Sorption Mechanism and Optimization Study for the Bioremediation of Pb(II) and Cd(II) Contamination by Two Novel Isolated Strains Q3 and Q5 of Bacillus sp.
Abstract
1. Introduction
2. Materials and Methods
- Isolation and identification of the new bacterial strains;
- Study of the critical growth conditions for the selected bacterial strains in an environment contaminated by Pb(II);
- Optimization of the operating conditions for the bioremediation process conducted by the selected bacterial strains;
- Study of the biotic and abiotic mechanisms involved in the bioremediation process conducted by the selected bacterial strains under the optimal operating conditions.
2.1. Isolation of Bacterial Strains
2.2. Molecular Identification of Isolated Bacterial Strains
2.3. Preliminary Study on Growth Rate of the Isolated Bacterial Strains
2.4. Optimization of Pb(II) and Cd(II) Bioremediation Process
2.5. FESEM and EDS Analyses
2.6. Data Analysis
3. Results
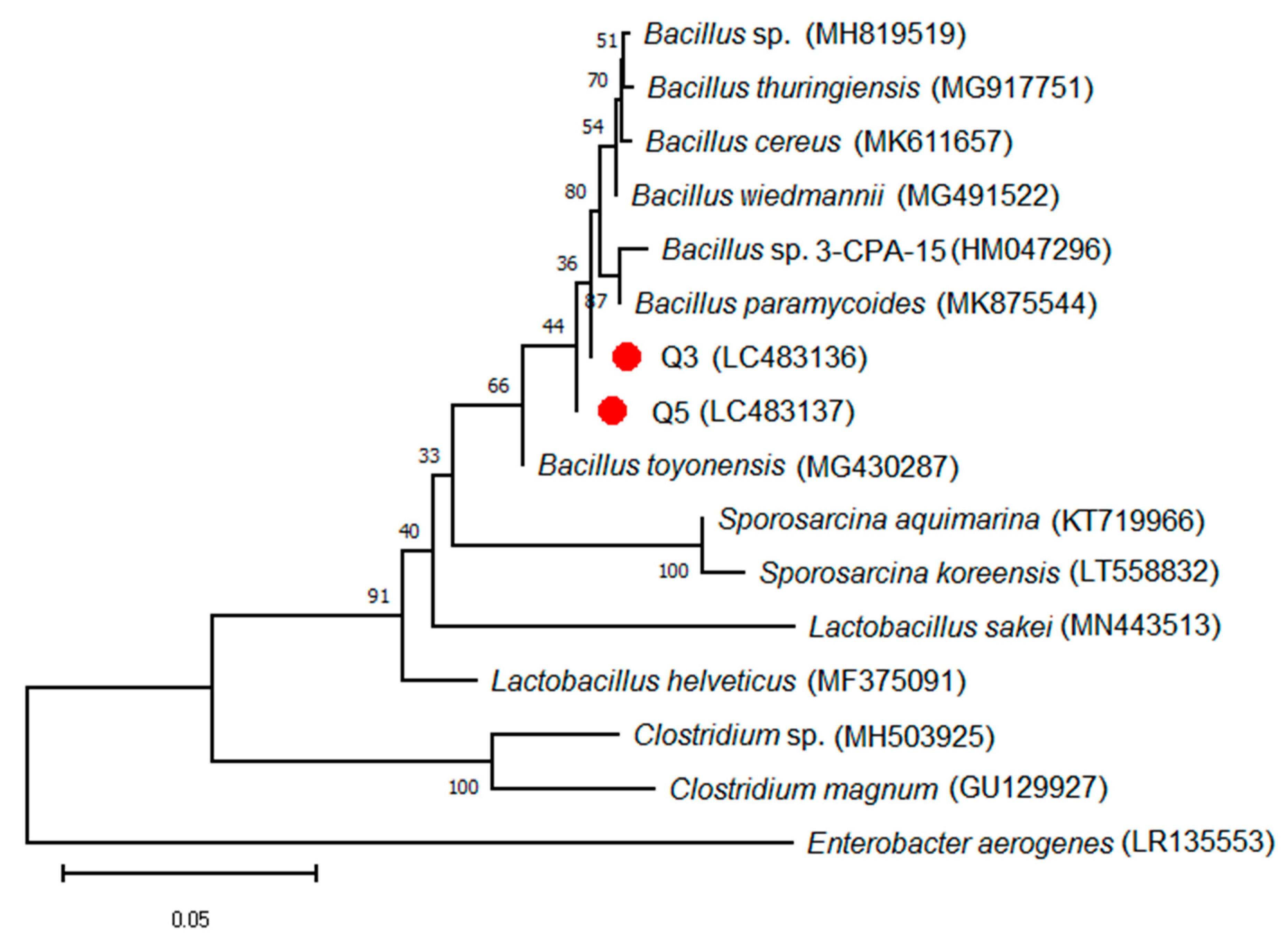
3.1. Evolutionary Analysis of Isolated Bacterial
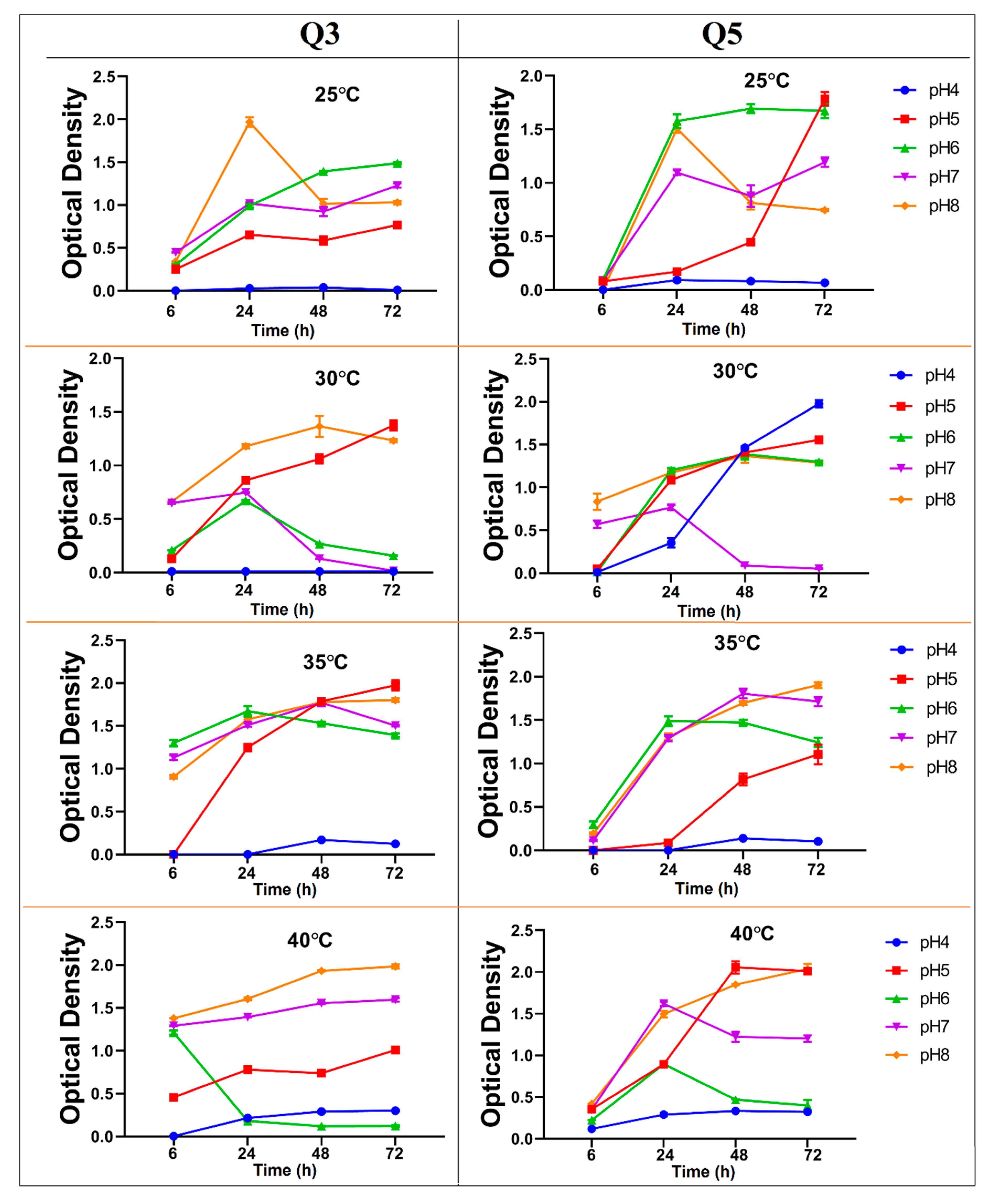
3.2. Growth Rate Profile of Q3 and Q5
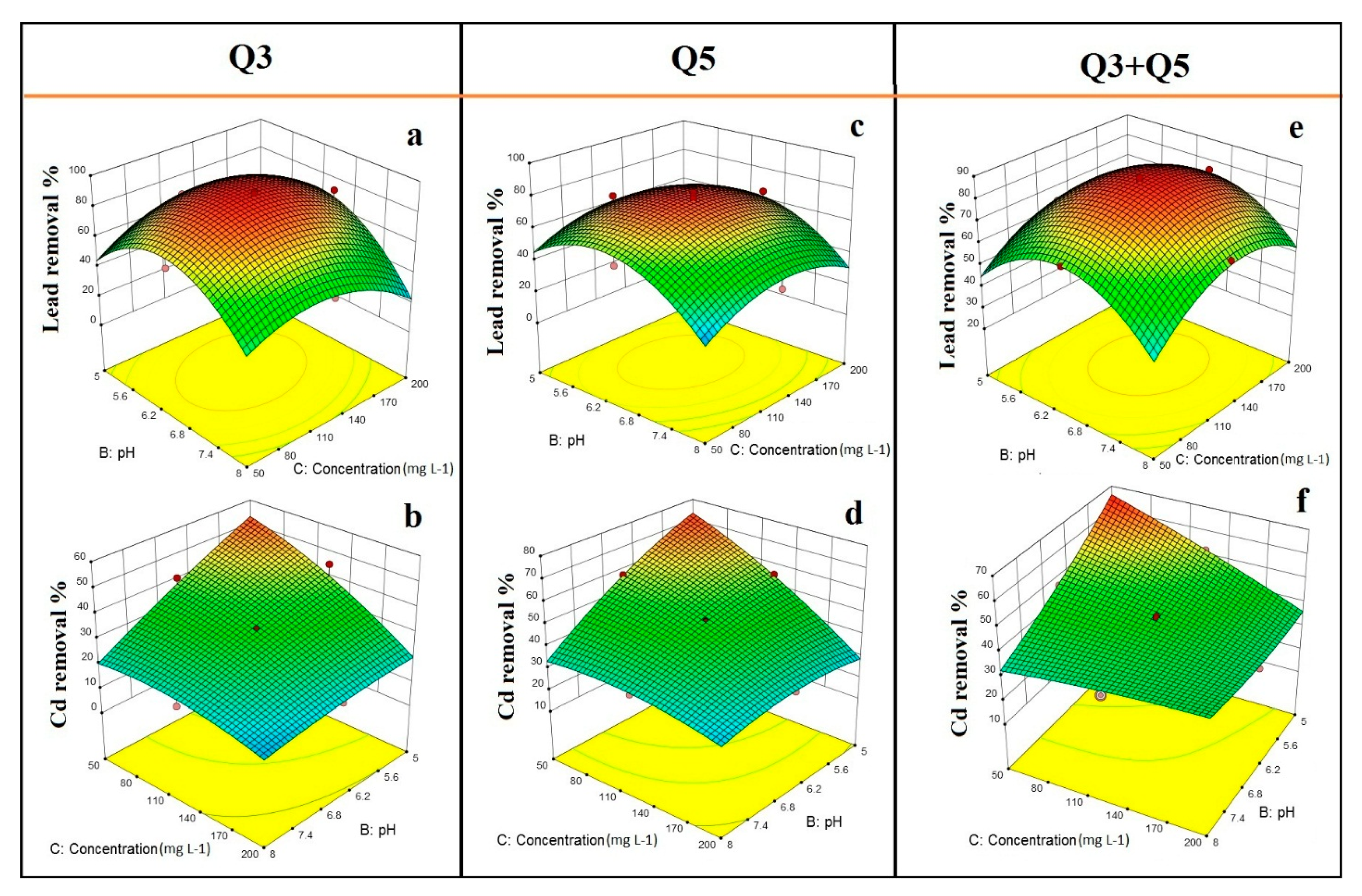
3.3. Optimization of Pb(II) and Cd(II) Removal Efficiency through RSM Using CCD
3.4. Effect of Temperature and Initial pH on Pb(II) and Cd(II) Bioremediation Process
3.5. Effect of Temperature and Initial Metal Concentration on Pb(II) and Cd(II) Bioremediation Process
3.6. Effect of Initial pH and Initial Metal Concentration on Pb(II) and Cd(II) Bioremediation Process
3.7. Optimization of Independent Variables for Pb(II) and Cd(II) Bioremediation Process
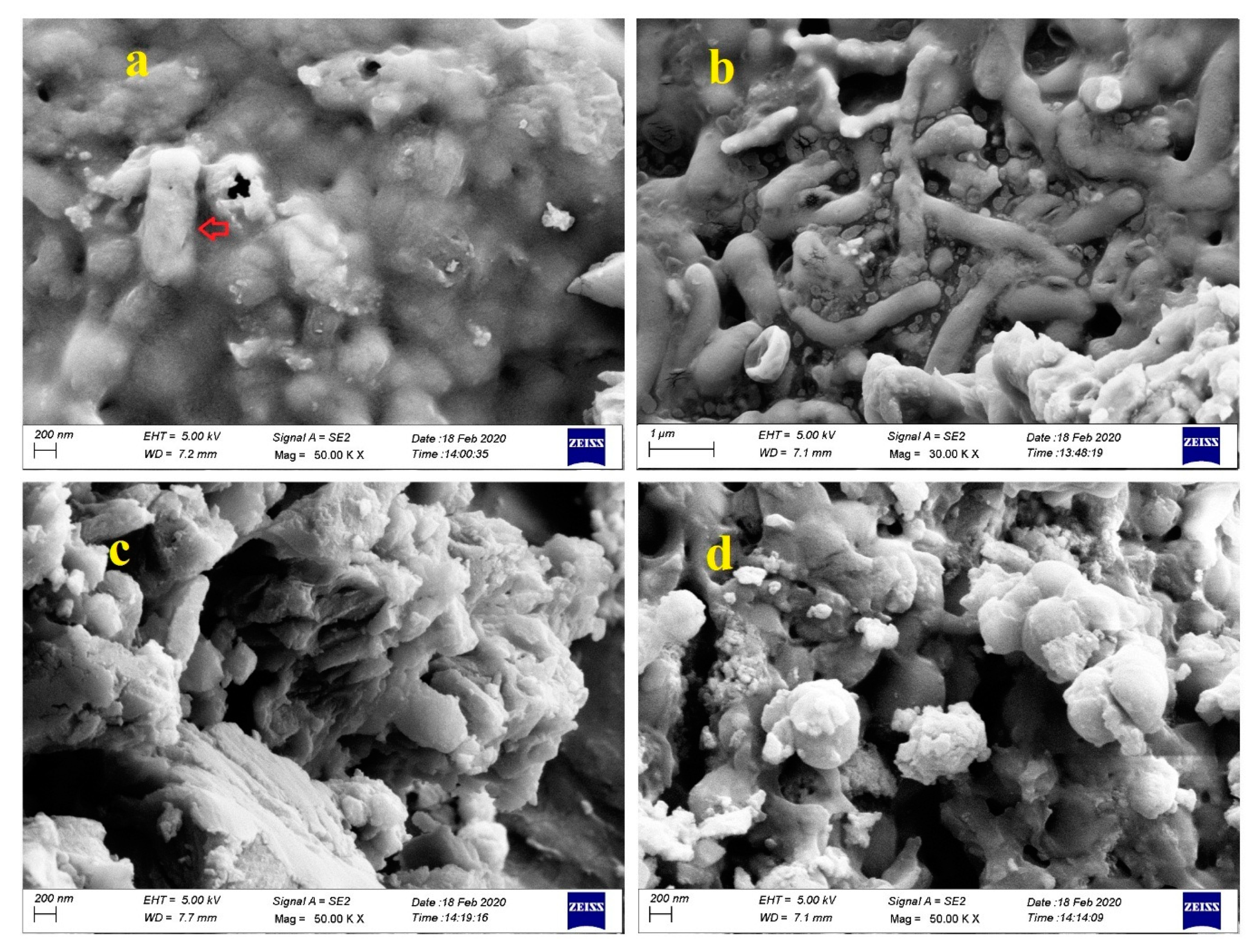
3.8. Analysis by FESEM and EDS
4. Discussion
4.1. Optimization of Pb(II) and Cd(II) Bioremediation Process
4.2. Mechanisms of Pb (II) and Cd (II) Bioremediation Used by Bacterial Strains Q3 and Q5
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Li, X.; Peng, W.; Jia, Y.; Lu, L.; Fan, W. Bioremediation of lead contaminated soil with Rhodobacter sphaeroides. Chemosphere 2016, 156, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Dabir, A.; Heidari, P.; Ghorbani, H.; Ebrahimi, A. Cadmium and lead removal by new bacterial isolates from coal and aluminum mines. Int. J. Environ. Sci. Technol. 2019, 16, 8297–8304. [Google Scholar] [CrossRef]
- Ferraro, A.; Fabbricino, M.; van Hullebusch, E.D.; Esposito, G. Investigation of different ethylenediamine-N,N′-disuccinic acid-enhanced washing configurations for remediation of a Cu-contaminated soil: Process kinetics and efficiency comparison between single-stage and multi-stage configurations. Environ. Sci. Pollut. Res. 2017, 24, 21960–21972. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.V.; Agovino, P.; Niu, X.; Roché, L. Linkage study of cancer risk among lead-exposed workers in New Jersey. Sci. Total Environ. 2007, 372, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Bueno, B.Y.M.; Torem, M.L.; Molina, F.; de Mesquita, L.M.S. Biosorption of lead(II), chromium(III) and copper(II) by R. opacus: Equilibrium and kinetic studies. Miner. Eng. 2008, 21, 65–75. [Google Scholar] [CrossRef]
- Kushwaha, A.; Hans, N.; Kumar, S.; Rani, R. A critical review on speciation, mobilization and toxicity of lead in soil-microbe-plant system and bioremediation strategies. Ecotoxicol. Environ. Saf. 2018, 147, 1035–1045. [Google Scholar] [CrossRef]
- Fabbricino, M.; Ferraro, A.; Luongo, V.; Pontoni, L.; Race, M. Soil washing optimization, recycling of the solution, and ecotoxicity assessment for the remediation of Pb-contaminated sites using EDDS. Sustainability 2018, 10, 636. [Google Scholar] [CrossRef]
- Järup, L.; Åkesson, A. Current status of cadmium as an environmental health problem. Toxicol. Appl. Pharmacol. 2009, 238, 201–208. [Google Scholar] [CrossRef]
- Seifpanahi-Shabani, K.; Eyvazkhani, A.; Heidari, P. Bioremediation of Textile Dyes Wastewater: Potential of Bacterial Isolates from a Mining Soils and Wetlands. Prog. Color. Color. Coat. J. 2019, 12, 155–161. [Google Scholar]
- Liu, Y.-G.; Ting, F.A.N.; Zeng, G.; Xin, L.I.; Qing, T.; Fei, Y.E.; Ming, Z.; Xu, W.; Huang, Y. Removal of cadmium and zinc ions from aqueous solution by living Aspergillus niger. Trans. Nonferrous Met. Soc. China 2006, 16, 681–686. [Google Scholar] [CrossRef]
- Guo, H.; Luo, S.; Chen, L.; Xiao, X.; Xi, Q.; Wei, W.; Zeng, G.; Liu, C.; Wan, Y.; Chen, J. Bioremediation of heavy metals by growing hyperaccumulaor endophytic bacterium Bacillus sp. L14. Bioresour. Technol. 2010, 101, 8599–8605. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, A.; van Hullebusch, E.D.; Huguenot, D.; Fabbricino, M.; Esposito, G. Application of an electrochemical treatment for EDDS soil washing solution regeneration and reuse in a multi-step soil washing process: Case of a Cu contaminated soil. J. Environ. Manage. 2015, 163, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Malik, A. Metal bioremediation through growing cells. Environ. Int. 2004, 30, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Wadgaonkar, S.L.; Ferraro, A.; Race, M.; Nancharaiah, Y.V.; Dhillon, K.S.; Fabbricino, M.; Esposito, G.; Lens, P.N.L. Optimization of Soil Washing to Reduce the Selenium Levels of Seleniferous Soil from Punjab, Northwestern India. J. Environ. Qual. 2018, 47, 1530–1537. [Google Scholar] [CrossRef] [PubMed]
- Race, M.; Ferraro, A.; Fabbricino, M.; La Marca, A.; Panico, A.; Spasiano, D.; Tognacchini, A.; Pirozzi, F. Ethylenediamine-N,N′-disuccinic acid (EDDS)—enhanced flushing optimization for contaminated agricultural soil remediation and assessment of prospective Cu and Zn transport. Int. J. Environ. Res. Public Health 2018, 15, 543. [Google Scholar] [CrossRef]
- de Alencar, F.L.S.; Navoni, J.L.; do Amaral, V.S. The use of bacterial bioremediation of metals in aquatic environments in the twenty-first century: A systematic review. Environ. Sci. Pollut. Res. 2017, 24, 16545–16559. [Google Scholar] [CrossRef]
- Potysz, A.; van Hullebusch, E.D.; Kierczak, J. Perspectives regarding the use of metallurgical slags as secondary metal resources—A review of bioleaching approaches. J. Environ. Manage. 2018, 219, 138–152. [Google Scholar] [CrossRef]
- Umar, M.; Kassim, K.A.; Chiet, K.T.P. Biological process of soil improvement in civil engineering: A review. J. Rock Mech. Geotech. Eng. 2016, 8, 767–774. [Google Scholar] [CrossRef]
- Arias, D.; Villca, G.; Panico, A.; Cisternas, L.A.; Jeldres, R.I.; González-Benito, G.; Rivas, M. Partial desalination of seawater for mining processes through a fluidized bed bioreactor filled with immobilized cells of Bacillus subtilis LN8B. Desalination 2020, 482, 114388. [Google Scholar] [CrossRef]
- Cui, Z.; Zhang, X.; Yang, H.; Sun, L. Bioremediation of heavy metal pollution utilizing composite microbial agent of Mucor circinelloides, Actinomucor sp. and Mortierella sp. J. Environ. Chem. Eng. 2017, 5, 3616–3621. [Google Scholar] [CrossRef]
- Choińska-Pulit, A.; Sobolczyk-Bednarek, J.; Łaba, W. Optimization of copper, lead and cadmium biosorption onto newly isolated bacterium using a Box-Behnken design. Ecotoxicol. Environ. Saf. 2018, 149, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Haferburg, G.; Sineriz, M.; Merten, D.; Büchel, G.; Kothe, E. Heavy metal resistance mechanisms in actinobacteria for survival in AMD contaminated soils. Chemie der Erde-Geochemistry 2005, 65, 131–144. [Google Scholar] [CrossRef]
- Wadgaonkar, S.L.; Ferraro, A.; Nancharaiah, Y.V.; Dhillon, K.S.; Fabbricino, M.; Esposito, G.; Lens, P.N.L. In situ and ex situ bioremediation of seleniferous soils from northwestern India. J. Soils Sediments 2019, 19, 762–773. [Google Scholar] [CrossRef]
- Chen, Z.; Pan, X.; Chen, H.; Guan, X.; Lin, Z. Biomineralization of Pb (II) into Pb-hydroxyapatite induced by Bacillus cereus 12-2 isolated from Lead–Zinc mine tailings. J. Hazard. Mater. 2016, 301, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Kalita, D.; Joshi, S.R. Study on bioremediation of Lead by exopolysaccharide producing metallophilic bacterium isolated from extreme habitat. Biotechnol. reports 2017, 16, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, K.; Mitra, S.; Sarkar, A.; Maiti, T.K. Alleviation of phytotoxic effects of cadmium on rice seedlings by cadmium resistant PGPR strain Enterobacter aerogenes MCC 3092. J. Hazard. Mater. 2018, 351, 317–329. [Google Scholar] [CrossRef]
- Huang, F.; Wang, Z.-H.; Cai, Y.-X.; Chen, S.-H.; Tian, J.-H.; Cai, K.-Z. Heavy metal bioaccumulation and cation release by growing Bacillus cereus RC-1 under culture conditions. Ecotoxicol. Environ. Saf. 2018, 157, 216–226. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y.R.; Zhang, L.M.; He, J.Z. Sorption mechanism and distribution of cadmium by different microbial species. J. Environ. Manage. 2019, 237, 552–559. [Google Scholar] [CrossRef]
- Hassan, S.H.A.; Kim, S.-J.; Jung, A.-Y.; Joo, J.H.; Oh, S.E.; Yang, J.E. Biosorptive capacity of Cd (II) and Cu (II) by lyophilized cells of Pseudomonas stutzeri. J. Gen. Appl. Microbiol. 2009, 55, 27–34. [Google Scholar] [CrossRef]
- Gupta, P.; Diwan, B. Bacterial Exopolysaccharide mediated heavy metal removal: A Review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 2017, 13, 58–71. [Google Scholar] [CrossRef]
- Li, W.-W.; Yu, H.-Q. Insight into the roles of microbial extracellular polymer substances in metal biosorption. Bioresour. Technol. 2014, 160, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Jaafari, J.; Yaghmaeian, K. Optimization of heavy metal biosorption onto freshwater algae (Chlorella coloniales) using response surface methodology (RSM). Chemosphere 2019, 217, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Barghouthi, S.A. A universal method for the identification of bacteria based on general PCR primers. Indian J. Microbiol. 2011, 51, 430–444. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hadiani, M.R.; Darani, K.K.; Rahimifard, N.; Younesi, H. Biosorption of low concentration levels of Lead (II) and Cadmium (II) from aqueous solution by Saccharomyces cerevisiae: Response surface methodology. Biocatal. Agric. Biotechnol. 2018, 15, 25–34. [Google Scholar] [CrossRef]
- Kang, C.H.; Kwon, Y.J.; So, J.S. Bioremediation of heavy metals by using bacterial mixtures. Ecol. Eng. 2016, 89, 64–69. [Google Scholar] [CrossRef]
- Arivalagan, P.; Singaraj, D.; Haridass, V.; Kaliannan, T. Removal of cadmium from aqueous solution by batch studies using Bacillus cereus. Ecol. Eng. 2014, 71, 728–735. [Google Scholar] [CrossRef]
- Choi, A.; Wang, S.; Lee, M. Biosorption of cadmium, copper, and lead ions from aqueous solutions by Ralstonia sp. and Bacillus sp. isolated from diesel and heavy metal contaminated soil. Geosci. J. 2009, 13, 331–341. [Google Scholar] [CrossRef]
- Hlihor, R.M.; Roşca, M.; Tavares, T.; Gavrilescu, M. The role of Arthrobacter viscosus in the removal of Pb (II) from aqueous solutions. Water Sci. Technol. 2017, 76, 1726–1738. [Google Scholar] [CrossRef]
- Igiri, B.E.; Okoduwa, S.I.R.; Idoko, G.O.; Akabuogu, E.P.; Adeyi, A.O.; Ejiogu, I.K. Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: A review. J. Toxicol. 2018, 2018. [Google Scholar] [CrossRef]
- Bandowe, B.A.M.; Bigalke, M.; Boamah, L.; Nyarko, E.; Saalia, F.K.; Wilcke, W. Polycyclic aromatic compounds (PAHs and oxygenated PAHs) and trace metals in fish species from Ghana (West Africa): Bioaccumulation and health risk assessment. Environ. Int. 2014, 65, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Brenner, K.; You, L.; Arnold, F.H. Engineering microbial consortia: A new frontier in synthetic biology. Trends Biotechnol. 2008, 26, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Roşca, M.; Hlihor, R.-M.; Cozma, P.; Drăgoi, E.N.; Diaconu, M.; Silva, B.; Tavares, T.; Gavrilescu, M. Comparison of Rhodotorula sp. and Bacillus megaterium in the removal of cadmium ions from liquid effluents. Green Process. Synth. 2018, 7, 74–88. [Google Scholar] [CrossRef]
- Tiquia-Arashiro, S.M. Lead absorption mechanisms in bacteria as strategies for lead bioremediation. Appl. Microbiol. Biotechnol. 2018, 102, 5437–5444. [Google Scholar] [CrossRef]
- Vijayaraghavan, K.; Yun, Y.-S. Bacterial biosorbents and biosorption. Biotechnol. Adv. 2008, 26, 266–291. [Google Scholar] [CrossRef]
- Pérez, J.A.M.; García-Ribera, R.; Quesada, T.; Aguilera, M.; Ramos-Cormenzana, A.; Monteoliva-Sánchez, M. Biosorption of heavy metals by the exopolysaccharide produced by Paenibacillus jamilae. World J. Microbiol. Biotechnol. 2008, 24, 2699. [Google Scholar] [CrossRef]
- Heidari, P.; Mazloomi, F.; Sanaeizade, S. Optimization Study of Nickel and Copper Bioremediation by Microbacterium oxydans Strain CM3 and CM7. Soil Sediment Contam. An Int. J. 2020, 29, 438–451. [Google Scholar] [CrossRef]
- Abbas, S.Z.; Rafatullah, M.; Hossain, K.; Ismail, N.; Tajarudin, H.A.; Khalil, H.P.S.A. A review on mechanism and future perspectives of cadmium-resistant bacteria. Int. J. Environ. Sci. Technol. 2018, 15, 243–262. [Google Scholar] [CrossRef]
- Jarosławiecka, A.; Piotrowska-Seget, Z. Lead resistance in micro-organisms. Microbiology 2014, 160, 12–25. [Google Scholar] [CrossRef]
- Özdemir, S.; Kilinc, E.; Poli, A.; Nicolaus, B.; Güven, K. Biosorption of Cd, Cu, Ni, Mn and Zn from aqueous solutions by thermophilic bacteria, Geobacillus toebii sub. sp. decanicus and Geobacillus thermoleovorans sub. sp. stromboliensis: Equilibrium, kinetic and thermodynamic studies. Chem. Eng. J. 2009, 152, 195–206. [Google Scholar] [CrossRef]
- Bai, H.-J.; Zhang, Z.-M.; Yang, G.-E.; Li, B.-Z. Bioremediation of cadmium by growing Rhodobacter sphaeroides: Kinetic characteristic and mechanism studies. Bioresour. Technol. 2008, 99, 7716–7722. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R.M.; Shetty, K.V.; Srinikethan, G. Cadmium (II) and nickel (II) biosorption by Bacillus laterosporus (MTCC 1628). J. Taiwan Inst. Chem. Eng. 2014, 45, 1628–1635. [Google Scholar] [CrossRef]
- Naik, M.M.; Dubey, S.K. Lead resistant bacteria: Lead resistance mechanisms, their applications in lead bioremediation and biomonitoring. Ecotoxicol. Environ. Saf. 2013, 98, 1–7. [Google Scholar] [CrossRef]
- De, J.; Ramaiah, N.; Vardanyan, L. Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar. Biotechnol. 2008, 10, 471–477. [Google Scholar] [CrossRef] [PubMed]


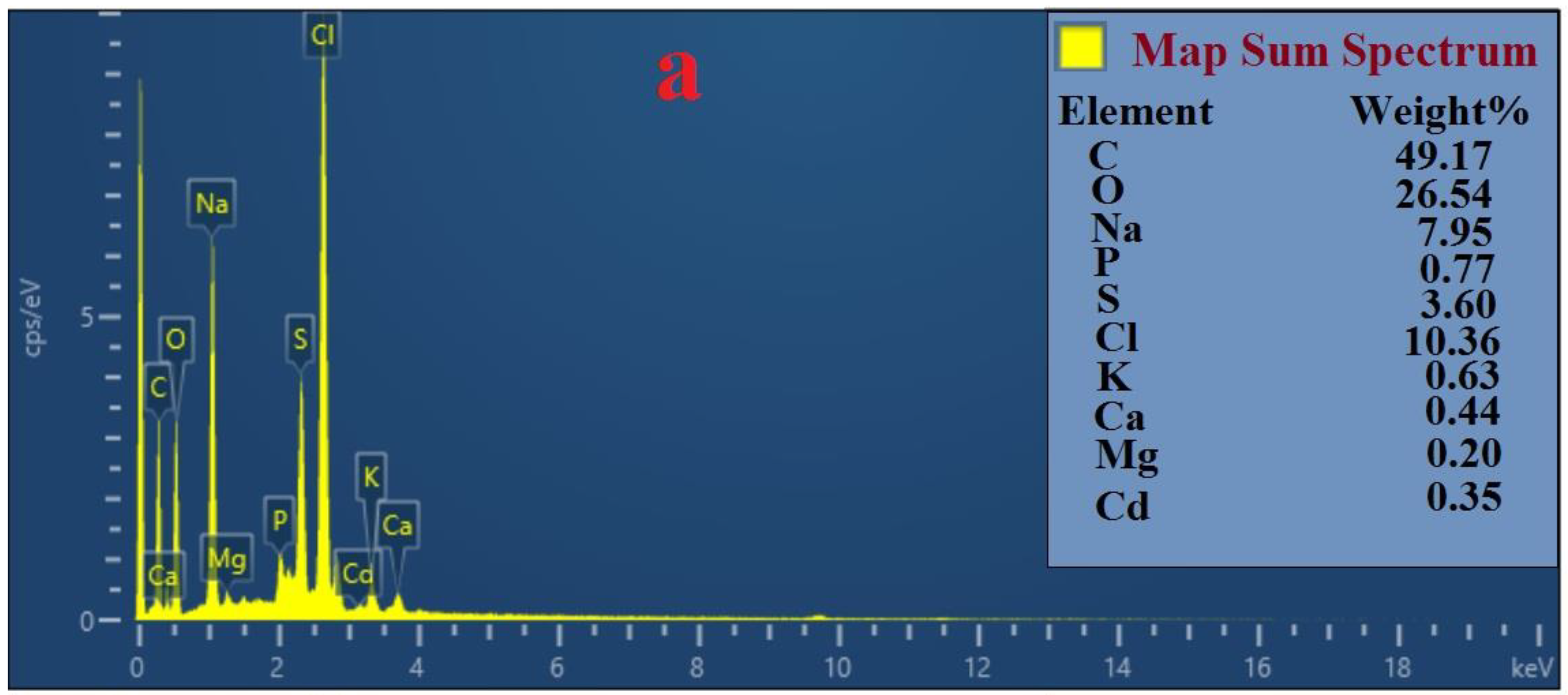
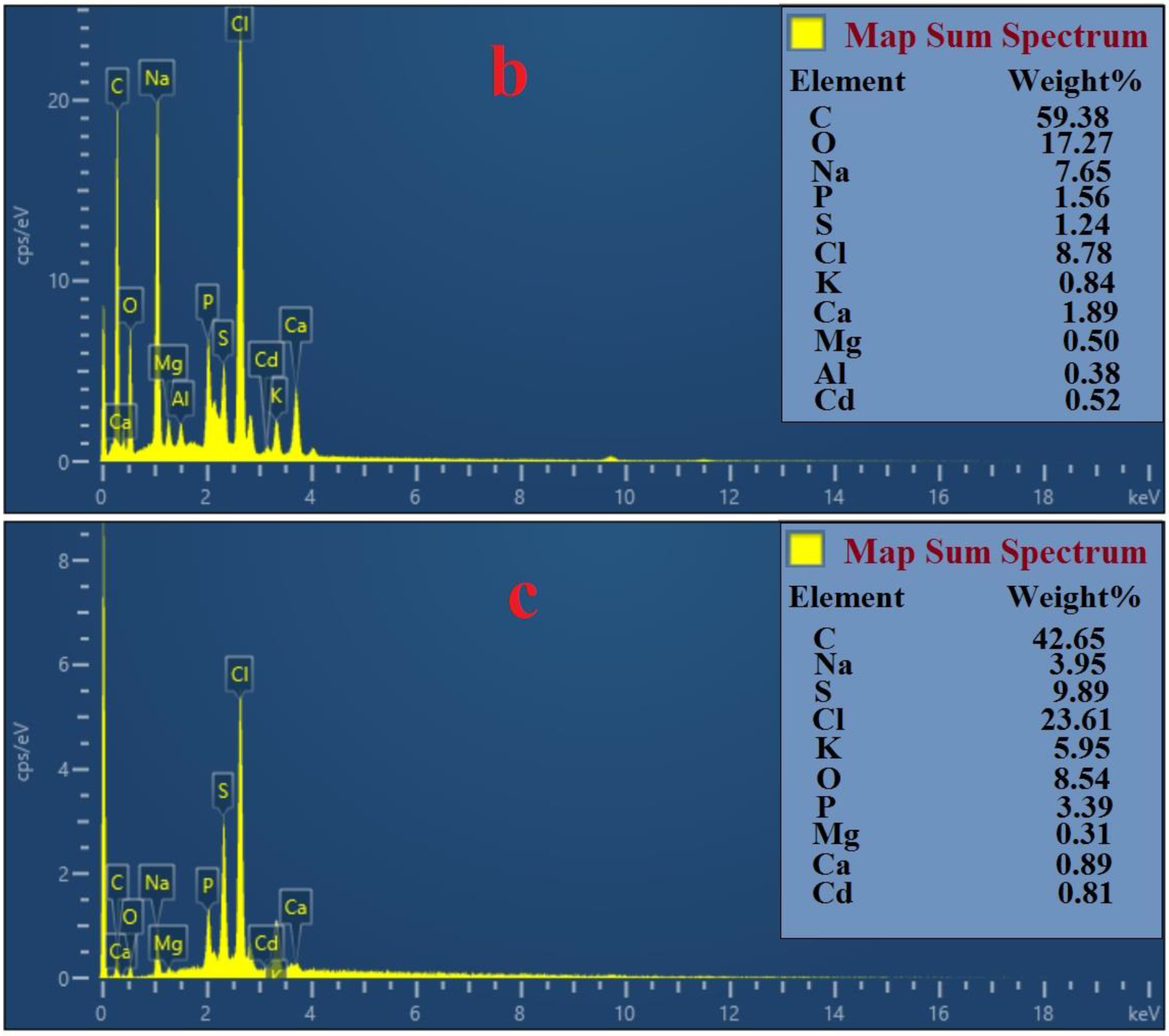
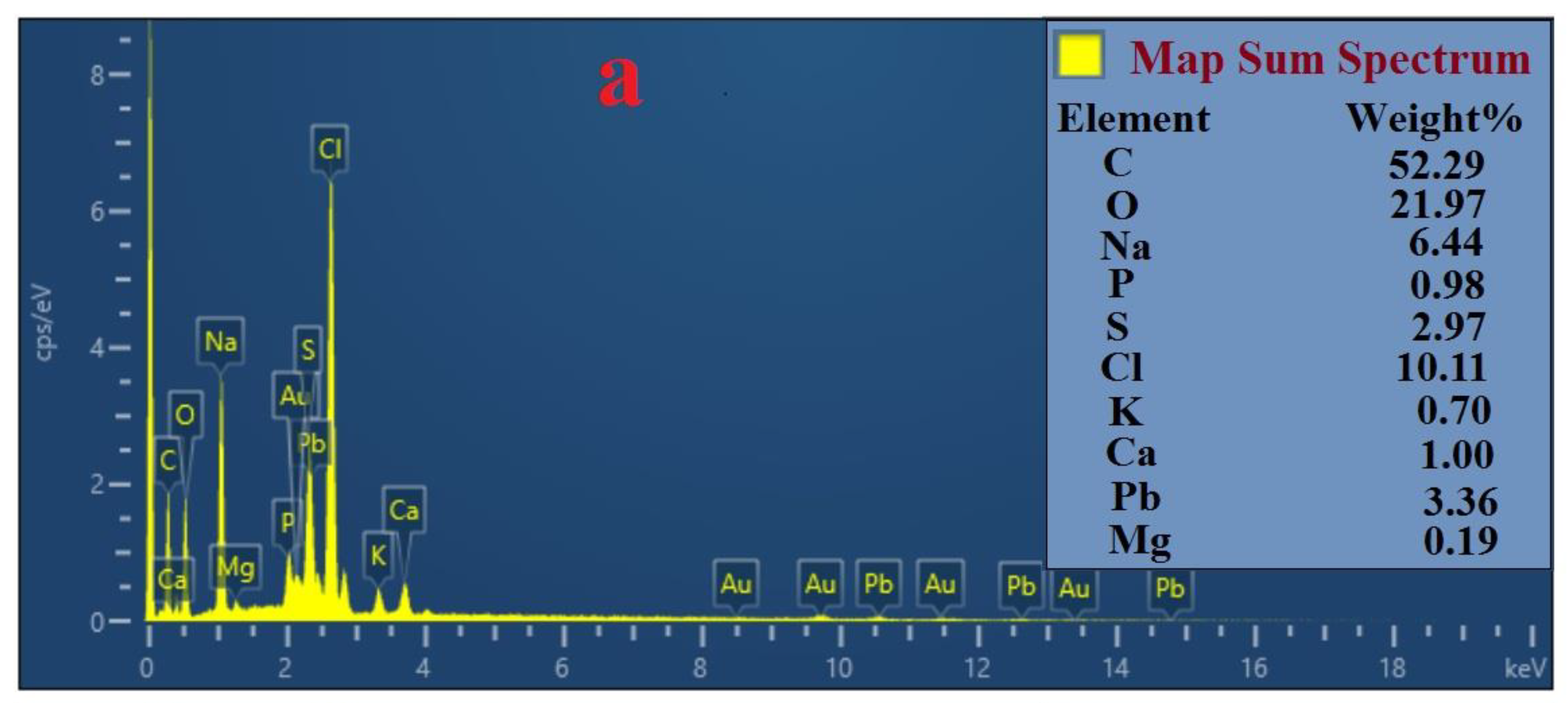

| Variables | Factor | Range and Level | ||
|---|---|---|---|---|
| −1 | 0 | +1 | ||
| Temperature (°C) | A | 30 | 35 | 40 |
| pH | B | 5 | 6.5 | 8 |
| Metal concentration (mg L−1) | C | 50 | 125 | 200 |
| Run no. | Variables | Removal Efficiency (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | Q3 | Q5 | Q3 + Q5 | ||||||||||
| Pb(II) | Cd(II) | Pb(II) | Cd(II) | Pb(II) | Cd(II) | ||||||||||
| Y exp | Ypre | Y exp | Ypre | Y exp | Ypre | Yexp | Ypre | Yexp | Ypre | Y xp | Ypre | ||||
| 1 | 40 | 6.5 | 125 | 92.07 | 93.29 | 35.58 | 32.60 | 66.29 | 66.68 | 48.41 | 46.82 | 73.63 | 71.75 | 37.13 | 37.62 |
| 2 | 35 | 6.5 | 125 | 87.53 | 89.51 | 32.02 | 32.00 | 75.37 | 75.45 | 48.64 | 48.40 | 86.08 | 85.74 | 43.08 | 42.42 |
| 3 | 40 | 8.0 | 200 | 19.78 | 20.60 | 12.97 | 10.97 | 24.92 | 24.04 | 20.70 | 20.17 | 31.13 | 34.19 | 29.90 | 28.52 |
| 4 | 30 | 6.5 | 125 | 71.90 | 71.10 | 17.86 | 18.72 | 69.85 | 70.51 | 30.12 | 31.85 | 70.18 | 70.34 | 24.52 | 25.82 |
| 5 | 30 | 8.0 | 50 | 31.32 | 30.74 | 8.95 | 6.67 | 25.27 | 22.64 | 16.66 | 15.46 | 26.32 | 28.51 | 16.08 | 14.59 |
| 6 | 40 | 5.0 | 50 | 56.00 | 55.21 | 57.73 | 58.21 | 35.38 | 35.38 | 77.54 | 77.51 | 33.69 | 35.82 | 68.11 | 67.23 |
| 7 | 35 | 6.5 | 125 | 90.17 | 89.51 | 30.82 | 32.00 | 76.58 | 75.45 | 47.54 | 48.40 | 87.08 | 85.74 | 43.62 | 42.42 |
| 8 | 30 | 8.0 | 200 | 10.05 | 10.73 | 5.81 | 5.85 | 24.07 | 23.99 | 16.04 | 16.39 | 44.53 | 42.82 | 27.51 | 27.94 |
| 9 | 40 | 8.0 | 50 | 36.55 | 34.82 | 19.67 | 21.35 | 16.29 | 14.60 | 32.07 | 33.06 | 25.36 | 23.85 | 28.50 | 29.22 |
| 10 | 35 | 6.5 | 50 | 69.48 | 73.51 | 40.52 | 38.09 | 55.93 | 59.83 | 57.60 | 57.35 | 65.31 | 65.07 | 46.66 | 47.37 |
| 11 | 35 | 6.5 | 125 | 89.51 | 89.51 | 31.93 | 32.00 | 73.26 | 75.45 | 48.82 | 48.40 | 85.23 | 85.74 | 42.24 | 42.42 |
| 12 | 40 | 5.0 | 200 | 65.34 | 65.82 | 17.61 | 20.42 | 37.10 | 39.46 | 24.75 | 25.91 | 41.78 | 40.02 | 29.16 | 30.20 |
| 13 | 30 | 5.0 | 50 | 21.61 | 20.69 | 33.04 | 35.57 | 42.29 | 42.90 | 50.87 | 51.36 | 26.90 | 24.32 | 43.28 | 44.21 |
| 14 | 35 | 6.5 | 125 | 89.98 | 89.51 | 32.93 | 32.00 | 75.56 | 75.45 | 48.96 | 48.40 | 83.64 | 85.74 | 44.50 | 42.42 |
| 15 | 30 | 5.0 | 200 | 23.87 | 25.50 | 8.50 | 7.34 | 37.65 | 39.08 | 14.60 | 13.58 | 30.61 | 32.54 | 22.39 | 21.23 |
| 16 | 35 | 6.5 | 200 | 72.40 | 68.80 | 18.47 | 18.78 | 65.49 | 62.65 | 31.63 | 32.02 | 75.80 | 74.33 | 34.45 | 35.53 |
| 17 | 35 | 5.0 | 125 | 67.87 | 67.48 | 44.97 | 40.30 | 64.43 | 60.21 | 55.46 | 54.86 | 63.63 | 63.90 | 52.33 | 52.39 |
| 18 | 35 | 6.5 | 125 | 91.22 | 89.51 | 30.49 | 32.00 | 75.90 | 75.45 | 48.53 | 48.40 | 85.10 | 85.74 | 42.01 | 42.42 |
| 19 | 35 | 6.5 | 125 | 89.52 | 89.51 | 29.58 | 32.00 | 78.14 | 75.45 | 48.17 | 48.40 | 83.86 | 85.74 | 42.66 | 42.42 |
| 20 | 35 | 8.0 | 125 | 49.07 | 49.89 | 18.57 | 21.12 | 37.10 | 42.37 | 33.30 | 34.04 | 65.04 | 63.06 | 35.00 | 36.73 |
| Metal Ions | Model Term | Tested Bacterial Strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Q3 | Q5 | Q3 + Q5 | ||||||||
| DF | Mean Square | p-Value | DF | Mean Square | p-Value | DF | Mean Square | p-Value | ||
| Pb(II) | Model | 9 | 1678.61 | <0.001 | 9 | 966.04 | <0.001 | 9 | 1192.88 | <0.001 |
| A | 1 | 1231.88 | <0.001 | 1 | 36.67 | 0.086 | 1 | 4.97 | 0.374 | |
| B | 1 | 772.99 | <0.001 | 1 | 795.66 | <0.001 | 1 | 1.79 | 0.589 | |
| C | 1 | 55.32 | 0.007 | 1 | 19.80 | 0.193 | 1 | 214.09 | <0.001 | |
| AB | 1 | 463.60 | <0.001 | 1 | 0.056 | 0.942 | 1 | 130.57 | <0.001 | |
| AC | 1 | 16.76 | 0.092 | 1 | 32.76 | 0.103 | 1 | 8.12 | 0.261 | |
| BC | 1 | 308.02 | <0.001 | 1 | 13.39 | 0.278 | 1 | 18.54 | 0.102 | |
| A2 | 1 | 147.10 | <0.001 | 1 | 128.97 | 0.005 | 1 | 593.22 | <0.001 | |
| B2 | 1 | 2613.61 | <0.001 | 1 | 1604.28 | <0.001 | 1 | 1362.31 | <0.001 | |
| C2 | 1 | 926.86 | <0.001 | 1 | 555.15 | <0.001 | 1 | 707.28 | <0.001 | |
| R2 = 0.997 | R2 = 0.988 | R2 = 0.995 | ||||||||
| Cd(II) | Model | 9 | 363.48 | <0.001 | 9 | 576.67 | <0.001 | 9 | 290.89 | <0.001 |
| A | 1 | 481.52 | <0.001 | 1 | 559.95 | <0.001 | 1 | 348.30 | <0.001 | |
| B | 1 | 919.22 | <0.001 | 1 | 1083.13 | <0.001 | 1 | 612.59 | <0.001 | |
| C | 1 | 931.94 | <0.001 | 1 | 1604.42 | <0.001 | 1 | 350.66 | <0.001 | |
| AB | 1 | 31.70 | 0.078 | 1 | 36.58 | <0.001 | 1 | 35.21 | 0.002 | |
| AC | 1 | 45.73 | 0.040 | 1 | 95.51 | <0.001 | 1 | 98.71 | <0.001 | |
| BC | 1 | 375.63 | <0.001 | 1 | 749.34 | <0.001 | 1 | 660.17 | <0.001 | |
| A2 | 1 | 110.57 | 0.004 | 1 | 225.70 | <0.001 | 1 | 314.77 | <0.001 | |
| B2 | 1 | 4.59 | 0.472 | 1 | 42.81 | <0.001 | 1 | 12.60 | 0.037 | |
| C2 | 1 | 35.01 | 0.066 | 1 | 37.88 | <0.001 | 1 | 2.59 | <0.001 | |
| R2 = 0.975 | R2 = 0.997 | R2 = 0.992 | ||||||||
| Metal Ions | Bacterial Strains | Temperature (°C) | Initial pH | Metal Concentration (mg L−1) | Removal Efficiency (%) |
|---|---|---|---|---|---|
| Pb(II) | Q3 | 38.8 | 5.8 | 115.4 | 93.8 |
| Q5 | 34.3 | 6.2 | 127.4 | 76.4 | |
| Q3 + Q5 | 35.1 | 6.5 | 135.8 | 86.1 | |
| Cd(II) | Q3 | 38.6 | 5.0 | 50.6 | 58.0 |
| Q5 | 38.3 | 5.0 | 50.0 | 78.0 | |
| Q3 + Q5 | 38.0 | 5.1 | 51.4 | 68.2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heidari, P.; Panico, A. Sorption Mechanism and Optimization Study for the Bioremediation of Pb(II) and Cd(II) Contamination by Two Novel Isolated Strains Q3 and Q5 of Bacillus sp. Int. J. Environ. Res. Public Health 2020, 17, 4059. https://doi.org/10.3390/ijerph17114059
Heidari P, Panico A. Sorption Mechanism and Optimization Study for the Bioremediation of Pb(II) and Cd(II) Contamination by Two Novel Isolated Strains Q3 and Q5 of Bacillus sp. International Journal of Environmental Research and Public Health. 2020; 17(11):4059. https://doi.org/10.3390/ijerph17114059
Chicago/Turabian StyleHeidari, Parviz, and Antonio Panico. 2020. "Sorption Mechanism and Optimization Study for the Bioremediation of Pb(II) and Cd(II) Contamination by Two Novel Isolated Strains Q3 and Q5 of Bacillus sp." International Journal of Environmental Research and Public Health 17, no. 11: 4059. https://doi.org/10.3390/ijerph17114059
APA StyleHeidari, P., & Panico, A. (2020). Sorption Mechanism and Optimization Study for the Bioremediation of Pb(II) and Cd(II) Contamination by Two Novel Isolated Strains Q3 and Q5 of Bacillus sp. International Journal of Environmental Research and Public Health, 17(11), 4059. https://doi.org/10.3390/ijerph17114059






