rRNA | [27] |
| aac families | Plasmid-mediated resistance genes | [27] | |
| ant families | Plasmid-mediated resistance genes | [27] | |
| aph families | Plasmid-mediated resistance genes | [27] | |
| AcrAB-TolC | Efflux pump systems; resistance to tobramycin and gentamicin | [28] | |
| kpnEF | Efflux pump systems; significant resistance to tobramycin and spectinomycin | [29] | |
| KpnO | Directly involved in aminoglycoside resistance; resistance of tobramycin, streptomycin and spectinomycin | [30] | |
| rrs or rpsL | rpsL mutations associated with high fitness costs and reduced virulence | [31] | |
| Quinolone resistance | DNA gyrase (gyrA-gyrB subunit) Topoisomerase IV (parC-parE subunit) | Resistance of nalidixic acid and ofloxacin | [32] |
| OmpK36 | Cell permeability | [33] | |
| acrAB | Cell permeability | [34] | |
| kdeA | Cell permeability | [35] | |
| OqxAB | Efflux pump; plasmid-mediated quinolone resistance | [36] | |
| qnr | Encoding a family of proteins that protect DNA gyrase and topoisomerase IV from quinolone inhibitory activity | [37] | |
| aa(6′)-Ib-cr | Quinolone modification | [37] | |
| β-lactam resistance | blaSHV-1 and blaTEM-1 | Penicillin resistance | [38] |
| blaSHV-2 | Extended-spectrum β-lactamase (ESBL) gene | [38] | |
| blaTEM-3 | plasmid-mediated ESBL variant | [38] | |
| blaCTX-M | ESBLs in K. pneumoniae caused by iatrogenic outbreaks | [39] | |
| ramA | Activating efflux pump; increasing acquired β-lactamase-mediated β-lactam resistance | [40] | |
| blaOXA, blaGES, blaSFO, blaPER, blaTLA, blaVEB and bla KLUC-5 | Horizontal gene transfer acquisition | [41,42,43,44] | |
| Polymyxin resistance | lpxM, ramA | Maturation of lipid A and lipid A neutralization | [45,46] |
| pbgP, pmrE | Combination of amino arabinose | [47,48] | |
| pmrC | Combination of phosphoethanolamine | [47,48] | |
| pagP | Combination of palmitate | [47,48] | |
| phoPQ, pmrA, pmrD and mgrB | LPS modified gene regulators | [49,50] | |
| RarA | High expression of efflux pumps AcrAB-TolC and KpnEF | [46] | |
| WcaJ | Non-mucus phenotype; increasing polymyxin resistance | [51] | |
| mcr-1 | Encoding a family of phosphoethanolamine transferases that can bind to phosphoethanolamine | [52] | |
| Tigecycline resistance | AcrAB-TolC, OqxAB | Overexpression of efflux pumps lead to tigecycline resistance | [53] |
| RarA, RamA, RamR and AcrR | Regulators of efflux pumps | [53] | |
| Lon and rpsJ | Encoding ribosome protein S10 | [54] | |
| ompK35K | Decreased transcript level of porin ompK35K can also lead to enhanced resistance | [55] | |
| tetA | Encoding tetracycline-resistant efflux pumps | [56] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Zhao, G.; Chao, X.; Xie, L.; Wang, H. The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health 2020, 17, 6278. https://doi.org/10.3390/ijerph17176278
Wang G, Zhao G, Chao X, Xie L, Wang H. The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. International Journal of Environmental Research and Public Health. 2020; 17(17):6278. https://doi.org/10.3390/ijerph17176278
Chicago/Turabian StyleWang, Guoying, Guo Zhao, Xiaoyu Chao, Longxiang Xie, and Hongju Wang. 2020. "The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae" International Journal of Environmental Research and Public Health 17, no. 17: 6278. https://doi.org/10.3390/ijerph17176278
APA StyleWang, G., Zhao, G., Chao, X., Xie, L., & Wang, H. (2020). The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. International Journal of Environmental Research and Public Health, 17(17), 6278. https://doi.org/10.3390/ijerph17176278





