Unexpected Prevalence of eae-Positive Escherichia coli in the Animas River, Durango, Colorado
Abstract
1. Introduction
2. Materials and Methods
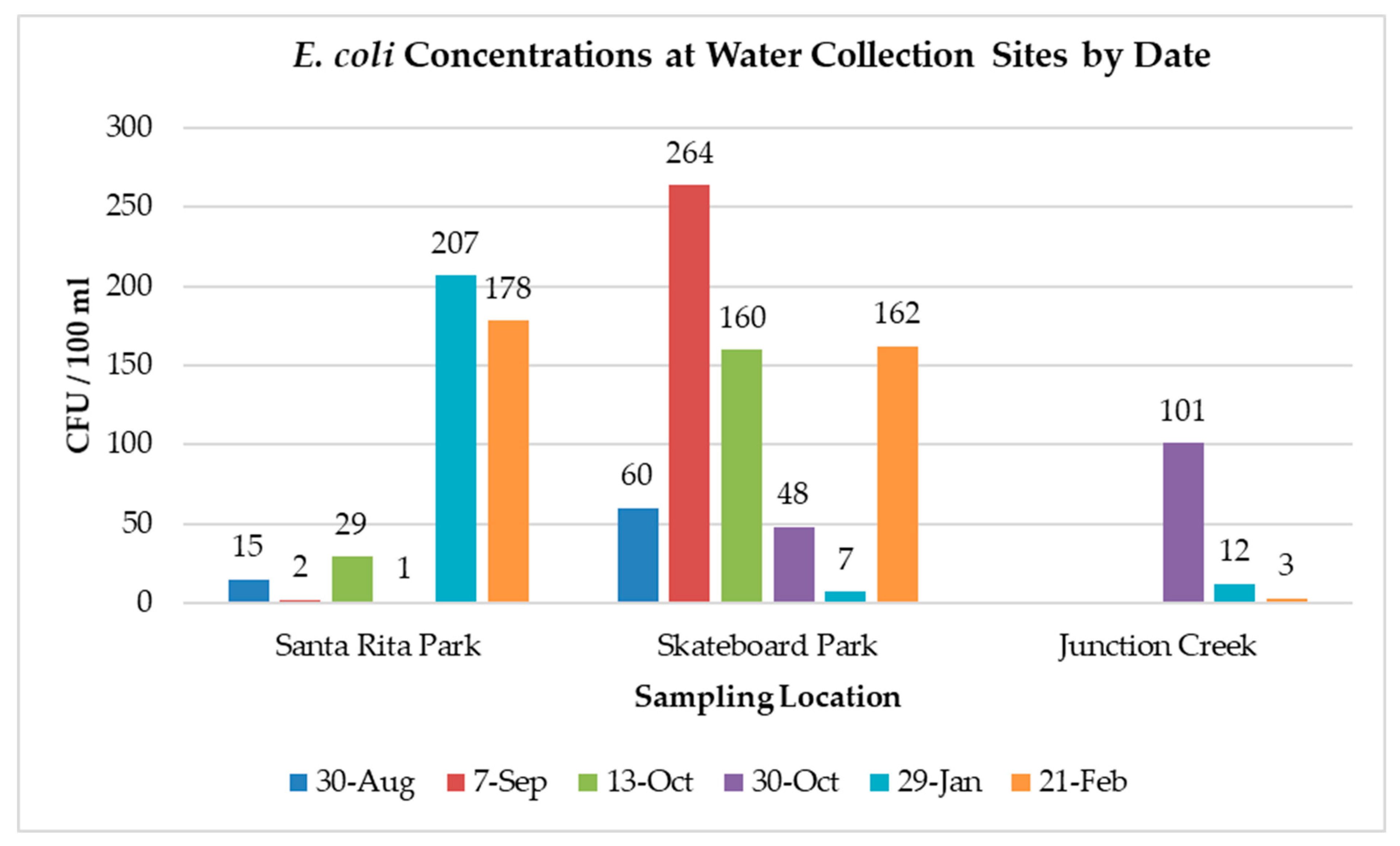
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Colorado Parks and Wildlife—2019 Colorado Fishing. 2019. Available online: https://cpw.state.co.us/Documents/RulesRegs/Brochure/fishing.pdf (accessed on 3 October 2019).
- RPI Consulting Inc. Economic Impacts of Whitewater Recreation—City of Durango. 2006. Available online: http://www.durangogov.org/DocumentCenter/View/57/FINAL-Economic-Impacts-of-Whitewater-Recreation---City-of-Durango-2006?bidId= (accessed on 3 October 2019).
- Church, S.E.; Kimball, B.A.; Fey, D.L.; Ferderer, D.A.; Yager, T.J.; Vaughn, R.B. Source, Transport, and Partitioning of Metals between Water, Colloids, and Bed Sediments of the Animas River, Colorado. U.S. Geological Survey Open-File Report 97-0151; 1997. Available online: http://pubs.usgs.gov/of/1997/ofr-97-0151/ (accessed on 3 October 2019).
- Courtney, L.A.; Clements, W.H. Assessing the influence of water and substratum quality on benthic macroinvertebrate communities in a metal-polluted stream: An experimental approach. Freshwater Biol. 2002, 47, 1766–1778. [Google Scholar] [CrossRef]
- Anderson, C.R. Effects of Mining on Benthic Macroinvertebrate Communities and Monitoring Strategies. In Integrated Investigations of Environmental Effects of Historical Mining in the Animas River Watershed, San Juan County, Colorado; Church, S.E., von Guerard, P., Finger, S.E., Eds.; Professional Paper 1651; U.S. Department of the Interior, U.S. Geological Survey: Reston, WV, USA, 1997. Available online: https://pubs.usgs.gov/pp/1651/downloads/Vol2_combinedChapters/vol2_chapE20.pdf (accessed on 3 October 2019).
- EPA. Draft Baseline Ecological Risk Assessment Upper Animas Mining District—Fact Sheet. 2015. Available online: https://www.epa.gov/sites/production/files/2015-06/documents/upper-animas-bera-fact-sheet-april-2015.pdf (accessed on 3 October 2019).
- EPA. Emergency Response to August 2015 Release from Gold King Mine. 2015. Available online: https://www.epa.gov/goldkingmine (accessed on 3 October 2019).
- RT News. Tribes Struggle with Toxic Spill as EPA is Accused of Deliberate Disaster. 2015. Available online: https://rt.com/usa/312499-toxic-spill-tribes-epa/ (accessed on 26 December 2019).
- The Guardian. Navajo Leader Feels Betrayed by EPA over ‘Contaminated’ Water Supply. 2015. Available online: https://www.theguardian.com/environment/2015/aug/21/navajo-nation-epa-contaminated-water (accessed on 3 October 2019).
- CBS News. EPA Says It Won’t Repay Claims for Spill That Caused Yellow Rivers. 2017. Available online: https://www.cbsnews.com/news/gold-king-mine-spill-colorado-rivers-epa-claims/ (accessed on 3 October 2019).
- Conservation Colorado. Two Years after the Gold King Spill. 2017. Available online: https://medium.com/@ConservationCO/two-years-after-the-gold-king-spill-2c8a83837d2b (accessed on 3 October 2019).
- Durango Herald. Is the Animas River Healthy? 2014. Available online: https://durangoherald.com/articles/72926-is-the-animas-river-healthy (accessed on 3 October 2019).
- Durango Herald. Animas River Health Alert Remains. 2011. Available online: https://durangoherald.com/articles/26537-animas-river-health-alert-remains (accessed on 3 October 2019).
- Albuquerque Journal. Sewage Leaks from Lift Station across Farmington Field into Animas River. 2014. Available online: https://www.abqjournal.com/494635/sewage-leaks-from-lift-station-across-farmington-field-into-animas-river.html (accessed on 3 October 2019).
- Durango Herald. Lightner Creek Mobile Home Park Owner Ordered to Shut down Sewage Pond. 2016. Available online: https://durangoherald.com/articles/103099 (accessed on 3 October 2019).
- EPA. Recreational Water Quality Criteria. EPA-820-F-12-061; 2012. Available online: https://www.epa.gov/wqc/2012-recreational-water-quality-criteria (accessed on 3 October 2019).
- Nataro, J.B.; Kaper, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar]
- Rangel, J.M.; Sparling, P.H.; Crowe, C.; Griffin, P.M.; Swerdlow, D.L. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 2015, 11, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Garmendia, J.; Frankel, G.; Crepin, V.F. Enteropathogenic and enterohemorrhagic Escherichia coli infections: Translocation, translocation, translocation. Infect. Immun. 2005, 73, 2573–2585. [Google Scholar] [CrossRef] [PubMed]
- Donnenberg, M.S.; Tzipori, S.; McKee, M.L.; O’Brien, A.D.; Alroy, J.; Kaper, J.B. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J. Clin. Investig. 1993, 92, 1418–1424. [Google Scholar] [CrossRef]
- WHO. Animal Waste, Water Quality, and Human Health. 2012. Available online: https://www.who.int/water_sanitation_health/publications/animal_waste/en/ (accessed on 3 October 2019).
- Callaway, T.R.; Carr, M.A.; Edrington, T.S.; Anderson, R.C.; Nisbet, D.J. Diet, Escherichia coli, and cattle: A review after 10 years. Curr. Issues Mol. Biol. 2009, 11, 67–79. [Google Scholar]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B.A. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef]
- Chee-Sanford, J.C.; Mackie, R.I.; Koike, S.; Krapac, I.G.; Lin, Y.F.; Yannarell, A.C.; Maxwell, S.; Aminov, R.I. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Qual. 2009, 38, 1086–1108. [Google Scholar] [CrossRef]
- McEachran, A.D.; Blackwell, B.R.; Hanson, J.D.; Wooten, K.J.; Mayer, G.D.; Cox, S.B.; Smith, P.N. Antibiotics, bacteria, and antibiotic resistance genes: Aerial transport from cattle feed yards via particulate matter. Environ. Health Perspect. 2015, 123, 337–343. [Google Scholar] [CrossRef]
- Hamner, S.; Broadaway, S.C.; Berg, E.; Stettner, S.; Pyle, B.H.; Big Man, N.; Old Elk, J.; Eggers, M.J.; Doyle, J.; Kindness, L.; et al. Detection and source tracking of Escherichia coli, harboring intimin and Shiga toxin genes, isolated from the Little Bighorn River, Montana. Int. J. Environ. Health Res. 2014, 24, 341–362. [Google Scholar] [CrossRef] [PubMed]
- Dockins, W.S.; McFeters, G.A. Fecal coliform elevated-temperature test: A physiological basis. Appl. Environ. Microbiol. 1978, 36, 341–348. [Google Scholar] [PubMed]
- Feng, P.; Weagent, S.D.; Grant, M.A.; Burkhardt, W. Bacteriological Analytical Method (BAM) Online: Enumeration of Escherichia Coli and the Coliform Bacteria. 2002. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-4-enumeration-escherichia-coli-and-coliform-bacteria (accessed on 3 October 2019).
- Ram, J.L.; Ritchie, R.P.; Fang, J.; Gonzales, F.S.; Selegean, J.P. Sequence-based source tracking of Escherichia coli based on genetic diversity of ß-glucuronidase. J. Environ. Qual. 2004, 33, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Deokule, J.S.; Garg, P.; Bhattacharya, S.K.; Nandy, S.K.; Nair, G.B.; Yamasaki, S.; Takeda, Y.; Ramamurthy, T. Concomitant infection of enterotoxigenic Escherichia coli in an outbreak of cholera caused by Vibrio cholerae O1 and O139 in Ahmedabad, India. J. Clin. Microbiol. 2001, 39, 3241–3246. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Path. 1966, 36, 493–496. [Google Scholar] [CrossRef]
- EPA. Quantitative Microbial Risk Assessment to Estimate Illness in Fresh Water Impacted by Agricultural Animal Sources of Fecal Contamination. EPA 822-R-10-005; 2010. Available online: https://www.epa.gov/sites/production/files/2015-11/documents/quantitiave-microbial-risk-fecal.pdf (accessed on 3 October 2019).
- Feng, P.; Lum, R.; Chang, G.W. Identification of uidA gene sequences in ß-D-glucuronidase-negative Escherichia coli. Appl. Environ. Microbiol. 1991, 57, 320–323. [Google Scholar]
- Martins, M.T.; Rivera, I.G.; Clark, D.L.; Stewart, M.H.; Wolfe, R.L.; Olson, B.H. Distribution of uidA gene sequences in Escherichia coli isolates in water sources and comparison with the expression of ß-glucuronidase activity in 4-methylumbelliferyl-ß-D-glucuronide media. Appl. Environ. Microbiol. 1993, 59, 2271–2276. [Google Scholar]
- EPA. Guidelines Establishing Test Procedures for the Analysis of Pollutants; Analytical Methods for Biological Pollutants in Ambient Water; Proposed Rule. 40 CFR Part 136. 2001. Available online: https://www.govinfo.gov/content/pkg/FR-2001-08-30/pdf/01-21813.pdf (accessed on 30 November 2019).
- Monday, S.R.; Whittam, T.S.; Feng, P.C.H. Genetic and evolutionary analysis of mutations in the gusA gene that cause absence of ß-glucuronidase activity in Escherichia coli O157:H7. J. Infect. Dis. 2001, 184, 918–921. [Google Scholar] [CrossRef]
- Díaz, S.; Vidal, D.; Herrera-León, S.; Sánchez, S. Sorbitol-fermenting, ß-glucuronidase-positive, Shiga toxin-negative Escherichia coli O157:H7 in free-ranging deer in south-central Spain. Foodborne Pathog. Dis. 2011, 8, 1313–1315. [Google Scholar] [CrossRef]
- Ogura, Y.; Seto, K.; Morimoto, Y.; Nakamura, K.; Sato, M.P.; Gotoh, Y.; Itoh, T.; Toyoda, A.; Ohnishi, M.; Hayashi, T. Genomic characterization of ß-glucuronidase-positive Escherichia coli O157:H7 producing Stx2a. Emerg. Infect. Dis. 2018, 24, 2219–2227. [Google Scholar] [CrossRef]
- Batchelor, M.; Prasannan, S.; Daniell, S.; Reece, S.; Connerton, I.; Bloomberg, G.; Dougan, G.; Frankel, G.; Matthews, S. Structural basis for recognition of the translocated intimin receptor (Tir) by intimin from enteropathogenic Escherichia coli. EMBO J. 2000, 19, 2452–2464. [Google Scholar] [CrossRef] [PubMed]
- WHO. Antimicrobial Resistance: Global Report on Surveillance. 2014. Available online: https://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed on 3 October 2019).
- Kuhnert, P.; Boerlin, P.; Frey, J. Target genes for virulence assessment of Escherichia coli isolates from water, food, and the environment. Microbiol. Rev. 2000, 24, 107–177. [Google Scholar]
- Radhouani, H.; Silva, N.; Poeta, P.; Torres, C.; Correia, S.; Igrejas, G. Potential impact of antimicrobial resistance in wildlife, environment, and human health. Front. Microbiol. 2014, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Gennari, M.; Ghidini, V.; Caburlotto, G.; Lleo, M.M. Virulence genes and pathogenicity islands in environmental Vibrio strains nonpathogenic to humans. Microbiol. Ecol. 2012, 82, 563–573. [Google Scholar] [CrossRef] [PubMed]
- EPA. Microbial Source Tracking Document. EPA/600/R-05/064; 2005. Available online: https://nepis.epa.gov/Adobe/PDF/2000D20V.PDF (accessed on 3 October 2019).

| Site Name and Brief Description | Geographic Coordinates | Sampling Dates |
|---|---|---|
| Animas River at Santa Rita Park, “south” in Durango | 37°15′35.18″ N 107°52′41.32″ W | Multiple dates during 2014–2019 |
| Skate Park drain feeding into Animas River, Durango | 37°16′39.70″ N 107°52′57.85″ W | Multiple dates during 2014–2019 |
| Junction Creek terminus near confluence with Animas River, Durango | 37°17′7.82″ N 107°52′20.17″ W | Multiple dates during 2014–2019 |
| “West” drain feeding into Animas River, Durango | 37°17′13.13″ N 107°52′18.61″ W | October 2015 only |
| “East” drain feeding into Animas River, Durango | 37°17′18.88″ N 107°52′13.29″ W | October 2015 only |
| Animas River “middle” site in Durango | 37°16′41.39″ N 107°52′53.87″ W | October 2015 only |
| Animas River “north” in Durango |
37°17′8.15″ N 107°52′19.28″ W | October 2015 only |
| Animas River, Baker’s Bridge, upstream and north of Durango |
37°27′30.08″ N 107°47′58.90″ W | October 2015 only |
| Junction Creek trailhead, upstream of Durango city limits |
37°19′51.89″ N 107°54′11.90″ W | October 2015 only |
| Lightner Creek terminus near confluence with Animas River |
37°16′05.10″ N 107°53′09.80″ W | 2018–2019 |
| Animas River Santa Rita Park Isolates | Skate Park Drain Isolates | Junction Creek Isolates | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| Ampicillin | ||||||||||||||||||
| Ciprofloxacin | ||||||||||||||||||
| Penicillin | ||||||||||||||||||
| Streptomycin | ||||||||||||||||||
| Tetracycline | ||||||||||||||||||
| Vancomycin | ||||||||||||||||||
| Site Name and Brief Description | E. coli Concentrations for Samples Collected on 16 October 2015 (Mean of Triplicate Counts) | Number of Isolates Positive for eae out of Total Isolates Tested (Sampling Conducted 30 October 2015) |
|---|---|---|
| Skate Park drain feeding into Animas River, Durango | 91 CFU per 100 mL | 2 of 19 |
| “West” drain feeding into Animas River, Durango | 149 CFU per 100 mL | 0 of 16 |
| “East” drain feeding into Animas River, Durango | Too numerous to count (about 630 colonies per 100 mL) | 3 of 16 |
| Animas River at Santa Rita Park, “south” in Durango | 84 CFU per 100 mL | 0 of 12 |
| Animas River “middle” site in Durango | 18 CFU per 100 mL | 0 of 2 |
| Animas River “north” in Durango | 18 CFU per 100 mL | Not tested |
| Animas River, Baker’s Bridge, upstream and north of Durango | 1 CFU per 100 mL | Not tested |
| Junction Creek trailhead, upstream of Durango city limits | 7 CFU per 100 mL | 0 of 2 |
| Junction Creek terminus near confluence with Animas River, Durango | Too numerous to count (about 280 colonies per 100 mL) | 0 of 10 |
| Site Name and Brief Description, and Dates of Testing | E. coli Concentrations for (Mean of Triplicate Counts) | Number of Isolates Positive for eae out of Total Isolates Tested |
|---|---|---|
| Junction Creek near confluence with Animas River, Durango (October 2018) | 8 CFU per 100 mL | 0 of 9 |
| Skate Park drain feeding into Animas River, Durango (October 2018) | 163 CFU per 100 mL | 2 of 15 |
| Lightner Creek near confluence with Animus River, Durango (October 2018) | 83 CFU per 100 mL | 1 of 10 |
| Animas River at Santa Rita Park, “south” in Durango (October 2018) | 226 CFU per 100 mL | 9 of 12 |
| Junction Creek near confluence with Animas River, Durango (March 2019) | 298 CFU per 100 mL | 0 of 4 |
| Skate Park drain feeding into Animas River, Durango (March 2019) | 83 CFU per 100 mL | 0 of 18 |
| Lightner Creek near confluence with Animus River, Durango (March 2019) | 108 CFU per 100 mL | 3 of 12 |
| Animas River at Santa Rita Park, “south” in Durango (March 2019) | 69 CFU per 100 mL | 2 of 11 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamner, S.; Fenster, S.D.; Nance, B.T.; McLain, K.A.; Parrish-Larson, K.S.; Morrow, M.W.; Ford, T.E. Unexpected Prevalence of eae-Positive Escherichia coli in the Animas River, Durango, Colorado. Int. J. Environ. Res. Public Health 2020, 17, 195. https://doi.org/10.3390/ijerph17010195
Hamner S, Fenster SD, Nance BT, McLain KA, Parrish-Larson KS, Morrow MW, Ford TE. Unexpected Prevalence of eae-Positive Escherichia coli in the Animas River, Durango, Colorado. International Journal of Environmental Research and Public Health. 2020; 17(1):195. https://doi.org/10.3390/ijerph17010195
Chicago/Turabian StyleHamner, Steve, Steven D. Fenster, Benjamin T. Nance, Katherine A. McLain, Kami S. Parrish-Larson, Michael W. Morrow, and Timothy E. Ford. 2020. "Unexpected Prevalence of eae-Positive Escherichia coli in the Animas River, Durango, Colorado" International Journal of Environmental Research and Public Health 17, no. 1: 195. https://doi.org/10.3390/ijerph17010195
APA StyleHamner, S., Fenster, S. D., Nance, B. T., McLain, K. A., Parrish-Larson, K. S., Morrow, M. W., & Ford, T. E. (2020). Unexpected Prevalence of eae-Positive Escherichia coli in the Animas River, Durango, Colorado. International Journal of Environmental Research and Public Health, 17(1), 195. https://doi.org/10.3390/ijerph17010195





