The Gut-Microbiome in Gulf War Veterans: A Preliminary Report
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Subjects
2.2. Demographics, Deployment Exposures and Health Symptom Surveys
2.3. Plasma Cytokine Analyses
2.4. Stool Sample Collection
2.5. Gut Microbiome Analysis
2.5.1. Sample Isolation
2.5.2. Library Preparation
2.5.3. Profiling Method
2.5.4. Data Analysis
3. Results
3.1. Human Subjects
3.1.1. GWIC Full Study Cohort
3.1.2. GWIC Microbiome Pilot Call Back Study Cohort
3.2. Cytokine Results
3.3. Gut Microbiome Results
3.3.1. Sample Diversity
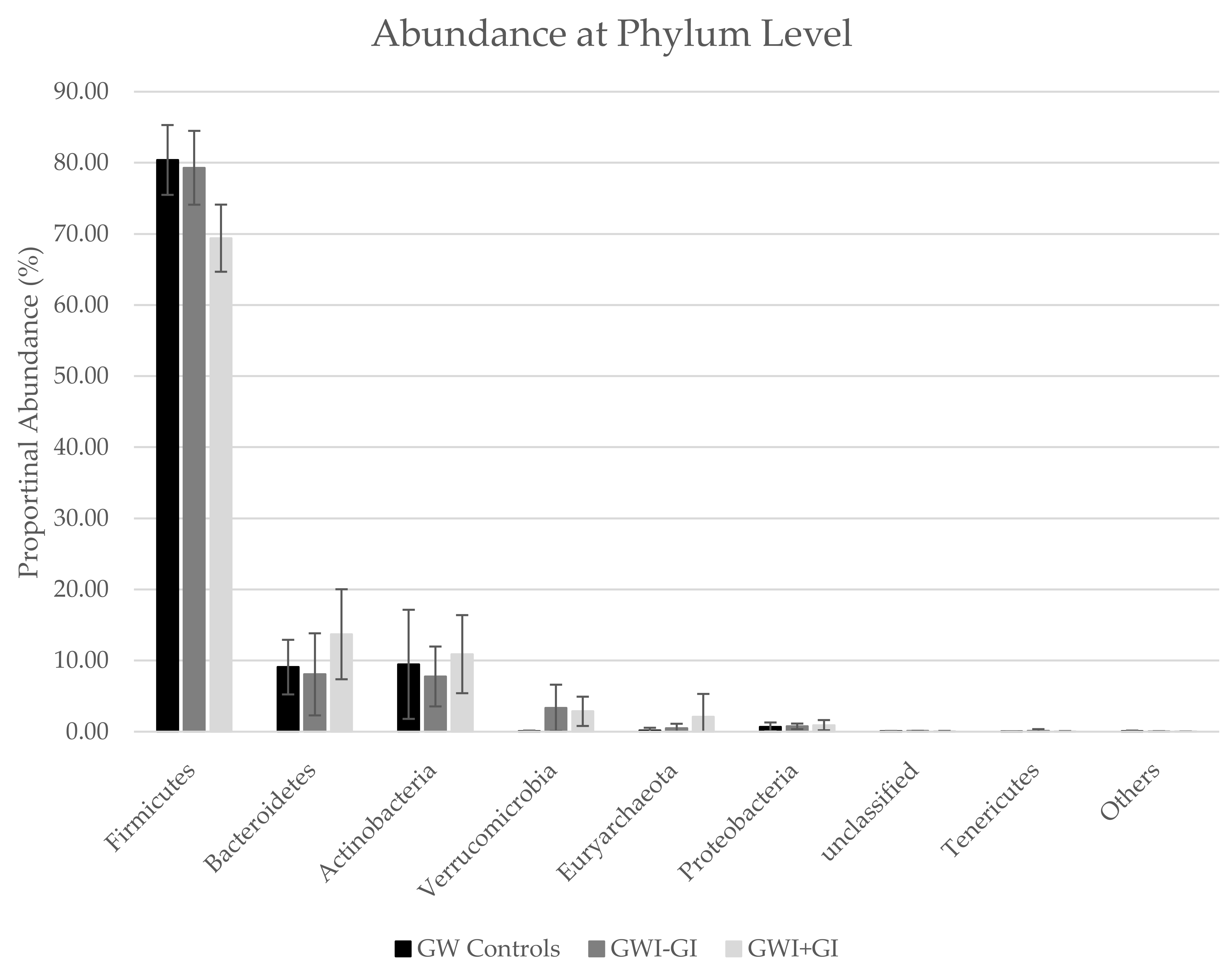
3.3.2. Phyla Distribution
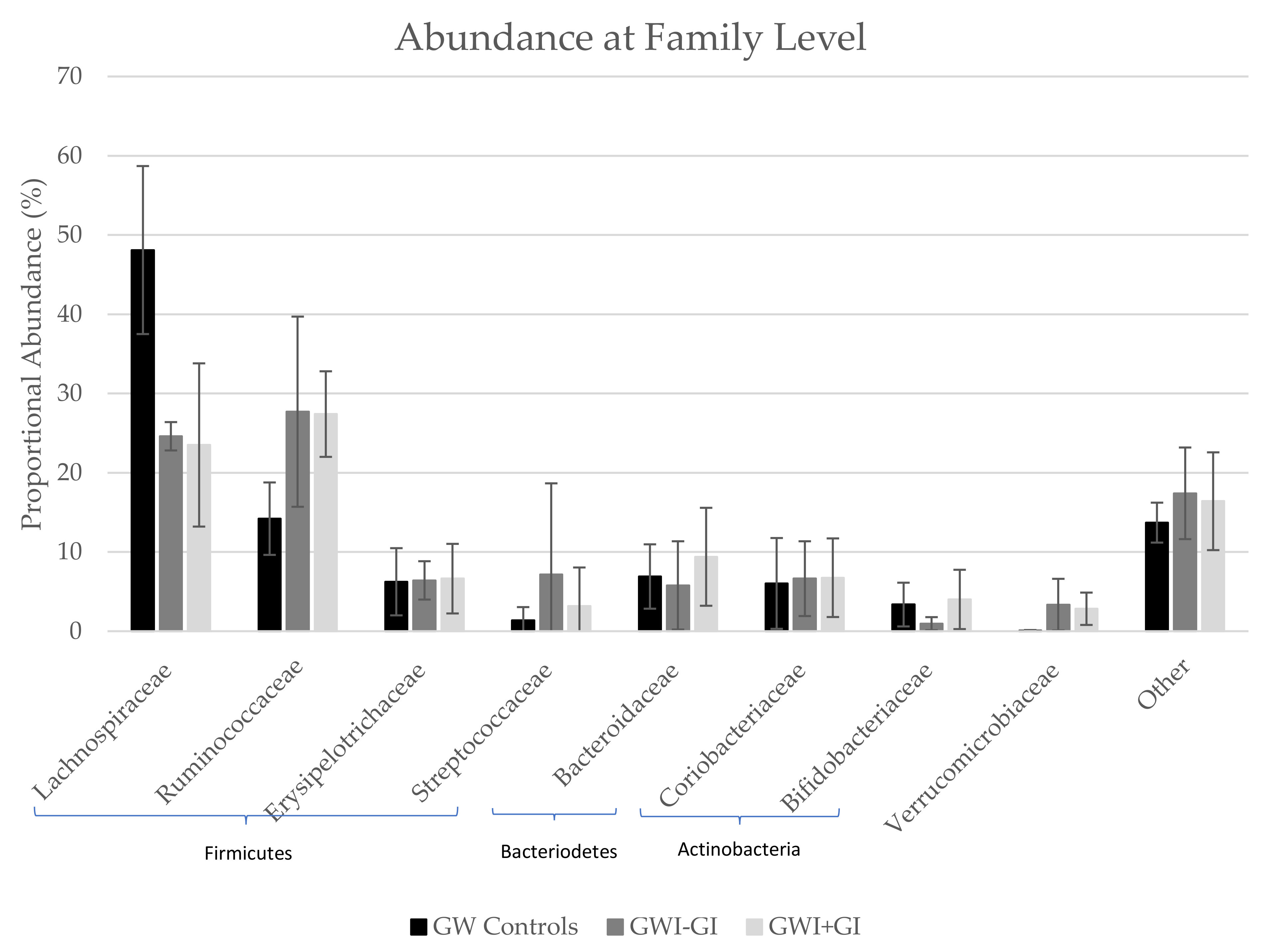
3.3.3. Family Distribution
3.3.4. Genus Level
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- White, R.F.; Steele, L.; O’Callaghan, J.P.; Sullivan, K.; Binns, J.H.; Golomb, B.A.; Bloom, F.E.; Bunker, J.A.; Crawford, F.; Graves, J.C.; et al. Recent research on Gulf War illness and other health problems in veterans of the 1991 Gulf War: Effects of toxicant exposures during deployment. Cortex 2016, 74, 449–475. [Google Scholar] [CrossRef] [PubMed]
- Research Advisory Committee on Gulf War Veterans’ Illnesses. Gulf War Illness and the Health of Gulf War Veterans: Scientific Findings and Recommendations; U.S. Government Printing Office: Washington, DC, USA, 2008.
- Sullivan, K.; Krengel, M.; Bradford, W.; Stone, C.; Thompson, T.A.; Heeren, T.; White, R.F. Neuropsychological functioning in military pesticide applicators from the Gulf War: Effects on information processing speed, attention and visual memory. Neurotoxicol. Teratol. 2018, 65, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Golomb, B.A. Acetylcholinesterase inhibitors and Gulf War illnesses. Proc. Natl. Acad. Sci. USA 2008, 105, 4295–4300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, K.; Krengel, M.; Proctor, S.P.; Devine, S.; Heeren, T.; White, R.F. Cognitive Functioning in Treatment-Seeking Gulf War Veterans: Pyridostigmine Bromide Use and PTSD. J. Psychopathol. Behav. Assess. 2003, 25, 95–103. [Google Scholar] [CrossRef]
- Zundel, C.G.; Krengel, M.H.; Heeren, T.; Yee, M.K.; Grasso, C.M.; Janulewicz Lloyd, P.A.; Coughlin, S.S.; Sullivan, K. Rates of Chronic Medical Conditions in 1991 Gulf War Veterans Compared to the General Population. Int. J. Environ. Res. Public Health 2019, 16, 949. [Google Scholar] [CrossRef] [PubMed]
- Steele, L. Prevalence and Patterns of Gulf War Illness in Kansas Veterans: Association of Symptoms with Characteristics of Person, Place, and Time of Military Service. Am. J. Epidemiol. 2000, 152, 992–1002. [Google Scholar] [CrossRef] [PubMed]
- Maule, A.L.; Janulewicz, P.A.; Sullivan, K.A.; Krengel, M.H.; Yee, M.K.; McClean, M.; White, R.F. Meta-analysis of self-reported health symptoms in 1990–1991 Gulf War and Gulf War-era veterans. BMJ Open 2018, 8, e016086. [Google Scholar] [CrossRef] [PubMed]
- Smylie, A.L.; Broderick, G.; Fernandes, H.; Razdan, S.; Barnes, Z.; Collado, F.; Sol, C.; Fletcher, M.A.; Klimas, N. A comparison of sex-specific immune signatures in Gulf War illness and chronic fatigue syndrome. BMC Immunol. 2013, 14, 29. [Google Scholar] [CrossRef]
- O’Callaghan, J.P.; Kelly, K.A.; Locker, A.R.; Miller, D.B.; Lasley, S.M. Corticosterone primes the neuroinflammatory response to DFP in mice: Potential animal model of Gulf War Illness. J. Neurochem. 2015, 133, 708–721. [Google Scholar] [CrossRef]
- Koo, B.-B.; Michalovicz, L.T.; Calderazzo, S.; Kelly, K.A.; Sullivan, K.; Killiany, R.J.; O’Callaghan, J.P. Corticosterone potentiates DFP-induced neuroinflammation and affects high-order diffusion imaging in a rat model of Gulf War Illness. Brain Behav. Immun. 2018, 67, 42–46. [Google Scholar] [CrossRef]
- Madhu, L.N.; Attaluri, S.; Kodali, M.; Shuai, B.; Upadhya, R.; Gitai, D.; Shetty, A.K. Neuroinflammation in Gulf War Illness is linked with HMGB1 and complement activation, which can be discerned from brain-derived extracellular vesicles in the blood. Brain Behav. Immun. 2019, 81, 430–443. [Google Scholar] [CrossRef]
- Joshi, U.; Pearson, A.; Evans, J.E.; Langlois, H.; Saltiel, N.; Ojo, J.; Klimas, N.; Sullivan, K.; Keegan, A.P.; Oberlin, S.; et al. A permethrin metabolite is associated with adaptive immune responses in Gulf War Illness. Brain Behav. Immun. 2019, 81, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Parihar, V.K.; Hattiangady, B.; Shuai, B.; Shetty, A.K. Mood and Memory Deficits in a Model of Gulf War Illness Are Linked with Reduced Neurogenesis, Partial Neuron Loss, and Mild Inflammation in the Hippocampus. Neuropsychopharmacology 2013, 38, 2348–2362. [Google Scholar] [CrossRef]
- Banks, C.N.; Lein, P.J. A review of experimental evidence linking neurotoxic organophosphorus compounds and inflammation. NeuroToxicology 2012, 33, 575–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alhasson, F.; Das, S.; Seth, R.; Dattaroy, D.; Chandrashekaran, V.; Ryan, C.N.; Chan, L.S.; Testerman, T.; Burch, J.; Hofseth, L.J.; et al. Altered gut microbiome in a mouse model of Gulf War Illness causes neuroinflammation and intestinal injury via leaky gut and TLR4 activation. PLoS ONE 2017, 12, e0172914. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, S.; Fried, D.E.; Grubišić, V.; McClain, J.L.; Gulbransen, B.D. Gastrointestinal neuroimmune disruption in a mouse model of Gulf War illness. FASEB J. 2019, 33, 6168–6184. [Google Scholar] [CrossRef]
- Emmerich, T.; Zakirova, Z.; Klimas, N.; Sullivan, K.; Shetty, A.K.; Evans, J.E.; Ait-Ghezala, G.; Laco, G.S.; Hattiangady, B.; Shetty, G.A.; et al. Phospholipid profiling of plasma from GW veterans and rodent models to identify potential biomarkers of Gulf War Illness. PLoS ONE 2017, 12, e0176634. [Google Scholar] [CrossRef] [PubMed]
- Golomb, B.A.; Allison, M.; Koperski, S.; Koslik, H.J.; Devaraj, S.; Ritchie, J.B. Coenzyme Q10 benefits symptoms in Gulf War veterans: Results of a randomized double-blind study. Neural. Comput. 2014, 26, 2594–2651. [Google Scholar] [CrossRef]
- Golomb, B.A. Oxidative Stress and Mitochondrial Injury in Chronic Multisymptom Conditions: From Gulf War Illness to Autism Spectrum Disorder. Nat. Preced. 2012. [Google Scholar] [CrossRef]
- Koslik, H.J.; Hamilton, G.; Golomb, B.A. Mitochondrial Dysfunction in Gulf War Illness Revealed by 31Phosphorus Magnetic Resonance Spectroscopy: A Case-Control Study. PLoS ONE 2014, 9, e92887. [Google Scholar] [CrossRef]
- Zhou, Q.; Verne, M.L.; Zhang, B.; Verne, G.N. Evidence for Somatic Hypersensitivity in Veterans With Gulf War Illness and Gastrointestinal Symptoms. Clin. J. Pain 2018, 34, 944. [Google Scholar] [CrossRef] [PubMed]
- Koch, T.R.; Emory, T.S. Evaluation of Chronic Gastrointestinal Symptoms following Persian Gulf War Exposure. Mil. Med. 2005, 170, 696–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunphy, R.C.; Bridgewater, L.; Price, D.D.; Robinson, M.E.; Zeilman, C.J.; Verne, G.N. Visceral and cutaneous hypersensitivity in Persian Gulf war veterans with chronic gastrointestinal symptoms. Pain 2003, 102, 79–85. [Google Scholar] [CrossRef]
- Rea, K.; Dinan, T.G.; Cryan, J.F. The microbiome: A key regulator of stress and neuroinflammation. Neurobiol. Stress 2016, 4, 23–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seth, R.K.; Kimono, D.; Alhasson, F.; Sarkar, S.; Albadrani, M.; Lasley, S.K.; Horner, R.; Janulewicz, P.; Nagarkatti, M.; Nagarkatti, P.; et al. Increased butyrate priming in the gut stalls microbiome associated-gastrointestinal inflammation and hepatic metabolic reprogramming in a mouse model of Gulf War Illness. Toxicol. Appl. Pharmacol. 2018, 350, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Janulewicz, P.; Krengel, M.; Quinn, E.; Heeren, T.; Toomey, R.; Killiany, R.; Zundel, C.; Ajama, J.; O’Callaghan, J.; Steele, L.; et al. The Multiple Hit Hypothesis for Gulf War Illness: Self-Reported Chemical/Biological Weapons Exposure and Mild Traumatic Brain Injury. Brain Sci. 2018, 8, 198. [Google Scholar] [CrossRef] [PubMed]
- Proctor, S. Health status of Persian Gulf War veterans: Self-reported symptoms, environmental exposures and the effect of stress. Int. J. Epidemiol. 1998, 27, 1000–1010. [Google Scholar] [CrossRef]
- Proctor, S.P. Development of a Structured Neurotoxicant Assessment Checklist (SNAC) for Clinical Use in Veteran Populations; Department of Veterans Affairs: Washington, DC, USA, 2006.
- Smets, E.M.A.; Garssen, B.; Bonke, B.; De Haes, J.C.J.M. The multidimensional Fatigue Inventory (MFI) psychometric qualities of an instrument to assess fatigue. J. Psychosom. Res. 1995, 39, 315–325. [Google Scholar] [CrossRef] [Green Version]
- Buysse, D.J.; Reynolds, C.F.; Monk, T.H.; Berman, S.R.; Kupfer, D.J. The Pittsburgh sleep quality index: A new instrument for psychiatric practice and research. Psychiatry Res. 1989, 28, 193–213. [Google Scholar] [CrossRef]
- Melzack, R. The McGill Pain Questionnaire: Major properties and scoring methods. PAIN 1975, 1, 277–299. [Google Scholar] [CrossRef]
- Fletcher, M.A.; Zeng, X.R.; Barnes, Z.; Levis, S.; Klimas, N.G. Plasma cytokines in women with chronic fatigue syndrome. J. Transl. Med. 2009, 7, 96. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. Available online: https://www.hindawi.com/journals/bmri/2017/9351507/ (accessed on 20 July 2019).
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microb. 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, S.H.; Belenguer, A.; Holtrop, G.; Johnstone, A.M.; Flint, H.J.; Lobley, G.E. Reduced Dietary Intake of Carbohydrates by Obese Subjects Results in Decreased Concentrations of Butyrate and Butyrate-Producing Bacteria in Feces. Appl. Environ. Microbiol. 2007, 73, 1073–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.D.; Wildenberg, M.E.; van den Brink, G.R. Mechanism of Action of Anti-TNF Therapy in Inflammatory Bowel Disease. J. Crohns Colitis 2016, 10, 989–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microb. 2019, 4, 623. [Google Scholar] [CrossRef]
- Picchianti-Diamanti, A.; Rosado, M.M.; D’Amelio, R. Infectious Agents and Inflammation: The Role of Microbiota in Autoimmune Arthritis. Front. Microbiol. 2018, 8, 2696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Azzouz, D.; Omarbekova, A.; Heguy, A.; Schwudke, D.; Gisch, N.; Rovin, B.H.; Caricchio, R.; Buyon, J.P.; Alekseyenko, A.V.; Silverman, G.J. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 2019, 78, 947–956. [Google Scholar] [CrossRef] [Green Version]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Acharya, C.; Bajaj, J.S. Gut Microbiota and Complications of Liver Disease. Gastroenterol. Clin. North Am. 2017, 46, 155–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism 2013, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
| Characteristic | GW Control n = 5 | GWI-GI n = 3 | GWI+GI n = 5 | p-Value |
|---|---|---|---|---|
| Age, years [Mean (SD 1)] | 52.8 (6.7) | 63.2 (15.5) | 53.3 (7.2) | 0.296 |
| Height, in [Mean (SD 1)] | 69.8 (1.0) | 67.5 (4.4) | 66.8 (2.7) | 0.238 |
| Weight, lbs [Mean (SD 1)] | 198.5 (18.3) | 207.0 (24.3) | 217.0 (99.7) | 0.904 |
| Gender [N (%)] | 0.420 | |||
| Male | 5 (100) | 2 (66.7) | 4 (80) | |
| Female | 0 (0) | 1 (33.3) | 1 (20) | |
| Race/Ethnicity [N (%)] | 0.420 | |||
| Black | 0 (0) | 0 (0) | 1 (20) | |
| White | 5 (100) | 3 (100) | 4 (80) | |
| Education [N (%)] | 0.482 | |||
| High School plus other training (technical/trade) | 1 (20) | 0 (0) | 0 (0) | |
| Associates Degree or 2 years of College | 0 (0) | 0 (0) | 2 (40) | |
| Some College | 0 (0) | 0 (0) | 1 (20) | |
| Bachelor’s Degree | 2 (40) | 1 (33.3) | 0 (0) | |
| Advanced or Professional Degree | 1 (20) | 2 (66.7) | 2 (40) | |
| Highest Grade [Mean (SD 1)] | 15 (2) | 15 (1) | 16 (3) | 0.957 |
| McGill Pain Score [Mean (SD)] | 12 (8) | 32 (7) | 39 (16) | 0.021 |
| Multi-dimensional Fatigue Inventory (MFI-20) [Mean (SD)] | 40 (8) | 53 (7) | 72 (10) | 0.0005 |
| Pittsburgh Sleep Quality Index (PSQI) [Mean (SD)] | 8 (3) | 7 (2) | 14 (2) | 0.003 |
| Body Mass Index (BMI) [Mean (SD)] | 28.6 (2.5) | 31.9 (0.7) | 34.0 (14.2) | 0.664 |
| Self-reported high sugar or diabetes [N (%)] No Yes | 5 (100.00)0 (0.0) | 2 (66.7)1 (33.3) | 4 (80.0)1 (20.0) | 0.420 |
| Exposed to chemical or biological warfare agents during military service [N (%)] | 0.049 | |||
| No | 2 (40.0) | 1 (33.3) | 0 (0.0) | |
| Yes | 0 (0.0) | 0 (0.0) | 4 (80.0) | |
| Not Sure | 3 (60.0) | 2 (66.7) | 1 (20.0) | |
| Taken pyridostigmine bromide (anti-nerve agent) pills [N (%)] | 0.420 | |||
| No | 1 (20.0) | 1 (33.3) | 0 (0.0) | |
| Yes | 4 (80.0) | 2 (66.7) | 5 (100.0) | |
| Wore a uniform treated with pesticides [N (%)] | 0.120 | |||
| No | 5 (100.0) | 2 (66.7) | 2 (40.0) | |
| Yes | 0 (0.0) | 1 (33.3) | 3 (60.0) | |
| Saw the area in which you lived fogged or sprayed with pesticides [N (%)] | 0.228 | |||
| No | 3 (60.0) | 3 (100.0) | 2 (40.0) | |
| Yes | 2 (40.0) | 0 (0.0) | 1 (20.0) | |
| Not Sure | 0 (0.0) | 0 (0.0) | 2 (40.0) |
| GW Controls | GWI–GI | GWI+GI | ||||
|---|---|---|---|---|---|---|
| Cytokine | Mean | SD | Mean | SD | Mean | SD |
| TNF-RI * | 452.6 | 183.1 | 819.2 | 250.3 | 852.7 | 244.0 |
| TNF-RII | 721.2 | 79.6 | 876.3 | 62.7 | 541.6 | 122.3 |
| GW Controls | GWI–GI | GWI+GI | ||||
|---|---|---|---|---|---|---|
| Family | Mean | SD | Mean | SD | Mean | SD |
| OTU Richness | 415 | 83.1 | 576 | 12.9 | 501 | 56.4 |
| Shannon Index | 3.79 | 0.23 | 4.03 | 0.15 | 3.94 | 0.16 |
| GW Controls | GWI–GI | GWI+GI | ||||
|---|---|---|---|---|---|---|
| Phylum | Mean | SD | Mean | SD | Mean | SD |
| Firmicutes | 80.40 | 4.91 | 79.30 | 5.19 | 69.40 | 4.72 |
| Bacteroidetes | 9.08 | 3.84 | 8.06 | 5.77 | 13.70 | 6.33 |
| Actinobacteria | 9.47 | 7.67 | 7.76 | 4.21 | 10.90 | 5.49 |
| Verrucomicrobia | 0.05 | 0.09 | 3.35 | 3.26 | 2.86 | 2.06 |
| Euryarchaeota | 0.17 | 0.38 | 0.49 | 0.63 | 2.09 | 3.22 |
| Proteobacteria | 0.68 | 0.61 | 0.75 | 0.39 | 0.91 | 0.72 |
| unclassified | 0.08 | 0.02 | 0.10 | 0.02 | 0.09 | 0.02 |
| Tenericutes | 0.00 | 0.00 | 0.13 | 0.22 | 0.02 | 0.06 |
| Others | 0.06 | 0.10 | 0.04 | 0.03 | 0.02 | 0.02 |
| GW Controls | GWI–GI | GWI+GI | ||||
|---|---|---|---|---|---|---|
| Family | Mean | SD | Mean | SD | Mean | SD |
| Lachnospiraceae | 48.10 | 10.60 | 24.60 | 1.79 | 23.50 | 10.30 |
| Ruminococcaceae | 14.20 | 4.57 | 27.70 | 12.00 | 27.40 | 5.40 |
| Erysipelotrichaceae | 6.24 | 4.24 | 6.41 | 2.42 | 6.63 | 4.39 |
| Streptococcaceae | 1.37 | 1.67 | 7.16 | 11.50 | 3.17 | 4.87 |
| Bacteroidaceae | 6.90 | 4.06 | 5.78 | 5.57 | 9.39 | 6.18 |
| Coriobacteriaceae | 6.03 | 5.73 | 6.63 | 4.72 | 6.75 | 4.96 |
| Bifidobacteriaceae | 3.36 | 2.76 | 0.95 | 0.83 | 4.01 | 3.74 |
| Verrucomicrobiaceae | 0.05 | 0.09 | 3.35 | 3.26 | 2.83 | 2.04 |
| Other | 13.70 | 2.52 | 17.40 | 5.78 | 16.40 | 6.17 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janulewicz, P.A.; Seth, R.K.; Carlson, J.M.; Ajama, J.; Quinn, E.; Heeren, T.; Klimas, N.; Lasley, S.M.; Horner, R.D.; Sullivan, K.; et al. The Gut-Microbiome in Gulf War Veterans: A Preliminary Report. Int. J. Environ. Res. Public Health 2019, 16, 3751. https://doi.org/10.3390/ijerph16193751
Janulewicz PA, Seth RK, Carlson JM, Ajama J, Quinn E, Heeren T, Klimas N, Lasley SM, Horner RD, Sullivan K, et al. The Gut-Microbiome in Gulf War Veterans: A Preliminary Report. International Journal of Environmental Research and Public Health. 2019; 16(19):3751. https://doi.org/10.3390/ijerph16193751
Chicago/Turabian StyleJanulewicz, Patricia A., Ratanesh K. Seth, Jeffrey M. Carlson, Joy Ajama, Emily Quinn, Timothy Heeren, Nancy Klimas, Steven M. Lasley, Ronnie D. Horner, Kimberly Sullivan, and et al. 2019. "The Gut-Microbiome in Gulf War Veterans: A Preliminary Report" International Journal of Environmental Research and Public Health 16, no. 19: 3751. https://doi.org/10.3390/ijerph16193751
APA StyleJanulewicz, P. A., Seth, R. K., Carlson, J. M., Ajama, J., Quinn, E., Heeren, T., Klimas, N., Lasley, S. M., Horner, R. D., Sullivan, K., & Chatterjee, S. (2019). The Gut-Microbiome in Gulf War Veterans: A Preliminary Report. International Journal of Environmental Research and Public Health, 16(19), 3751. https://doi.org/10.3390/ijerph16193751






