Association of Genetic and Environmental Factors with Non-Alcoholic Fatty Liver Disease in a Chinese Han Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Demographic Information and Epidemiological Investigation
2.3. Isolation of Genomic DNA
2.4. SNP Selection and Genotyping
2.5. Statistical Analysis
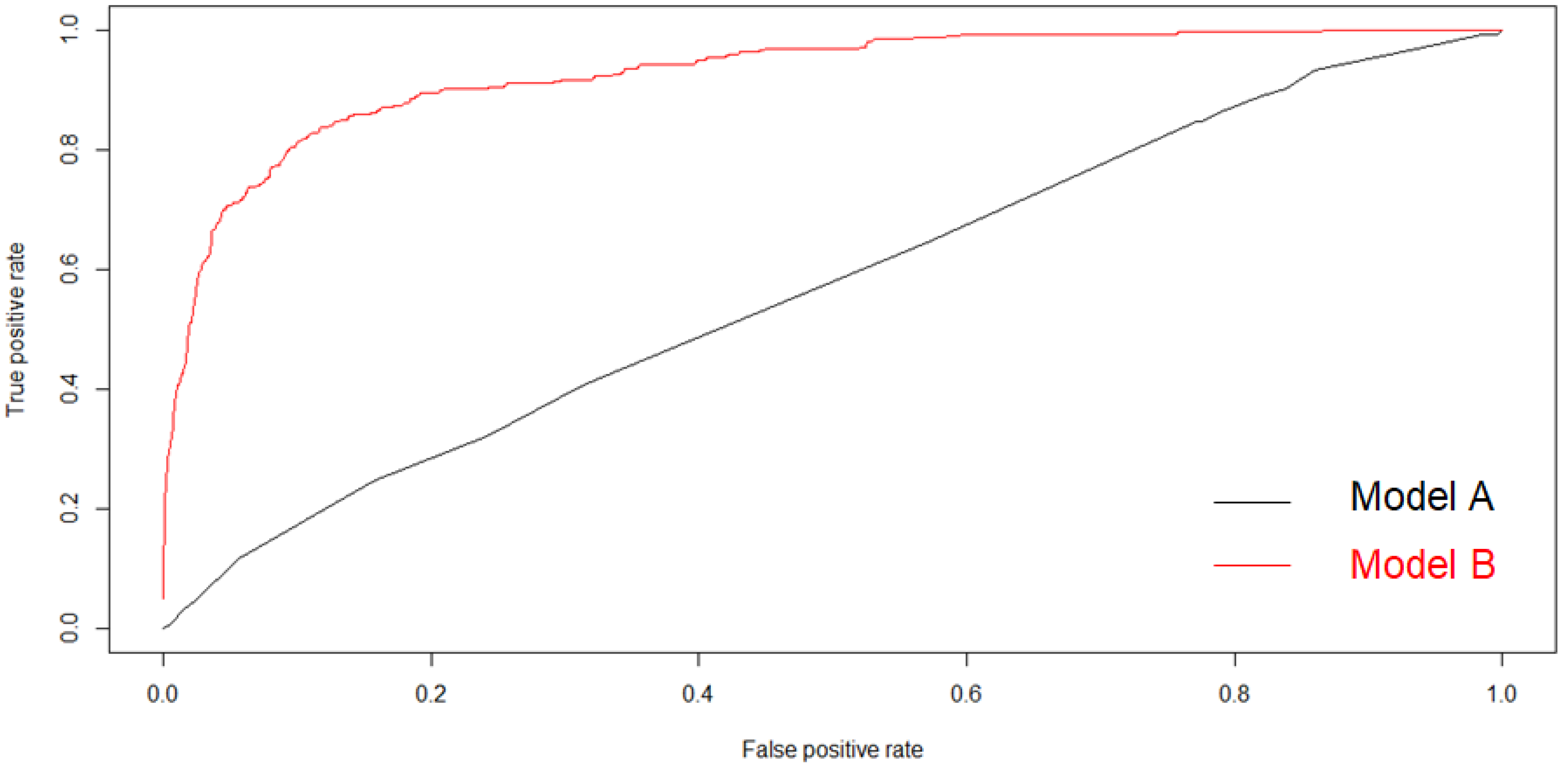
3. Results
3.1. General Characteristics
3.2. Gene-Based Model: SNPs Associated with NAFLD
3.3. All Covariance-Based Model
3.4. Interactions between Gene Polymorphism and Other Covariance Estimators for the Risk of NAFLD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mary, E.; Rinella, M.D. Nonalcoholic fatty liver disease: A systematic review. JAMA 2015, 313, 2263–2273. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence andoutcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.B.; McCullough, A.J. Natural history of nonalcoholic fatty liver disease. Dig. Dis. Sci. 2016, 61, 1226–1233. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Blissett, D.; Blissett, R.; Henry, L.; Stepanova, M.; Younossi, Y.; Racila, A.; Hunt, S.; Beckerman, R. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 2016, 64, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Marchisello, S.; Di Pino, A.; Scicali, R.; Urbano, F.; Piro, S.; Purrello, F.; Rabuazzo, A.M. Pathophysiological, molecular and therapeutic issues of nonalcoholic fatty liver disease: An overview. Int. J. Mol. Sci. 2019, 20, 1948. [Google Scholar] [CrossRef]
- Adams, L.A.; Lindor, D.L. Nonalcoholic fatty liver disease. Ann. Epidemiol. 2007, 17, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xue, J.; Chen, P.; Chen, L.; Yan, S.; Liu, L. Prevalence of nonalcoholic fatty liver disease in mainland of China: A meta-analysis of published studies. J. Gastroenterol. Hepatol. 2014, 29, 42–51. [Google Scholar] [CrossRef]
- Kasturiratne, A.; Weerasinghe, S.; Dassanayake, A.S.; Rajindrajith, S.; de Silva, A.P.; Kato, N.; Wickremasinghe, A.R.; de Silva, H.J. Influence of non-alcoholic fatty liver disease on the development of diabetes mellitus. J. Gastroenterol. Hepatol. 2013, 28, 142–147. [Google Scholar] [CrossRef]
- Yamazaki, H.; Tsuboya, T.; Tsuji, K.; Dohke, M.; Maguchi, H. Independent association between improvement of nonalcoholic fatty liver disease and reduced incidence of type 2 diabetes mellitus. Diabetes Care 2015, 38, 1673–1679. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver (Easl). EASL-EASD-EASO clinical practice guidelines for the management of non-alcoholic fatty liver disease. Obes. Facts 2016, 65–90. [Google Scholar] [CrossRef]
- Abenavoli, L.; Milic, N.; Peta, V.; Alfieri, F.; De Lorenzo, A.; Bellentani, S. Alimentary regimen in nonalcoholic fatty liver disease: Mediterranean diet. World J. Gastroenterol. 2014, 20, 16831–16840. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; George, J. Genetic and epigenetic mechanisms of NASH. Hepatol. Int. 2016, 10, 394–406. [Google Scholar] [CrossRef]
- Dongiovanni, P.; Anstee, Q.M.; Valenti, L. Genetic predisposition in NAFLD and NASH: Impact on severity of liver disease and response to treatment. Curr. Pharm. Des. 2013, 19, 5219–5238. [Google Scholar] [CrossRef] [PubMed]
- Macaluso, F.S.; Maida, M.; Petta, S. Genetic background in nonalcoholic fatty liver disease: A comprehensive review. World J. Gastroenterol. 2015, 21, 11088–11111. [Google Scholar] [CrossRef]
- Devarajan, A.; Bourquard, N.; Hama, S.; Navab, M.; Grijalva, R.; Morvardi, S.; Clarke, C.F.; Vergnes, L.; Reue, K.; Teiber, J.F.; et al. Paraoxonase 2 deficiency alters mitochondrial function and exacerbates the development of atherosclerosis. Antioxid. Redox Signal 2011, 14, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Horke, S.; Witte, I.; Wilgenbus, P.; Kruger, M.; Strand, D.; Forstermann, U. Paraoxonase-2 reduces oxidative stress in vascular cells and decreases endoplasmic reticulum stress-induced caspase activation. Circulation 2007, 115, 2055–2064. [Google Scholar] [CrossRef]
- Stergiakouli, E.; Gaillard, R.; Tavaré, J.M.; Balthasar, N.; Loos, R.J.; Taal, H.R.; Evans, D.M.; Rivadeneira, F.; St Pourcain, B.; Uitterlinden, A.G.; et al. Genome-wide association study of height-adjusted BMI in childhood identifies functional variant in ADCY3. Obesity (Silver Spring) 2014, 22, 2252–2259. [Google Scholar] [CrossRef] [PubMed]
- Siljee, J.E.; Wang, Y.; Bernard, A.A.; Ersoy, B.A.; Zhang, S.M.; Marley, A.; Von Zastrow, M.; Reiter, J.F.; Vaisse, C. Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway. Nat. Genet. 2018, 50, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Grarup, N.; Moltke, I.; Andersen, M.K.; Dalby, M.; Vitting-Seerup, K.; Kern, T.; Mahendran, Y.; Jørsboe, E.; Larsen, C.V.L.; Dahl-Petersen, I.K.; et al. Loss-of-function variants in ADCY3 increase risk of obesity and type 2 diabetes. Nat. Genet. 2018, 50, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Lai, Q.; Giralt, A.; Le May, C.; Zhang, L.; Cariou, B.; Denechaud, P.D.; Fajas, L. E2F1 inhibits circulating cholesterol clearance by regulating Pcsk9 expression in the liver. JCI Insight. 2017, 18, 2. [Google Scholar] [CrossRef]
- Theocharidou, E.; Papademetriou, M.; Reklou, A.; Sachinidis, A.; Boutari, C.; Giouleme, O. The role of PCSK9 in the pathogenesis of non-alcoholic fatty liver disease and the effect ofPCSK9 inhibitors. Curr. Pharm. Des. 2018, 24, 3654–3657. [Google Scholar] [CrossRef] [PubMed]
- Ruscica, M.; Ferri, N.; Macchi, C.; Meroni, M.; Lanti, C.; Ricci, C.; Maggioni, M.; Fracanzani, A.L.; Badiali, S.; Fargion, S.; et al. Liver fat accumulation is associated with circulating PCSK9. Ann. Med. 2016, 48, 384–391. [Google Scholar] [CrossRef]
- Beer, N.L.; Tribble, N.D.; Mcculloch, L.J.; Roos, C.; Johnson, P.R.; Orho-melander, M.; Gloyn, A.L. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum. Mol. Genet. 2009, 18, 4081–4088. [Google Scholar] [CrossRef]
- Santoro, N.; Zhang, C.K.; Zhao, H.; Pakstis, A.J.; Kim, G.; Kursawe, R.; Dykas, D.J.; Bale, A.E.; Giannini, C.; Pierpont, B.; et al. Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology 2011, 55, 781–789. [Google Scholar] [CrossRef]
- Bugianesi, E.; Manzini, P.; D’antico, S.; Vanni, E.; Longo, F.; Leone, N.; Massarenti, P.; Piga, A.; Marchesini, G.; Rizzetto, M. Relative contribution of iron burden, HFE mutations, and insulin resistance to fibrosis in nonalcoholic fatty liver. Hepatology 2004, 39, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Wieckowska, A.; Feldstein, A.E. Diagnosis of nonalcoholic fatty liver disease: Invasive versus noninvasive. Semin. Liver Dis. 2008, 28, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.Y.; Lei, S.; Huang, L.; Wang, Y.N.; Wang, X.N.; Zhou, P.P.; Xu, X.J.; Zhang, L.; Xu, L.W.; Yang, L. Associations of genetic variations in ABCA1 and lifestyle factors with coronary artery disease in a southern Chinese population with dyslipidemia: A nested case-control study. Int. J. Environ. Res. Public Health 2019, 16, 786. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 7–10. [Google Scholar] [CrossRef]
- Deist, T.M.; Dankers, F.J.W.M.; Valdes, G.; Wijsman, R.; Hsu, I.C.; Oberije, C.; Lustberg, T.; van Soest, J.; Hoebers, F.; Jochems, A.; et al. Machine learning algorithms for outcome prediction in (chemo) radiotherapy: An empirical comparison of classifiers. Med. Phys. 2018, 45, 3359–3449. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Hu, C.; Bell, M.L.; Billheimer, D.; Guerra, S.; Roe, D.; Vasquez, M.M.; Bedrick, E.J. Regularized continuous-time Markov Model via elastic net. Biometrics 2018, 74, 1045–1054. [Google Scholar] [CrossRef]
- Knol, M.J.; VanderWeele, T.J. Recommendations for presenting analyses of effect modification and interaction. Int. J. Epidemiol. 2012, 41, 514–520. [Google Scholar] [CrossRef]
- Shih, D.M.; Meng, Y.; Sallam, T.; Vergnes, L.; Shu, M.L.; Reue, K.; Tontonoz, P.; Fogelman, A.M.; Lusis, A.J.; Reddy, S.T. PON2 deficiency leads to increased susceptibility to diet-induced obesity. Antioxidants 2019, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Johnson, C.; Mincey, K.D. Obesity Sepidemiology worldwide. gastroenterol. Clin. N. Am. 2016, 45, 571–579. [Google Scholar] [CrossRef]
- Ng, C.J.; Bourquard, N.; Grijalva, V.; Hama, S.; Shih, D.M.; Navab, M.; Fogelman, A.M.; Lusis, A.J.; Young, S.; Reddy, S.T. Paraoxonase-2 deficiency aggravates atherosclerosis in mice despite lower apolipoprotein-B-containing lipoproteins: Anti-atherogenic role for paraoxonase-2. J. Biol. Chem. 2006, 281, 29491–29500. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.F.; Leroux, M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 2017, 18, 533–547. [Google Scholar] [CrossRef]
- Slimani, A.; Harira, Y.; Trabelsi, I.; Jomaa, W.; Maatouk, F.; Hamda, K.B.; Slimane, M.N. Effect of E670G polymorphism in PCSK9 gene on the risk and severity of coronary heart disease and ischemic stroke in a Tunisian cohort. J. Mol. Neurosci. 2014, 53, 150–157. [Google Scholar] [CrossRef]
- Wang, L.; Smith, J.; Breton, C.; Clark, P.; Zhang, J.; Ying, L.; Che, Y.; Lape, J.; Bell, P.; Calcedo, R.; et al. Meganuclease targeting of PCSK9 in macaque liver leads to stable reduction in serum cholesterol. Nat Biotechnol. 2018, 36, 717–725. [Google Scholar] [CrossRef]
- Makkonen, J.; Pietilainen, K.H.; Rissanen, A.; Kaprio, J.; Yki-Jarvinen, H. Genetic factors contribute to variation in serum alanine aminotransferase activity independent of obesity and alcohol: A study in monozygotic and dizygotic twins. J. Hepatol. 2009, 50, 1035–1042. [Google Scholar] [CrossRef]
- Petit, J.M.; Masson, D.; Guiu, B.; Rollot, F.; Duvillard, L.; Bouillet, B.; Brindisi, M.-C.; Buffier, P.; Hillon, P.; Cercueil, J.-P.; et al. GCKR polymorphism influences liver fat content in patients with type 2 diabetes. Acta Diabetol. 2016, 53, 237–242. [Google Scholar] [CrossRef]
- Park, S.H.; Jeon, W.K.; Kim, S.H.; Park, D.I.; Cho, Y.K.; Sung, I.K.; Sohn, C.I.; Keum, D.K.; Kim, B.I. Prevalence and risk factors of non-alcoholic fatty liver disease among Korean adults. J. Gastroenterol. Hepatol. 2006, 21, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kutateladze, T.G. Diet and the epigenome. Nat. Commun. 2018, 9, 3375. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Valenti, L.; Romeo, S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J. Hepatol. 2018, 68, 268–279. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | NAFLD (+) | NAFLD (−) | t/z/χ2 | p |
|---|---|---|---|---|
| Total | 327 | 1836 | ||
| Age, Median (IQR), year | 64 (14.50) | 62 (17.00) | 0.13 | 0.717 |
| Sex, n (%) | 3.02 | 0.082 | ||
| Male | 49 (49.00) | 917 (45.50) | ||
| Female | 51 (51.00) | 1096 (54.5) | ||
| Weight, Median (IQR), year | 63.2 (5.05) | 59.4 (12.82) | 158.93 | <0.001 |
| BMI, mean ± SD, kg/m2 | 24.06 (4.55) | 23.15 (3.84) | 194.48 | <0.001 |
| Waistline, Median (IQR), cm | 82.87 (11.00) | 126.40 | <0.001 | |
| SBP, Median (IQR), mmHg | 145 (16.50) | 136 (30.00) | 16.62 | <0.001 |
| DBP, Median (IQR), mmHg | 90 (15.00) | 80 (16.00) | 17.27 | <0.001 |
| TC, Median (IQR), mmol/L | 4.99 (1.39) | 4.85 (1.27) | 28.68 | <0.001 |
| TG, Median (IQR), mmol/L | 1.50 (0.65) | 1.28 (0.87) | 101.84 | <0.001 |
| HDL-C, Median (IQR), mmol/L | 1.24 (0.32) | 1.26 (0.38) | 33.34 | <0.001 |
| LDL-C, Median (IQR), mmol/L | 3.18 (1.37) | 2.98 (1.11) | 5.11 | 0.0238 |
| The history of diabetes | 0.0082 | |||
| + | 23 (7.00) | 70 (3.80) | 7.00 | |
| − | 304 (93.00) | 1766 (96.20) | ||
| The intake of vegetables, n (%) | 16.63 | 0.0023 | ||
| <45 g/day | 107 (32.70) | 1499 (81.60) | ||
| ≥45 g/day | 220 (67.30) | 337 (18.40) | ||
| The intake of egg, n (%) | 18.46 | 0.0004 | ||
| <4 eggs/week | 86 (26.30) | 1560 (85.00) | ||
| ≥4 eggs/week | 241 (73.70) | 276 (15.00) | ||
| The intake of fried, n (%) | 34.82 | <0.001 | ||
| Never | 175 (53.50) | 679 (37.00) | ||
| Regularly | 152 (46.50) | 1157 (63.00) | ||
| The intake of sweet, n (%) | 55.58 | <0.001 | ||
| <4 times/week | 177 (54.10) | 1715 (93.40) | ||
| ≥4 times/week | 150 (45.90) | 121 (6.60) |
| SNP | Genotype | Unadjusted OR (95%CI) | Unadjusted p | Adjusted OR (95%CI) | Adjusted p |
|---|---|---|---|---|---|
| rs7493 | |||||
| additive | CG/CC | 1.27 (0.98–1.64) | 0.065 | 1.26 (0.97–1.62) | 0.088 |
| GG/CC | 1.82 (0.10–3.29) | 0.049 | 1.87 (1.01–3.48) | 0.046 | |
| dominant | GG + CG/CC | 1.31 (1.03–1.68) | 0.026 | 1.31 (1.01–1.69) | 0.036 |
| recessive | GG/CG + CC | 1.68 (0.93–3.03) | 0.083 | 1.75 (0.95–3.22) | 0.074 |
| rs7593130 | |||||
| additive | CT/TT | 1.03 (0.79–1.35) | 0.800 | 0.95 (0.72–1.25) | 0.706 |
| CC/TT | 1.57 (1.12–2.19) | 0.009 | 1.46 (1.04–2.07) | 0.031 | |
| dominant | CC + CT/TT | 1.15 (0.90–1.48) | 0.252 | 1.07 (0.82–1.38) | 0.626 |
| recessive | CC/CT + TT | 1.54 (1.14–2.07) | 0.005 | 1.51 (1.11–2.05) | 0.008 |
| rs1260326 | |||||
| additive | CT/CC | 0.31 (0.11–0.84) | 0.133 | 1.31 (0.93–1.84) | 0.123 |
| TT/CC | 1.48 (1.04–2.10) | 0.031 | 1.48 (1.03–2.13) | 0.033 | |
| dominant | TT + CT/CC | 1.35 (0.99–1.86) | 0.057 | 1.37 (0.99–1.90) | 0.055 |
| recessive | TT/CT + CC | 1.23 (0.95–1.58) | 0.112 | 1.22 (0.94–1.58) | 0.133 |
| rs11583680 | |||||
| additive | CT/CC | 0.63 (0.45–0.87) | 0.005 | 0.61(0.424–086) | 0.005 |
| TT/CC | 0.35 (0.08–1.49) | 0.156 | 0.47 (0.11–2.06) | 0.311 | |
| dominant | TT + CT/CC | 0.61 (0.44–0.83) | 0.002 | 0.61 (0.44–0.85) | 0.003 |
| recessive | TT/CT + CC | 0.38 (0.09–1.62) | 0.19 | 0.37 (0.12–2.20) | 0.368 |
| Characters | OR | 95%CI | p |
|---|---|---|---|
| gene score | 1.49 | 1.23–1.81 | <0.001 |
| Dyslipidemia | 2.42 | 1.84–3.19 | <0.001 |
| Sex | 1.24 | 0.98–1.57 | 0.083 |
| The intake of egg | 6.52 | 5.23–8.11 | <0.001 |
| Hypertension | 1.02 | 1.01–1.02 | <0.001 |
| The intake of sweet | 4.36 | 3.51–5.41 | <0.001 |
| The intake of vegetable | 0.29 | 0.24–0.35 | <0.001 |
| SNPs | Adequate Vegetables | Inadequate Vegetables | OR (95%CI) for Hypertension Patients within Strata of Genotype | RERI (95%CI) | p | ||
|---|---|---|---|---|---|---|---|
| Case/Control (n) | OR (95%CI) | Case/Control (n) | OR (95%CI) | ||||
| rs11583680 | |||||||
| Non-risk allele carriers (TT + CT) | 20/348 | 30/72 | |||||
| 1 | 7.25 (3.90–13.48) | 7.25 (3.90–13.48) | |||||
| p < 0.001 | p < 0.001 | ||||||
| Risk allele carriers (CC) | 87/1151 | 190/265 | 4.91 (0.66–9.17) | 0.024 | |||
| 1.32 (0.80–2.17) | 12.99 (8.77–19.22) | 9.49 (7.12–12.64) | |||||
| p = 0.283 | p < 0.001 | p < 0.001 | |||||
| OR (95%CI) for risk allele carriers within strata of vegetable intake | 1.32 (0.80–2.17) | 1.31 (1.04–1.66) | |||||
| p = 0.283 | p = 0.022 | ||||||
| rs7593130 | |||||||
| Non-risk allele carriers (CT + TT) | 85/1272 | 174/296 | |||||
| 1 | 8.80 (6.59–11.74) | 8.80 (6.59–11.74) | |||||
| p < 0.001 | p < 0.001 | ||||||
| Risk allele carriers (CC) | 22/227 | 46/41 | 7.55 (0.13–14.97) | 0.046 | |||
| 1.45 (0.90–2.37) | 16.79 (10.44–26.99) | 11.58 (6.31–21.25) | |||||
| p = 0.137 | p < 0.001 | p < 0.001 | |||||
| OR (95%CI) for risk allele carriers within strata of vegetable intake | 1.45 (0.90–2.37) | 1.38 (1.10–1.74) | |||||
| p = 0.137 | p = 0.006 | ||||||
| rs7493 | |||||||
| Non-risk allele carriers (CC) | 62/1022 | 142/238 | |||||
| 1 | 9.84 (7.07–13.68) | 9.84 (7.07–13.68) | |||||
| p < 0.001 | p < 0.001 | ||||||
| Risk allele carriers (GG + CG) | 45/477 | 78/99 | 2.60 (−1.81–7.00) | 0.248 | |||
| 1.56 (1.04–2.32) | 12.99 (8.77–19.22) | 8.35 (5.46–12.79) | |||||
| p = 0.03 | p < 0.001 | p < 0.001 | |||||
| OR (95%CI) for risk allele carriers within strata of vegetable intake | 1.56 (1.04–2.32) p = 0.03 | 1.15 (0.96–1.38) p = 0.133 | |||||
| Other Lifestyles | Dyslipidemia (−) | Dyslipidemia (+) | OR (95%CI) for Hypertension Patients within Strata of Genotype | RERI (95%CI) | p | ||
|---|---|---|---|---|---|---|---|
| Case/Control (n) | OR (95%CI) | Case/Control (n) | OR (95%CI) | ||||
| Hypertension | |||||||
| Hypertension (−) | 33/459 | 90/423 | |||||
| 1 | 2.96 (1.94–4.51) | 2.96 (1.94–4.51) | |||||
| p < 0.001 | p < 0.001 | ||||||
| Hypertension (+) | 41/302 | 163/652 | −0.37 (−1.56–0.82) | 0.541 | |||
| 1.89 (1.17–3.05) | 3.48 (2.35–5.15) | 1.84 (1.27–2.66) | |||||
| p = 0.01 | p < 0.001 | p = 0.001 | |||||
| OR (95%CI) for risk allele carriers within strata of dyslipidemia | 1.89 (1.17–3.05) | 1.08 (0.94–1.25) | |||||
| p = 0.01 | p = 0.267 | ||||||
| The intake of vegetables | |||||||
| Adequately | 19/613 | 88/886 | |||||
| 1 | 3.20 (1.93–5.32) | 3.20 (1.93–5.32) | |||||
| p < 0.001 | p < 0.001 | ||||||
| Inadequately | 55/148 | 165/189 | 13.97 (4.81–23.12) | 0.002 | |||
| 11.99 (6.91–20.81) | 28.17 (17.05–46.53) | 2.35 (1.62–3.41) | |||||
| p < 0.001 | p < 0.001 | p < 0.001 | |||||
| OR (95%CI) for risk allele carriers within strata of dyslipidemia | 11.99 (6.91–20.81) | 2.96 (2.55–3.45) | |||||
| p < 0.001 | p < 0.001 | ||||||
| The intake of egg | |||||||
| Adequately | 18/658 | 68/902 | |||||
| 1 | 2.76 (1.62–4.68) | 2.76 (1.62–4.68) | |||||
| p < 0.001 | p < 0.001 | ||||||
| Inadequately | 56/103 | 185/173 | 17.47 (4.55–30.39) | 0.008 | |||
| 19.88 (11.24–35.12) | 39.09 (24.43–65.23) | 1.97 (1.34–2.89) | |||||
| p < 0.001 | p < 0.001 | p = 0.001 | |||||
| OR (95%CI) for risk allele carriers within strata of dyslipidemia | 11.99 (6.91–20.81) p < 0.001 | 3.77 (3.20–4.24) p < 0.001 | |||||
| The intake of sweetmeat | |||||||
| Adequately | 10/615 | 41/690 | |||||
| 1 | 2.38 (1.66–3.42) | 2.38 (1.66–3.42) | |||||
| p < 0.001 | p < 0.001 | ||||||
| Inadequately | 10/370 | 32/395 | 12.08 (1.77–22.38) | 0.022 | |||
| 13.12 (7.57–22.74) | 26.57 (17.35–40.70) | 2.03 (1.19–3.46) | |||||
| p < 0.001 | p < 0.001 | p = 0.010 | |||||
| OR (95%CI) for risk allele carriers within strata of dyslipidemia | 1.66 (0.69–4.03) p = 0.261 | 3.34 (2.82–3.96) p < 0.001 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Ye, C.-Y.; Wang, L.; Li, J.-M.; Yang, L. Association of Genetic and Environmental Factors with Non-Alcoholic Fatty Liver Disease in a Chinese Han Population. Int. J. Environ. Res. Public Health 2020, 17, 5217. https://doi.org/10.3390/ijerph17145217
Li Z, Ye C-Y, Wang L, Li J-M, Yang L. Association of Genetic and Environmental Factors with Non-Alcoholic Fatty Liver Disease in a Chinese Han Population. International Journal of Environmental Research and Public Health. 2020; 17(14):5217. https://doi.org/10.3390/ijerph17145217
Chicago/Turabian StyleLi, Zheng, Cheng-Yin Ye, Li Wang, Jin-Mei Li, and Lei Yang. 2020. "Association of Genetic and Environmental Factors with Non-Alcoholic Fatty Liver Disease in a Chinese Han Population" International Journal of Environmental Research and Public Health 17, no. 14: 5217. https://doi.org/10.3390/ijerph17145217
APA StyleLi, Z., Ye, C.-Y., Wang, L., Li, J.-M., & Yang, L. (2020). Association of Genetic and Environmental Factors with Non-Alcoholic Fatty Liver Disease in a Chinese Han Population. International Journal of Environmental Research and Public Health, 17(14), 5217. https://doi.org/10.3390/ijerph17145217






