Time Length of Negativization and Cycle Threshold Values in 182 Healthcare Workers with Covid-19 in Milan, Italy: An Observational Cohort Study
Abstract
1. Introduction
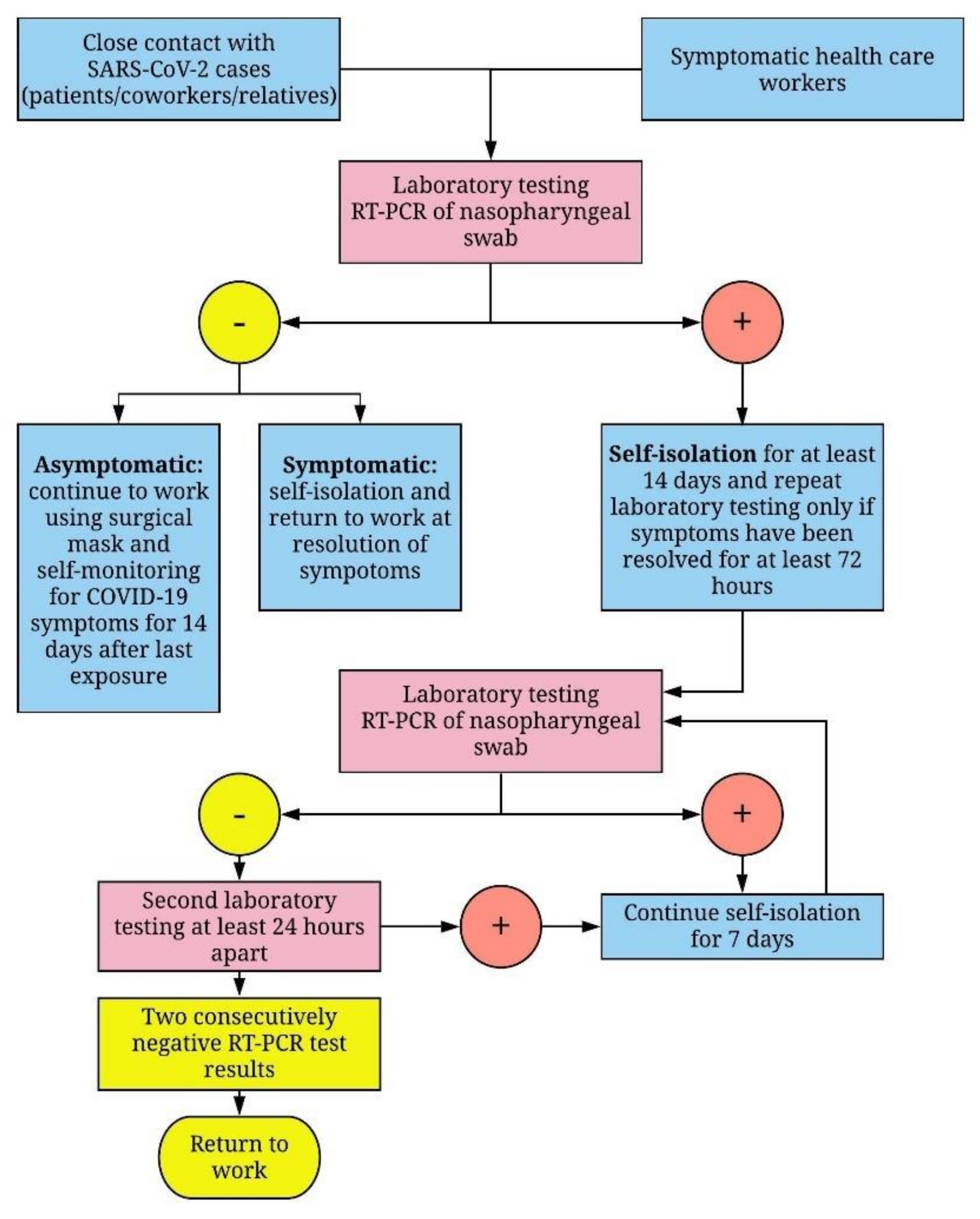
2. Materials and Methods
2.1. Study Population
2.2. RT-PCR Detection
2.3. Statistical Analysis
3. Results
3.1. General Description
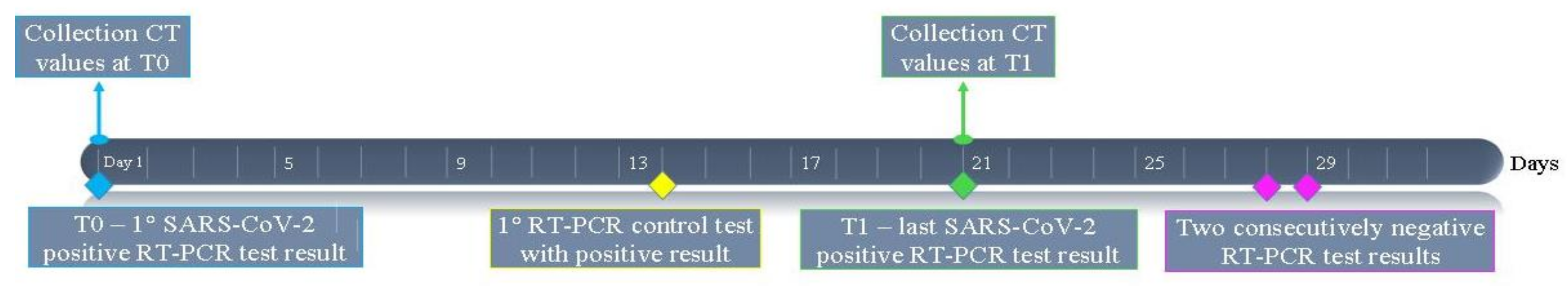
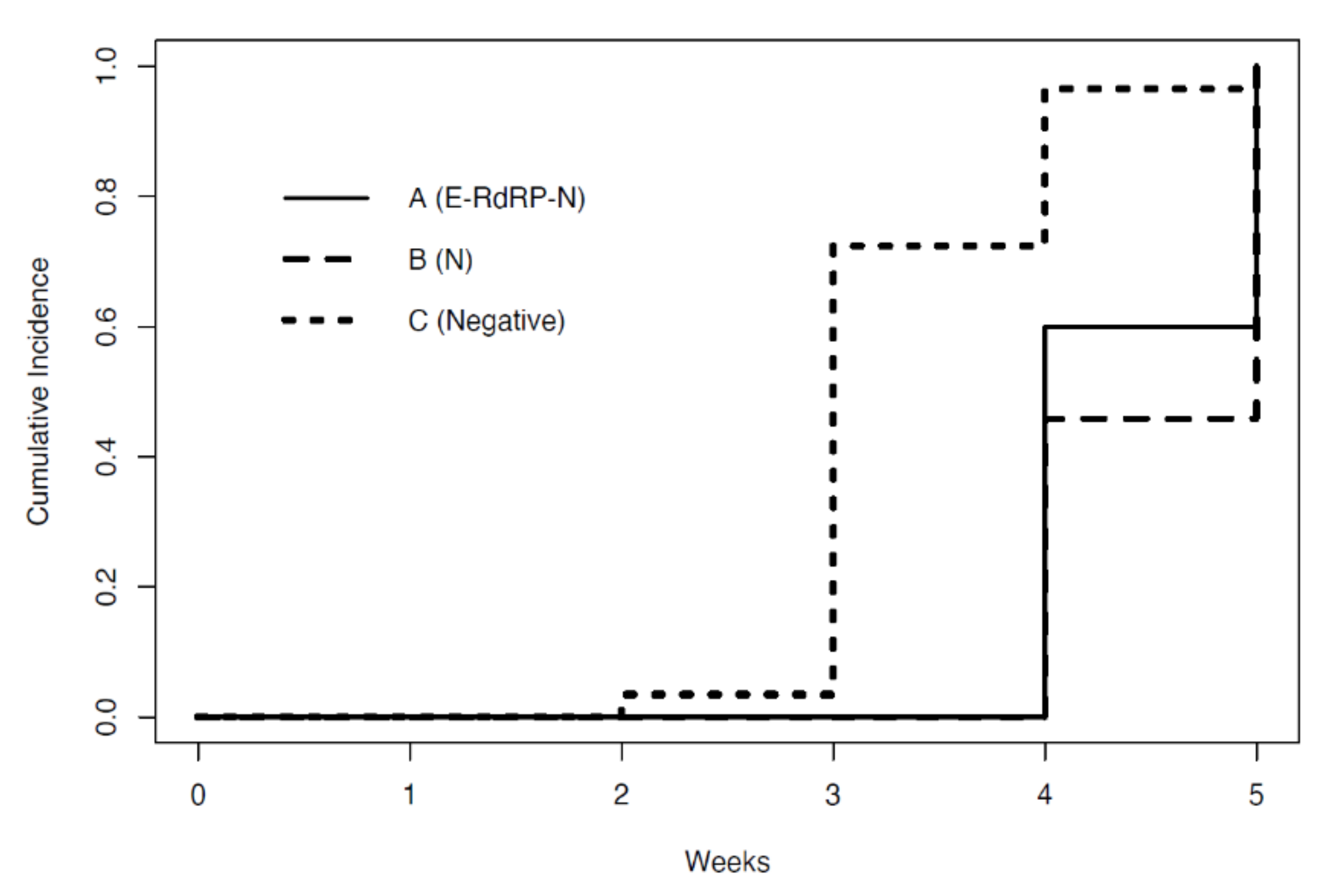
3.2. Time at Which Subjects Became Test-Negative
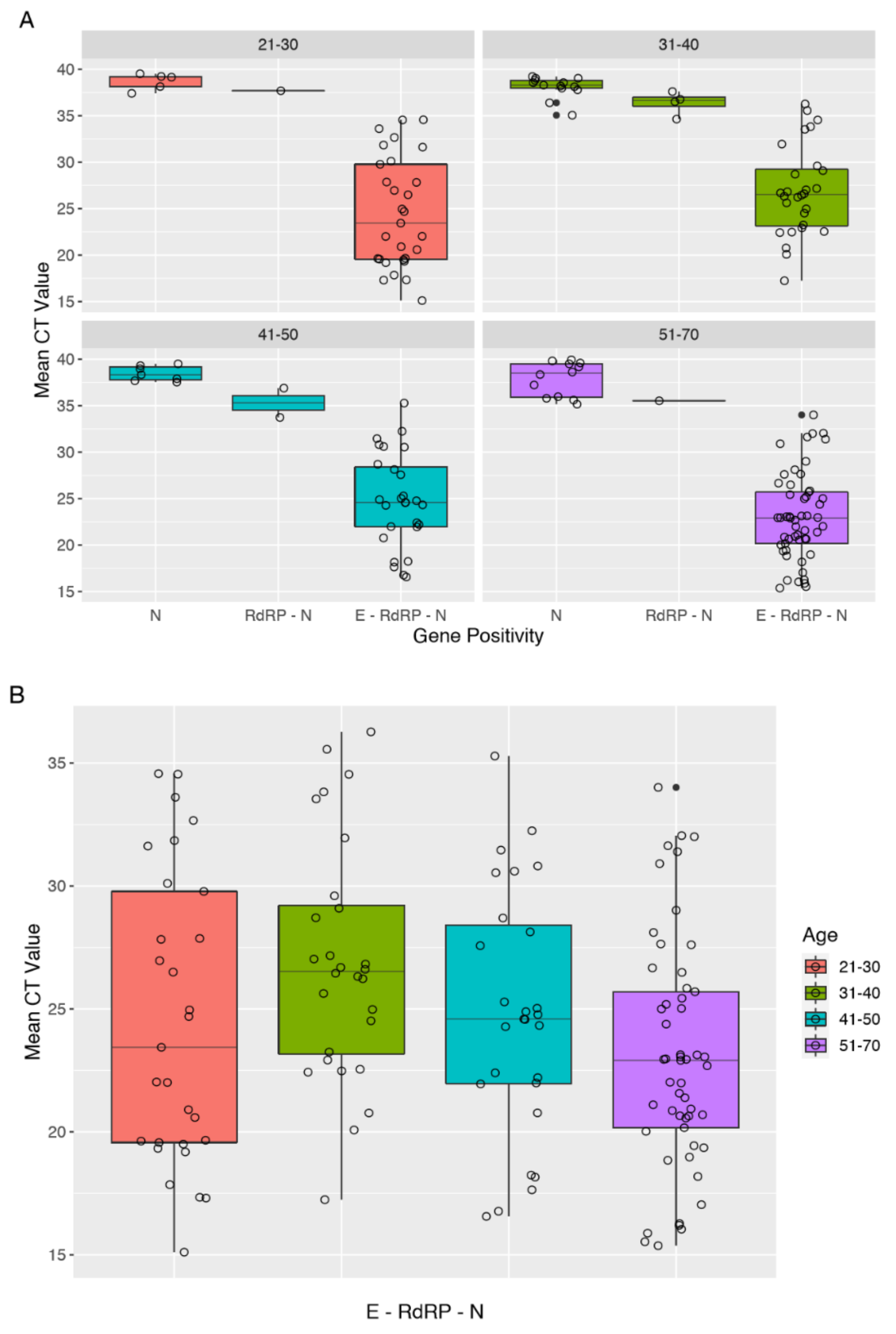
3.3. Analysis of CT Values of RT-PCR Tests
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef]
- WHO. Coronavirus Disease 2019 (COVID-19). Situation Report-108. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 8 May 2020).
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Loeffelholz, M.J.; Tang, Y.W. Laboratory diagnosis of emerging human coronavirus infections—The state of the art. Emerg. Microbes Infect. 2020, 9, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Rothan, H.A.; Byrareddy, S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020, 109, 102433. [Google Scholar] [CrossRef] [PubMed]
- Istituto Superiore di Sanità. Sorveglianza Integrata COVID-19 in Italia—6 May 2020. Available online: https://www.epicentro.iss.it/coronavirus/bollettino/Infografica_6maggio%20ITA.pdf (accessed on 8 May 2020).
- Mizumoto, K.; Kagaya, K.; Zarebski, A.; Chowell, G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020, 25, 2000180. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Jiang, F.; Su, W.; Chen, C.; Chen, J.; Mei, W.; Zhan, L.Y.; Jia, Y.; Zhang, L.; Liu, D.; et al. Clinical characteristics and outcomes of patients undergoing surgeries during the incubation period of COVID-19 infection. EclinicalMedicine 2020. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Nishiura, H.; Kobayashi, T.; Miyama, T.; Suzuki, A.; Jung, S.M.; Hayashi, K.; Kinoshita, R.; Yang, Y.; Yuan, B.; Akhmetzhanov, A.R.; et al. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19). Int. J. Infect. Dis. 2020, 94, 154–155. [Google Scholar] [CrossRef]
- Rovetta, A.; Bhagavathula, A.S. Modelling the epidemiological trend and behaviour of COVID-19 in Italy. Medrxiv 2020. [Google Scholar] [CrossRef]
- Anelli, F.; Leoni, G.; Monaco, R.; Nume, C.; Rossi, R.C.; Marinoni, G.; Spata, G.; De Giorgi, D.; Peccarisi, L.; Miani, A.; et al. Italian doctors call for protecting healthcare workers and boosting community surveillance during covid-19 outbreak. BMJ 2020, 368, 1254. [Google Scholar] [CrossRef]
- Liu, R.; Han, H.; Liu, F.; Lv, Z.; Wu, K.; Liu, Y.; Feng, Y.; Zhu, C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from January to February 2020. Clin. Chim. Acta 2020, 505, 172–175. [Google Scholar] [CrossRef] [PubMed]
- WHO. Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases. Available online: https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 (accessed on 21 April 2020).
- Chang, C.K.; Hou, M.H.; Chang, C.F.; Hsiao, C.D.; Huang, T.H. The SARS coronavirus nucleocapsid protein forms and functions. Antiviral. Res. 2014, 103, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Schoeman, D.; Fielding, B.C. Corona virus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Song, W.; Huang, H.; Sun, Q. Pharmacological Therapeutics Targeting RNA-Dependent RNA Polymerase, Proteinase and Spike Protein: From Mechanistic Studies to Clinical Trials for COVID-19. J. Clin. Med. 2020, 9, 1131. [Google Scholar] [CrossRef] [PubMed]
- Mönkemüller, K.; Fry, L.; Rickes, S. Covid-19, Coronavirus, SARS-CoV-2 and the small bowel. Rev. Esp. Enferm. Dig. 2020. [Google Scholar] [CrossRef] [PubMed]
- Farfour, E.; Lesprit, P.; Visseaux, B.; Pascreau, T.; Jolly, E.; Houhou, N.; Mazaux, L.; Asso-Bonnet, M.; Vasse, M.; SARS-CoV-2 Foch Hospital study group. The Allplex 2019-nCoV (Seegene) assay: Which performances are for SARS-CoV-2 infection diagnosis? Eur. J. Clin. Microbiol. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Ferioli, M.; Cisternino, C.; Leo, V.; Pisani, L.; Palange, P.; Nava, S. Protecting Healthcare Workers From SARS-CoV-2 Infection: Practical Indications. Eur. Respir. Rev. 2020, 29, 200068. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients With COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 95, 1054–1062. [Google Scholar] [CrossRef]
- He, X.; Lau, E.H.Y.; Wu, P.; Deng, X.; Wang, J.; Hao, X.; Lau, Y.C.; Wong, J.Y.; Guan, Y.; Tan, X.; et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020. [Google Scholar] [CrossRef]
- Xiao, A.T.; Tong, Y.X.; Zhang, S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Dramé, M.; Teguo, M.T.; Proye, E.; Hequet, F.; Hentzien, M.; Kanagaratnam, L.; Godaert, L. Should RT-PCR be considered a gold standard in the diagnosis of Covid-19? J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
| Positive HCW | Healed HCW at 30 April 2020 | |
|---|---|---|
| n | 182 | 58 |
| Age, year | 43.5 (12.9) | 42.2 (13.8) |
| Sex | ||
| Male | 77 (42.3%) | 31 (53.4%) |
| Female | 105 (57.7%) | 27 (46.6%) |
| Professional role | ||
| Physicians | 71 (39%) | 25 (43%) |
| Nurses | 48 (26.5%) | 13 (22%) |
| Non-medical workers | 33 (18%) | 9 (16%) |
| Health technicians | 22 (12%) | 8 (14%) |
| Sanitary Operator Partners | 8 (4.5%) | 3 (5%) |
| Group | n | Mean CT T0 | Mean CT T1 |
|---|---|---|---|
| A: Positive (E–RdRP–N) | 5 | 22.60 (8.29) | 33.60 (2.18) |
| B: Single-gene positive (N) | 24 | 22.06 (4.40) | 38.03 (1.43) |
| C: Negative | 29 | 25.08 (6.02) | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cariani, L.; Orena, B.S.; Ambrogi, F.; Gambazza, S.; Maraschini, A.; Dodaro, A.; Oggioni, M.; Orlandi, A.; Pirrone, A.; Uceda Renteria, S.; et al. Time Length of Negativization and Cycle Threshold Values in 182 Healthcare Workers with Covid-19 in Milan, Italy: An Observational Cohort Study. Int. J. Environ. Res. Public Health 2020, 17, 5313. https://doi.org/10.3390/ijerph17155313
Cariani L, Orena BS, Ambrogi F, Gambazza S, Maraschini A, Dodaro A, Oggioni M, Orlandi A, Pirrone A, Uceda Renteria S, et al. Time Length of Negativization and Cycle Threshold Values in 182 Healthcare Workers with Covid-19 in Milan, Italy: An Observational Cohort Study. International Journal of Environmental Research and Public Health. 2020; 17(15):5313. https://doi.org/10.3390/ijerph17155313
Chicago/Turabian StyleCariani, Lisa, Beatrice Silvia Orena, Federico Ambrogi, Simone Gambazza, Anna Maraschini, Antonella Dodaro, Massimo Oggioni, Annarosa Orlandi, Alessia Pirrone, Sara Uceda Renteria, and et al. 2020. "Time Length of Negativization and Cycle Threshold Values in 182 Healthcare Workers with Covid-19 in Milan, Italy: An Observational Cohort Study" International Journal of Environmental Research and Public Health 17, no. 15: 5313. https://doi.org/10.3390/ijerph17155313
APA StyleCariani, L., Orena, B. S., Ambrogi, F., Gambazza, S., Maraschini, A., Dodaro, A., Oggioni, M., Orlandi, A., Pirrone, A., Uceda Renteria, S., Bernazzani, M., Cantù, A. P., Ceriotti, F., & Lunghi, G. (2020). Time Length of Negativization and Cycle Threshold Values in 182 Healthcare Workers with Covid-19 in Milan, Italy: An Observational Cohort Study. International Journal of Environmental Research and Public Health, 17(15), 5313. https://doi.org/10.3390/ijerph17155313





