In Silico Subtractive Proteomics Approach for Identification of Potential Drug Targets in Staphylococcus saprophyticus
Abstract
1. Introduction
2. Methodology
2.1. Proteome Retrieval
2.2. Removal of Paralog Sequences
2.3. Retrieval of Essential Proteins
2.4. Essential Non-Homologous Protein Identification
2.5. Unique Pathway Identification
2.6. Subcellular Localization Prediction
2.7. Druggability Analysis
3. Results and Discussion
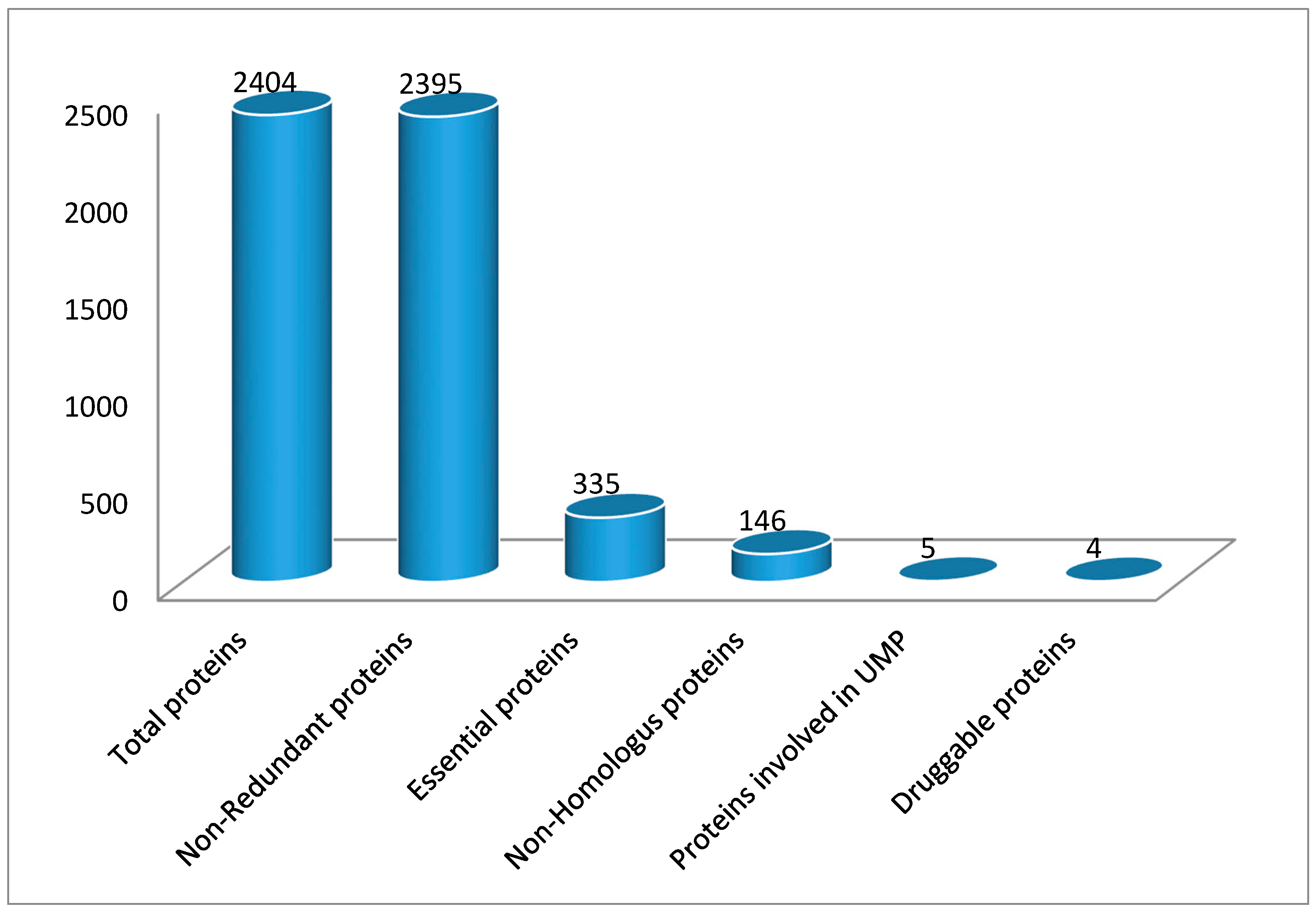
3.1. Identification of Essential Proteins
3.2. Essential Non-Homologous Protein Identification
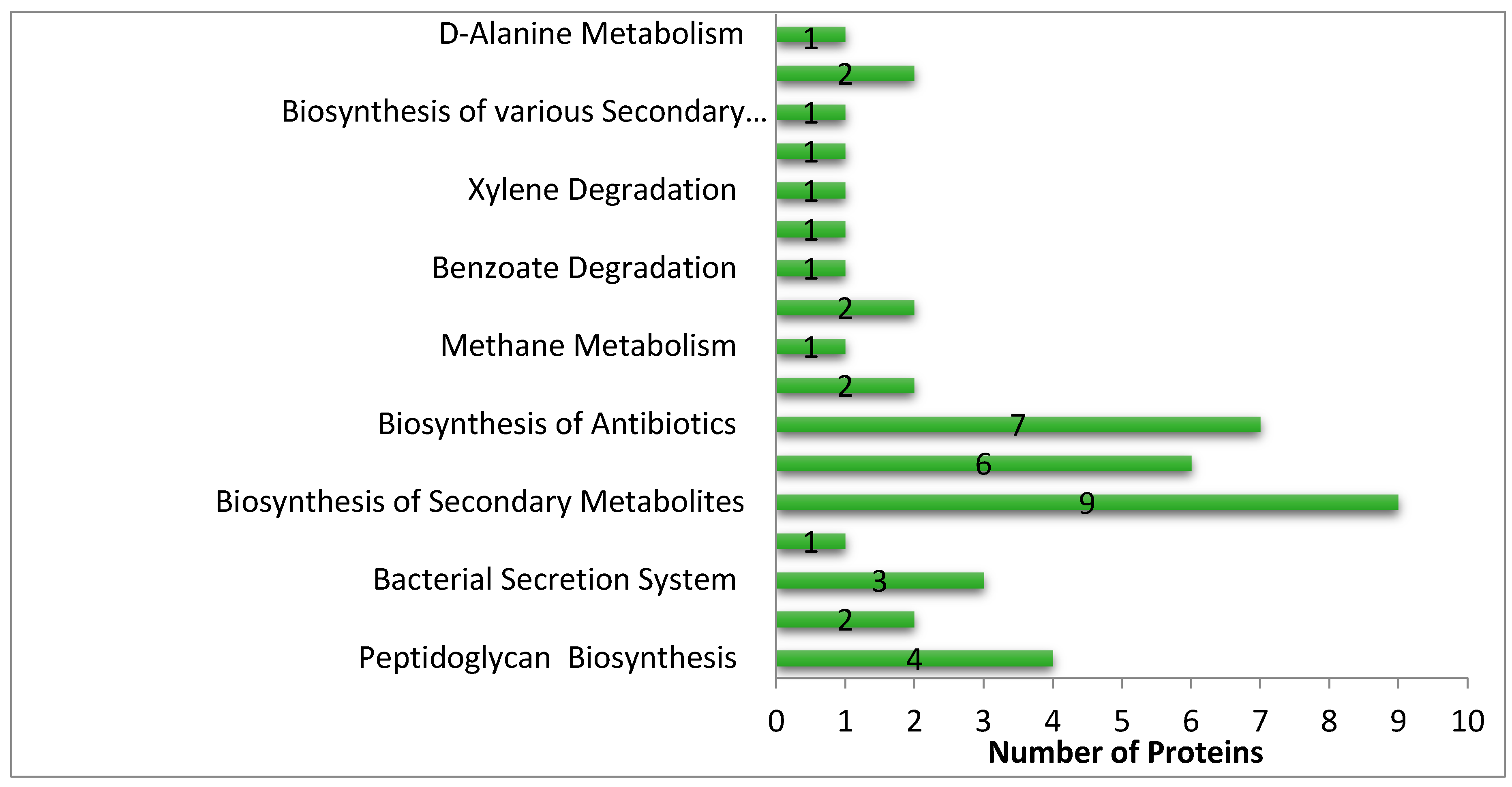
3.3. Metabolic Pathway Analysis
3.4. Subcellular Localization Prediction
3.5. Druggability of Therapeutic Targets
3.6. Pathways Specific to S. saprophyticus in Comparison with H. sapiens
3.7. Peptidoglycan Biosynthesis
3.8. Vancomycin Resistance
3.9. D-Alanine Metabolism
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pereira, A.T. Coagulase-negative strains of staphylococcus possessing antigen 51 as agents of urinary infection. J. Clin. Pathol. 1962, 15, 252–253. [Google Scholar] [CrossRef]
- Maskell, R. Importance of coagulase-negative staphylococci as pathogens in the urinary tract. Lancet 1974, 303, 1155–1158. [Google Scholar] [CrossRef]
- Fowler, J.E. Staphylococcus saprophyticus as the cause of infected urinary calculus. Ann. Intern. Med. 1985, 102, 342–343. [Google Scholar] [CrossRef] [PubMed]
- Raz, R.; Colodner, R.; Kunin, C.M. Who are you—Staphylococcus saprophyticus? Clin. Infect. Dis. 2005, 40, 896–898. [Google Scholar] [CrossRef]
- Natsis, N.E.; Cohen, P.R. Coagulase-negative staphylococcus skin and soft tissue infections. Am. J. Clin. Dermatol. 2018, 19, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Lala, V.; Minter, D.A. Acute Cystitis; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2020. [Google Scholar]
- Argemi, X.; Hansmann, Y. Coagulase-negative staphylococci pathogenomics. Int. J. Mol. Sci. 2019, 20, 1215. [Google Scholar] [CrossRef]
- Tolaymat, A.; Al-Jayousi, Z. Staphylococcus saprophyticus urinary-tract infection in male children. Child Nephrol. Urol. 1991, 11, 100–102. [Google Scholar]
- Hovelius, B.; Colleen, S.; Mardh, P.-A. Urinary tract infections in men caused by Staphylococcus saprophyticus. Scand. J. Infect. Dis. 1984, 16, 37–41. [Google Scholar] [CrossRef]
- Kauffman, C.A.; Hertz, C.S.; Sheagren, J.N. Staphylococcus saprophyticus: Role in urinary tract infections in men. J. Urol. 1983, 130, 493–494. [Google Scholar] [CrossRef]
- Widerstrom, M.; Wistrom, J.; Sjostedt, A.; Monsen, T. Coagulase-negative staphylococci: Update on the molecular epidemiology and clinical presentation, with a focus on Staphylococcus epidermidis and Staphylococcus saprophyticus. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2012, 31, 7–20. [Google Scholar] [CrossRef]
- Pailhoriès, H.; Cassisa, V.; Chenouard, R.; Kempf, M.; Eveillard, M.; Lemarié, C. Staphylococcus saprophyticus: Which beta-lactam? Int. J. Infect. Dis. 2017, 65, 63–66. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ghosh, S.; Prava, J.; Samal, H.B.; Suar, M.; Mahapatra, R.K. Comparative genomics study for the identification of drug and vaccine targets in Staphylococcus aureus: MurA ligase enzyme as a proposed candidate. J. Microbiol. Methods 2014, 101, 1–8. [Google Scholar] [CrossRef]
- Kumar, A.; Thotakura, P.L.; Tiwary, B.K.; Krishna, R. Target identification in Fusobacterium nucleatum by subtractive genomics approach and enrichment analysis of host-pathogen protein-protein interactions. BMC Microbiol. 2016, 16, 84. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.I.; Ferdous, S.; Jewel, N.A.; Akter, A.; Mahmud, Z.; Islam, M.M.; Afrin, T.; Karim, N. Identification of potential drug targets by subtractive genome analysis of Escherichia coli O157:H7: An in silico approach. Adv. Appl. Bioinform. Chem. AABC 2015, 8, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Uddin, R.; Jamil, F. Prioritization of potential drug targets against P. aeruginosa by core proteomic analysis using computational subtractive genomics and Protein-Protein interaction network. Comput. Biol. Chem. 2018, 74, 115–122. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Wen, Q.-F.; Liu, S.; Dong, C.; Guo, H.-X.; Gao, Y.-Z.; Guo, F.-B. Geptop 2.0: An updated, more precise, and faster geptop server for identification of prokaryotic essential genes. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Anishetty, S.; Pulimi, M.; Pennathur, G. Potential drug targets in Mycobacterium tuberculosis through metabolic pathway analysis. Comput. Biol. Chem. 2005, 29, 368–378. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Amineni, U.; Pradhan, D.; Marisetty, H. In silico identification of common putative drug targets in Leptospira interrogans. J. Chem. Biol. 2010, 3, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef] [PubMed]
- Gardy, J.L.; Spencer, C.; Wang, K.; Ester, M.; Tusnady, G.E.; Simon, I.; Hua, S.; deFays, K.; Lambert, C.; Nakai, K.; et al. PSORT-B: Improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 2003, 31, 3613–3617. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Uddin, R.; Siddiqui, Q.N.; Sufian, M.; Azam, S.S.; Wadood, A. Proteome-wide subtractive approach to prioritize a hypothetical protein of XDR-Mycobacterium tuberculosis as potential drug target. Genes Genom. 2019, 41, 1281–1292. [Google Scholar] [CrossRef]
- Sakharkar, K.R.; Sakharkar, M.K.; Chow, V.T. A novel genomics approach for the identification of drug targets in pathogens, with special reference to Pseudomonas aeruginosa. Silico Biol. 2004, 4, 355–360. [Google Scholar]
- Goyal, M.; Citu, S. In silico identification of novel drug targets in Acinetobacter baumannii by subtractive genomic approach. Asian J. Pharm. Clin. Res. 2018, 11, 230–236. [Google Scholar] [CrossRef]
- Cheng, A.C.; Coleman, R.G.; Smyth, K.T.; Cao, Q.; Soulard, P.; Caffrey, D.R.; Salzberg, A.C.; Huang, E.S. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 2007, 25, 71–75. [Google Scholar] [CrossRef]
- Keller, T.H.; Pichota, A.; Yin, Z. A practical view of ‘druggability’. Curr. Opin. Chem. Biol. 2006, 10, 357–361. [Google Scholar] [CrossRef]
- Butt, A.M.; Nasrullah, I.; Tahir, S.; Tong, Y. Comparative genomics analysis of Mycobacterium ulcerans for the identification of putative essential genes and therapeutic candidates. PLoS ONE 2012, 7, e43080. [Google Scholar] [CrossRef]
- Hasan, M.A.; Khan, M.A.; Sharmin, T.; Hasan Mazumder, M.H.; Chowdhury, A.S. Identification of putative drug targets in Vancomycin-resistant Staphylococcus aureus (VRSA) using computer aided protein data analysis. Gene 2016, 575, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.S.; Chowdhury, C.; Nelson, D.E. Physiological functions of D-alanine carboxypeptidases in Escherichia coli. Trends Microbiol. 2008, 16, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D.; Walsh, C.T. D-Alanyl-D-alanine ligases and the molecular mechanism of vancomycin resistance. Acc. Chem. Res. 1992, 25, 468–473. [Google Scholar] [CrossRef]
| Protein Name (Protein ID) | Common Pathway | Unique Pathway |
|---|---|---|
| UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-L-lysine ligase (Q49WE7) | ssp00550-Peptidoglycan biosynthesis | |
| Penicillin-binding protein 1 (Q49WW3) | ssp01100-Metabolic pathways | ssp00550-Peptidoglycan biosynthesis ssp01501-beta-Lactam resistance |
| Protein translocase subunit SecE (Q49V45) | ssp03060-Protein export | ssp03070-Bacterial secretion system |
| Aspartokinase (Q49XJ5) | ssp01210-2-Oxocarboxylic acid metabolism ssp01230-Biosynthesis of amino acids ssp00260-Glycine, serine, and threonine metabolism ssp00270-Cysteine and methionine metabolisms ssp00300-Lysine biosynthesis ssp01100-Metabolic pathways | ssp0026-Lobactam biosynthesis ssp01110-Biosynthesis of secondary metabolites ssp01120-Microbial metabolism in diverse environments ssp01130-Biosynthesis of antibiotics |
| Penicillin-binding protein 2 (Q49XQ6) | ssp01100-Metabolic pathways | ssp00550-Peptidoglycan biosynthesis ssp01501-beta-Lactam resistance |
| Homoserine dehydrogenase (Q49XB5) | ssp01230-Biosynthesis of amino acids ssp00260-Glycine, serine, and threonine metabolism ssp00270-Cysteine and methionine metabolisms ssp00300-Lysine biosynthesis ssp01100-Metabolic pathways | ssp01110-Biosynthesis of secondary metabolites ssp01120-Microbial metabolism in diverse environments ssp01130-Biosynthesis of antibiotics |
| Protein translocase subunit SecY (Q49ZE8) | ssp03060-Protein export | ssp03070-Bacterial secretion system |
| Fructose-bisphosphate aldolase (Q49Z72) | ssp01200-Carbon metabolism ssp01100-Metabolic pathways ssp01230-Biosynthesis of amino acids ssp00051-Fructose and mannose metabolism ssp00010-Glycolysis/Gluconeogenesis ssp00030-Pentose phosphate pathway | ssp01120-Microbial metabolism in diverse environments ssp01110-Biosynthesis of secondary metabolites ssp01130-Biosynthesis of antibiotics ssp00680-Methane metabolism |
| Riboflavin synthase alpha chain (Q49YJ8) | ssp00740-Riboflavin metabolism ssp01100-Metabolic pathways | ssp01110-Biosynthesis of secondary metabolites |
| Glutamine synthetase (Q49XA2) | ssp01100-Metabolic pathways ssp01230-Biosynthesis of amino acids ssp00220-Arginine biosynthesis ssp00250-Alanine, aspartate, and glutamate metabolism ssp00630-Glyoxylate and dicarboxylate metabolism ssp00910-Nitrogen metabolism | ssp01120-Microbial metabolism in diverse environments ssp02020-Two-component system |
| Riboflavin biosynthesis protein RibD (Q49YJ9) | ssp00740-Riboflavin metabolism ssp01100-Metabolic pathways | ssp01120-Microbial metabolism in diverse environments ssp02024-Quorum sensing |
| Beta sliding clamp (Q4A179) | ssp00220-Arginine biosynthesis ssp00310-Lysine degradation ssp00360-Phenylalanine metabolism ssp00630-Glyoxylate and dicarboxylate metabolism ssp00010-Glycolysis/Gluconeogenesis ssp00020-Citrate cycle (TCA cycle) ssp00040-Pentose and glucuronate interconversions ssp00053-Ascorbate and aldarate metabolism ssp00061-Fatty acid biosynthesis ssp00071-Fatty acid degradation ssp00250-Alanine, aspartate, and glutamate metabolism ssp00260-Glycine, serine, and threonine metabolism ssp00280-Valine, leucine, and isoleucine degradation ssp00330-Lysine biosynthesis ssp00350-Arginine and proline metabolism ssp00361-Tyrosine metabolism ssp00562-D-Glutamine and D-glutamate metabolism ssp00620-Inositol phosphate metabolism ssp00622-Pyruvate metabolism ssp00650-Propanoate metabolism ssp00660-Butanoate metabolism ssp00997-Nicotinate and nicotinamide metabolism ssp01100-Metabolic pathways ssp01210-2-Oxocarboxylic acid metabolism ssp01212-Fatty acid metabolism ssp03030-DNA replication ssp03430-Mismatch repair ssp03440-Homologous recombination | ssp00362-Benzoate degradation ssp00300-Chlorocyclohexane and chlorobenzene degradation ssp00471-Xylene degradation ssp00640-C5-Branched dibasic acid metabolism ssp00760-Biosynthesis of various secondary metabolites-Part 3 ssp01110-Biosynthesis of secondary metabolites ssp01120-Microbial metabolism in diverse environments ssp01130-Biosynthesis of antibiotics |
| Putative preprotein translocase subunit (Q49Y76) | ssp03060-Protein Export | ssp02024-Quorum sensing ssp03070-Bacterial secretion system |
| DAHP synthetase-chorismate mutase (Q49YG8) | ssp01230-Biosynthesis of amino acids ssp01100-Metabolic pathways ssp00400-Phenylalanine, tyrosine, and tryptophan biosynthesis | ssp01110-Biosynthesis of secondary metabolites ssp01130-Biosynthesis of antibiotics |
| Putative heptaprenyl diphosphate synthase component (Q49XS4) | ssp00900-Terpenoid backbone biosynthesis | ssp01110-Biosynthesis of secondary metabolites |
| Malonyl CoA-acyl carrier protein transacylase (Q49X14) | ssp00061-Fatty acid biosynthesis ssp01212-Fatty acid metabolism ssp01100-Metabolic pathways | ssp01110-Biosynthesis of secondary metabolites ssp01130-Biosynthesis of antibiotics |
| Enoyl-[acyl-carrier-protein] reductase [NADPH](Q49WE0) | ssp00061-Fatty acid biosynthesis ssp01212-Fatty acid metabolism ssp01100-Metabolic pathways | ssp01110-Biosynthesis of secondary metabolites ssp01130-Biosynthesis of antibiotics |
| Chromosomal replication initiator protein DnaA (Q4A180) | ssp02020-Two-component system | |
| UDP-N-acetylenolpyruvoylglucosamine reductase (Q49VT7) | ssp00550-Peptidoglycan biosynthesis | |
| D-alanine-D-alanine ligase (Q49Z31) | ssp01502-Vancomycin resistance ssp00473-D-Alanine metabolism ssp00550-Peptidoglycan biosynthesis | |
| UDP-N-acetylmuramoylalanine-D-glutamate ligase (Q49WW5) | ssp00550-Peptidoglycan biosynthesis | |
| Alanine racemase (Q49Z24) | ssp01502-Vancomycin resistance |
| Protein ID | Subcellular Localization | Whether Druggable |
|---|---|---|
| Q4A180 | Cytoplasmic | No |
| Q49VT7 | Cytoplasmic | Yes |
| Q49Z31 | Cytoplasmic | Yes |
| Q49WW5 | Cytoplasmic | Yes |
| Q49Z24 | Cytoplasmic | Yes |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahid, F.; Ashfaq, U.A.; Saeed, S.; Munir, S.; Almatroudi, A.; Khurshid, M. In Silico Subtractive Proteomics Approach for Identification of Potential Drug Targets in Staphylococcus saprophyticus. Int. J. Environ. Res. Public Health 2020, 17, 3644. https://doi.org/10.3390/ijerph17103644
Shahid F, Ashfaq UA, Saeed S, Munir S, Almatroudi A, Khurshid M. In Silico Subtractive Proteomics Approach for Identification of Potential Drug Targets in Staphylococcus saprophyticus. International Journal of Environmental Research and Public Health. 2020; 17(10):3644. https://doi.org/10.3390/ijerph17103644
Chicago/Turabian StyleShahid, Farah, Usman Ali Ashfaq, Sania Saeed, Samman Munir, Ahmad Almatroudi, and Mohsin Khurshid. 2020. "In Silico Subtractive Proteomics Approach for Identification of Potential Drug Targets in Staphylococcus saprophyticus" International Journal of Environmental Research and Public Health 17, no. 10: 3644. https://doi.org/10.3390/ijerph17103644
APA StyleShahid, F., Ashfaq, U. A., Saeed, S., Munir, S., Almatroudi, A., & Khurshid, M. (2020). In Silico Subtractive Proteomics Approach for Identification of Potential Drug Targets in Staphylococcus saprophyticus. International Journal of Environmental Research and Public Health, 17(10), 3644. https://doi.org/10.3390/ijerph17103644






