Biomedical Holistic Ontology for People with Rare Diseases
Abstract
1. Introduction
2. Materials and Methods
2.1. Rare Diseases Scenarios
2.2. Methodology to Build the Ontology and to Perform the Sentiment Analysis
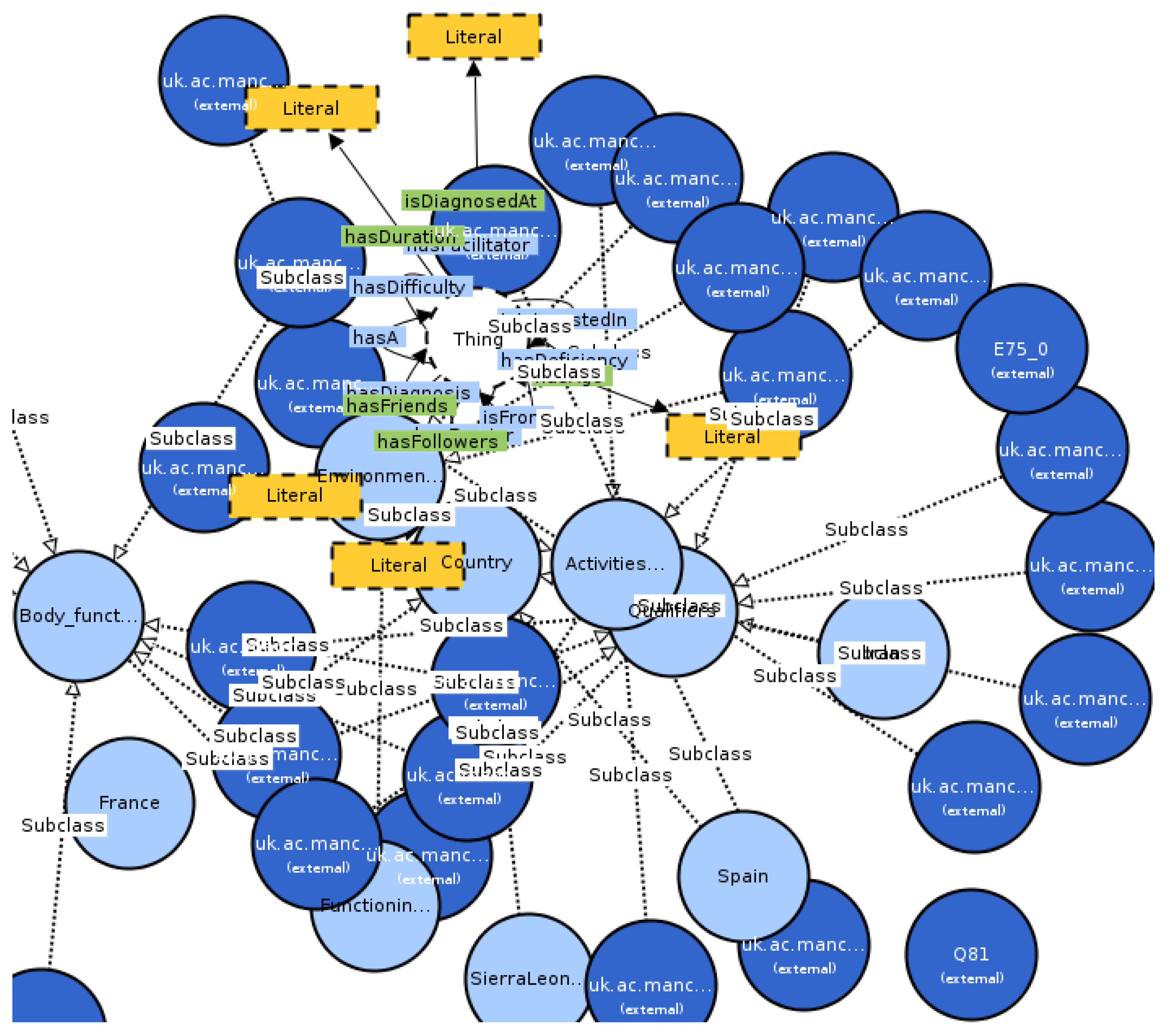
3. Results
- Number of classes: 14,558;
- Number of individuals: 85;
- Number of object properties: 69;
- Number of data properties: 32;
- Total classes MIEROTed: 27;
- Maximum depth: 4;
- Maximum number of children: 35;
- Average number of children: 7;
- Classes with a single child: 11;
- Classes with more than 25 children: 6.
- S: An abbreviation for ALC with transitive roles.
- AL: Attribute language. This is the base language which allows: atomic negation (negation of concepts that do not appear on the left-hand side of axioms), concept intersection, universal restrictions and limited existential quantification (restrictions that only have fillers of things).
- C: Complex concept negation.
- H: Role hierarchy (subproperties: rdfs:subPropertyOf).
- I: Inverse properties.
- (D): Use of datatype properties, data values or datatypes.
4. Discussion
5. Conclusions and Future Work
- A new holistic ontology about rare diseases has been built and shared. This ontology was composed by the integration of existing ontologies (medical and contextual) and includes information about 25 scenarios of people with rare diseases. The ontology has been validated and usefulness assessed. Depending on the user (a patient, a health professional, a policy maker, etc.) some parts of the ontology may be more interesting than others; thus, several views of the ontology should be generated.
- Code is shared openly to the community so that this research is partially reproducible.
- People are informed about the importance of supporting rare diseases and the problems of this collective. It is an objective to disseminate this study in Biomedical repositories such as Bioportal in order to inform the general public about problems involved in rare diseases. Therefore, these efforts aim to engage other people to work in this domain, helping the collective and providing it with more information.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADO | Alzheimer’s disease |
| DALY | Disability-adjusted life year |
| DMTO | Diabetes Mellitus treatment ontology |
| FEDER | Spanish Federation of Rare Diseases |
| HL7 | Health Level Seven International |
| HORD | Holistic Ontology of people with Rare Diseases |
| ICD | International Classification of Diseases |
| ICF | International Classification of Functioning, Disability and Health |
| ICHI | International Classification of Health Interventions |
| MESH | Medical Subject Headings |
| MIREOT | Minimum Information to Reference an External Ontology Term |
| MSO | Multiple Sclerosis Ontology |
| NLTK | Natural Language Toolkit |
| OOPS | OntOlogy Pitfall Scanner |
| ORDO | Orphanet Rare Disease Ontology |
| PDON | Parkinson’s disease |
| PMR | Physical Medicine and Rehabilitation Ontology |
| QALY | Quality-adjusted life year |
| RD | Rare disease |
| SHA | Secure hash algorithm |
| SIOC | Semantically-Interlinked Online Communities |
| SNOMED CT | Systematized Nomenclature of Medicine—Clinical Terms |
| WHO | World Health Organization |
References
- Henrard, S.; Arickx, F. Negotiating Prices of Drugs for Rare Diseases. Available online: http://www.who.int/bulletin/volumes/94/10/15-163519/en (accessed on 9 August 2020).
- Lassila, O.; McGuinness, D. The role of frame-based representation on the semantic web. Linköping Electron. Artic. Comput. Inf. Sci. 2001, 6, 2001. [Google Scholar]
- Donnelly, K. SNOMED-CT: The advanced terminology and coding system for eHealth. Stud. Health Technol. Inf. 2006, 121, 279–290. [Google Scholar]
- Bodenreider, O. The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D267–D270. [Google Scholar] [CrossRef] [PubMed]
- Kibbe, W.A.; Arze, C.; Felix, V.; Mitraka, E.; Bolton, E.; Fu, G.; Mungall, C.J.; Binder, J.X.; Malone, J.; Vasant, D.; et al. Disease Ontology 2015 update: An expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015, 43, D1071–D1078. [Google Scholar] [CrossRef]
- Tudorache, T.; Nyulas, C.I.; Noy, N.F.; Musen, M.A. Using Semantic Web in ICD-11: Three Years Down the Road. In Proceedings of the Semantic Web—ISWC 2013: 12th International Semantic Web Conference, Sydney, Australia, 21–25 October 2013; pp. 195–211. [Google Scholar]
- Solli, H.M.; da Silva, A.B. The Holistic Claims of the Biopsychosocial Conception of WHO’s International Classification of Functioning, Disability, and Health (ICF): A Conceptual Analysis on the Basis of a Pluralistic–Holistic Ontology and Multidimensional View of the Human being. J. Med. Philos. A Forum Bioeth. Philos. Med. 2012, 37, 277–294. [Google Scholar] [CrossRef]
- El-Sappagh, S.; Elmogy, M. A fuzzy ontology modeling for case base knowledge in diabetes mellitus domain. Eng. Sci. Technol. Int. J. 2017, 20, 1025–1040. [Google Scholar] [CrossRef]
- El-Sappagh, S.; Kwak, D.; Ali, F.; Kwak, K.S. DMTO: A realistic ontology for standard diabetes mellitus treatment. J. Biomed. Semant. 2018, 9, 8. [Google Scholar] [CrossRef]
- Subirats, L.; Gil, R.; García, R. Personalization of Ontologies Visualization: Use Case of Diabetes. In Current Trends in Semantic Web Technologies: Theory and Practice; Springer: Berlin, Germany, 2019; Volume 815, pp. 3–24. [Google Scholar]
- Malhotra, A.; Younesi, E.; Gündel, M.; Müller, B.; Heneka, M.T.; Hofmann-Apitius, M. ADO: A disease ontology representing the domain knowledge specific to Alzheimer’s disease. Alzheimers Dementia J. Alzheimers Assoc. 2014, 10, 238–246. [Google Scholar] [CrossRef]
- Younesi, E.; Malhotra, A.; Gündel, M.; Scordis, P.; Kodamullil, A.T.; Page, M.; Müller, B.; Springstubbe, S.; Wüllner, U.; Scheller, D.; et al. PDON: Parkinson’s disease ontology for representation and modeling of the Parkinson’s disease knowledge domain. Theor. Biol. Med. Model. 2015, 12, 20. [Google Scholar] [CrossRef]
- Malhotra, A.; Gündel, M.; Rajput, A.; Mevissen, H.; Saiz, A.; Pastor, X.; Lozano-Rubi, R.; Martinez-Lapiscina, E.H.; Zubizarreta, I.; Mueller, B.; et al. Knowledge Retrieval from PubMed Abstracts and Electronic Medical Records with the Multiple Sclerosis Ontology. PLoS ONE 2015, 10, e0116718. [Google Scholar] [CrossRef]
- Subirats, L.; Ceccaroni, L.; Lopez-Blazquez, R.; Miralles, F.; García-Rudolph, A.; Tormos, J.M. Circles of Health: Towards an advanced social network about disabilities of neurological origin. J. Biomed. Inform. 2013, 46, 1006–1029. [Google Scholar] [CrossRef] [PubMed]
- Guarino, N. Formal ontology and information systems. In Proceedings of the FOIS’98 Conference, Trento, Italy, 6–8 June 1998; Volume 98, pp. 81–97. [Google Scholar]
- Calvo, M.; Subirats, L.; Ceccaroni, L.; Maroto, J.M.; de Pablo, C.; Miralles, F. Automatic assessment of socioeconomic impact in cardiac rehabilitation. Int. J. Environ. Res. Public Health 2013, 10, 5266–5283. [Google Scholar] [CrossRef] [PubMed]
- Sarntivijai, S.; Vasant, D.; Jupp, S.; Saunders, G.; Bento, A.P.; Gonzalez, D.; Betts, J.; Hasan, S.; Koscielny, G.; Dunham, I.; et al. Linking rare and common disease: Mapping clinical disease-phenotypes to ontologies in therapeutic target validation. J. Biomed. Semant. 2016, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Groza, T.; Köhler, S.; Moldenhauer, D.; Vasilevsky, N.; Baynam, G.; Zemojtel, T.; Schriml, L.M.; Kibbe, W.A.; Schofield, P.N.; Beck, T.; et al. The Human Phenotype Ontology: Semantic Unification of Common and Rare Disease. Am. J. Hum. Genet. 2015, 97, 111–124. [Google Scholar] [CrossRef]
- Kotsilieris, T.; Pavlaki, A.; Christopoulou, S.; Anagnostopoulos, I. The impact of social networks on health care. Soc. Netw. Anal. Min. 2017, 7, 18. [Google Scholar] [CrossRef]
- Subirats, L.; Reguera, N.; Bañón, A.M.; Gómez-Zúñiga, B.; Minguillón, J.; Armayones, M. Mining Facebook Data of People with Rare Diseases: A Content-Based and Temporal Analysis. Int. J. Environ. Res. Public Health 2018, 15, 1877. [Google Scholar] [CrossRef]
- Syed-Abdul, S.; Fernandez-Luque, L.; Jian, W.S.; Li, Y.C.; Crain, S.; Hsu, M.H.; Wang, Y.C.; Khandregzen, D.; Chuluunbaatar, E.; Nguyen, P.A.; et al. Misleading Health-Related Information Promoted Through Video-Based Social Media: Anorexia on YouTube. J. Med. Internet Res. 2013, 15, e30. [Google Scholar] [CrossRef]
- Nguyen, T.; Larsen, M.E.; O’Dea, B.; Nguyen, D.T.; Yearwood, J.; Phung, D.; Venkatesh, S.; Christensen, H. Kernel-based features for predicting population health indices from geocoded social media data. Decis. Support Syst. 2017, 102, 22–31. [Google Scholar] [CrossRef]
- Bateman, D.R.; Brady, E.; Wilkerson, D.; Yi, E.H.; Karanam, Y.; Callahan, C.M. Comparing Crowdsourcing and Friendsourcing: A Social Media-Based Feasibility Study to Support Alzheimer Disease Caregivers. JMIR Res. Protoc. 2017, 6, e56. [Google Scholar] [CrossRef]
- Breslin, J.; Decker, S.; Harth, A.; Bojars, U. SIOC: An approach to connect web-based communities. Int. J. Web Based Communities 2006, 2, 133–142. [Google Scholar] [CrossRef]
- Vandenbussche, P.; Atemezing, G.; Poveda-Villalón, M.; Vatant, B. Linked Open Vocabularies (LOV): A Gateway to Reusable Semantic Vocabularies on the Web; IOS Press: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Bizer, C.; Heath, T.; Berners-Lee, T. Linked data-the story so far. In Semantic Services, Interoperability and Web Applications: Emerging Concepts; IGI Global: Hershey, PA, USA, 2011; pp. 205–227. [Google Scholar]
- Hoehndorf, R.; Dumontier, M.; Gkoutos, G.V. Evaluation of research in biomedical ontologies. Brief Bioinform. 2012, 14, 696–712. [Google Scholar] [CrossRef]
- World Health Organization. The World Health Report 2000: Health Systems: Improving Performance; Technical Report; WHO: Geneva, Switzerland, 2000. [Google Scholar]
- Uschold, M.; Gruninger, M. Ontologies: Principles, methods and applications. Knowl. Eng. Rev. 1996, 2, 93–136. [Google Scholar] [CrossRef]
- Courtot, M.; Gibson, F.; Lister, A.; Malone, J.; Schöber, D.; Brinkman, R.; Ruttenberg, A. MIREOT: The minimum information to reference an external ontology term. Appl Ontol. 2011, 6, 23–33. [Google Scholar] [CrossRef]
- Poveda-Villalón, M.; Suárez-Figueroa, M.C.; Gómez-Pérez, A. Validating ontologies with oops! In Knowledge Engineering and Knowledge Management; Springer: Berlin, Germany, 2012; pp. 267–281. [Google Scholar]
- Aymé, S.; Bellet, B.; Rath, A. Rare diseases in ICD11: Making rare diseases visible in health information systems through appropriate coding. Orphanet J. Rare Dis. 2015, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Balahur, A.; Turchi, M. Comparative experiments using supervised learning and machine translation for multilingual sentiment analysis. Comput. Speech Lang. 2014, 28, 56–75. [Google Scholar] [CrossRef]
- De Vries, E.; Schoonvelde, M.; Schumacher, G. No Longer Lost in Translation: Evidence that Google Translate Works for Comparative Bag-of-Words Text Applications. Political Anal. 2018, 26, 417–430. [Google Scholar] [CrossRef]

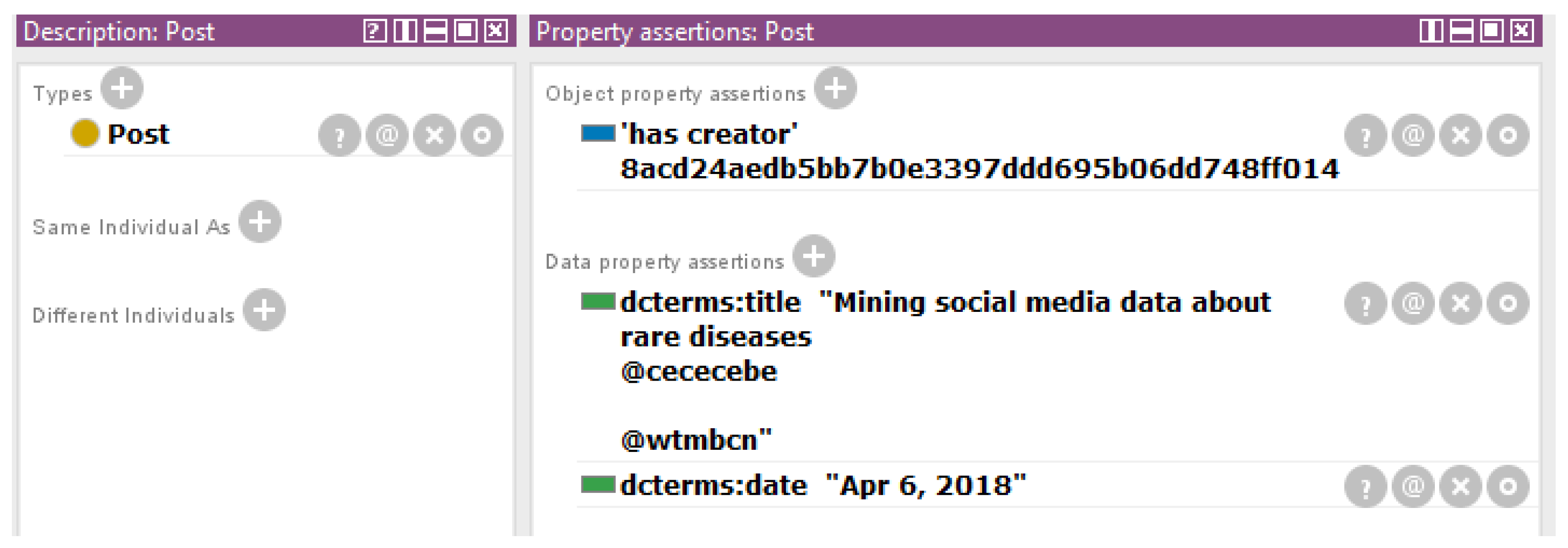
| Attribute | Description |
|---|---|
| Time | UTC time when a Tweet was created |
| Represents the geographic location of a Tweet as reported by the user or client application. The inner coordinates array is formatted as geoJSON (longitude first, then latitude) | |
| Language used in the Tweet | |
| SHA1 (Secure Hash Algorithm 1) of | If the represented Tweet is a reply, this field contains the string representation of the original Tweet’s author ID. This will not necessarily always be the user directly mentioned in the Tweet. |
| SHA1 of | If the represented Tweet is a reply, this field contains the screen name of the original Tweet’s author. |
| If the represented Tweet is a reply, this field contains the string representation of the original Tweet’s ID. | |
| SHA1 of | Identifier of the user who authored the Tweet |
| Count of the followers of the user | |
| Count of the friends of the user | |
| Location of the user | |
| Hashtags, indices and other information of the user |
| Category of Attribute | Attributes |
|---|---|
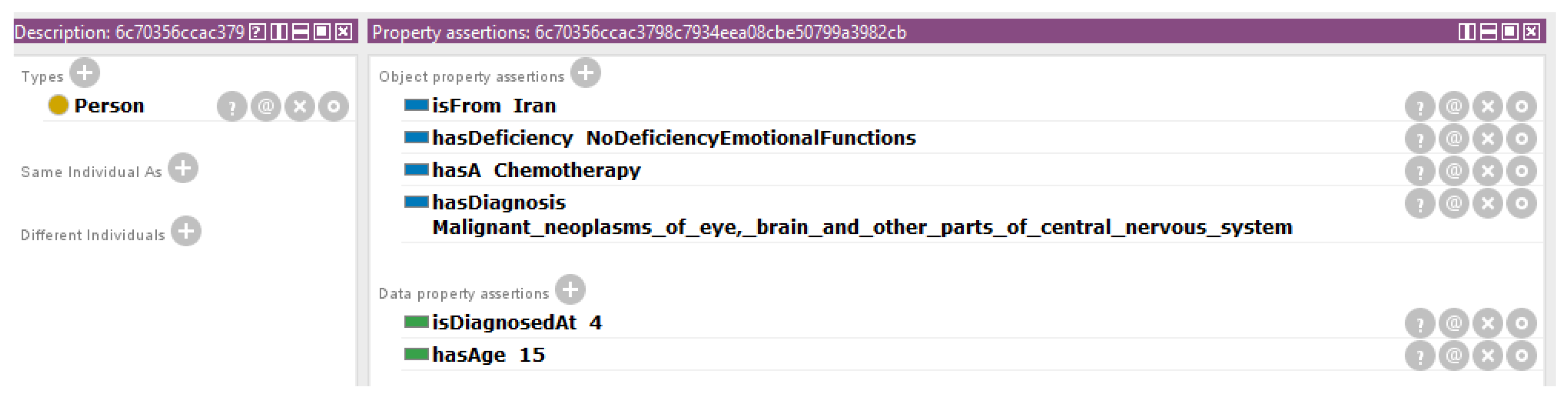
| Demographic and clinical information | Name, age, country, disease, age of diagnosis and treatment. |
| Body functions | Emotional functions, consciousness, vomiting, respiratory functions, skin functions, hearing and vestibular functions, cognitive functions, and pain in head and neck. |
| Activities and participation | Interests, remunerative employment, non-remunerative employment, higher education, sports, arts and culture, and walking. |
| Environmental factors (facilitators and barriers) | Technological facilitators for communication, barrier regarding health professionals, barrier in financial assets, and barrier in health systems. |
| Attribute [min, max] Mean (std) | Polarity | Subjectivity |
|---|---|---|
| Age [1, 45] 23 (11.2) | −0.15 | −0.02 |
| Spain [0, 1] 0.7 (0.5) | 0.13 | −0.01 |
| Iran [0, 1] 0.1 (0.3) | −0.12 | −0.23 |
| Age of Diagnosis [0, 31] 9.3 (8.2) | −0.31 | 0.40 |
| Emotional Functions [0, 4] 0.7 (1.0) | −0.47 | −0.07 |
| Remunerative employment [0, 4] 0.7 (0.7) | −0.30 | 0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subirats, L.; Conesa, J.; Armayones, M. Biomedical Holistic Ontology for People with Rare Diseases. Int. J. Environ. Res. Public Health 2020, 17, 6038. https://doi.org/10.3390/ijerph17176038
Subirats L, Conesa J, Armayones M. Biomedical Holistic Ontology for People with Rare Diseases. International Journal of Environmental Research and Public Health. 2020; 17(17):6038. https://doi.org/10.3390/ijerph17176038
Chicago/Turabian StyleSubirats, Laia, Jordi Conesa, and Manuel Armayones. 2020. "Biomedical Holistic Ontology for People with Rare Diseases" International Journal of Environmental Research and Public Health 17, no. 17: 6038. https://doi.org/10.3390/ijerph17176038
APA StyleSubirats, L., Conesa, J., & Armayones, M. (2020). Biomedical Holistic Ontology for People with Rare Diseases. International Journal of Environmental Research and Public Health, 17(17), 6038. https://doi.org/10.3390/ijerph17176038







