Detection of Hepatitis A Virus and Other Enteric Viruses in Shellfish Collected in the Gulf of Naples, Italy
Abstract
1. Introduction
2. Materials and Methods
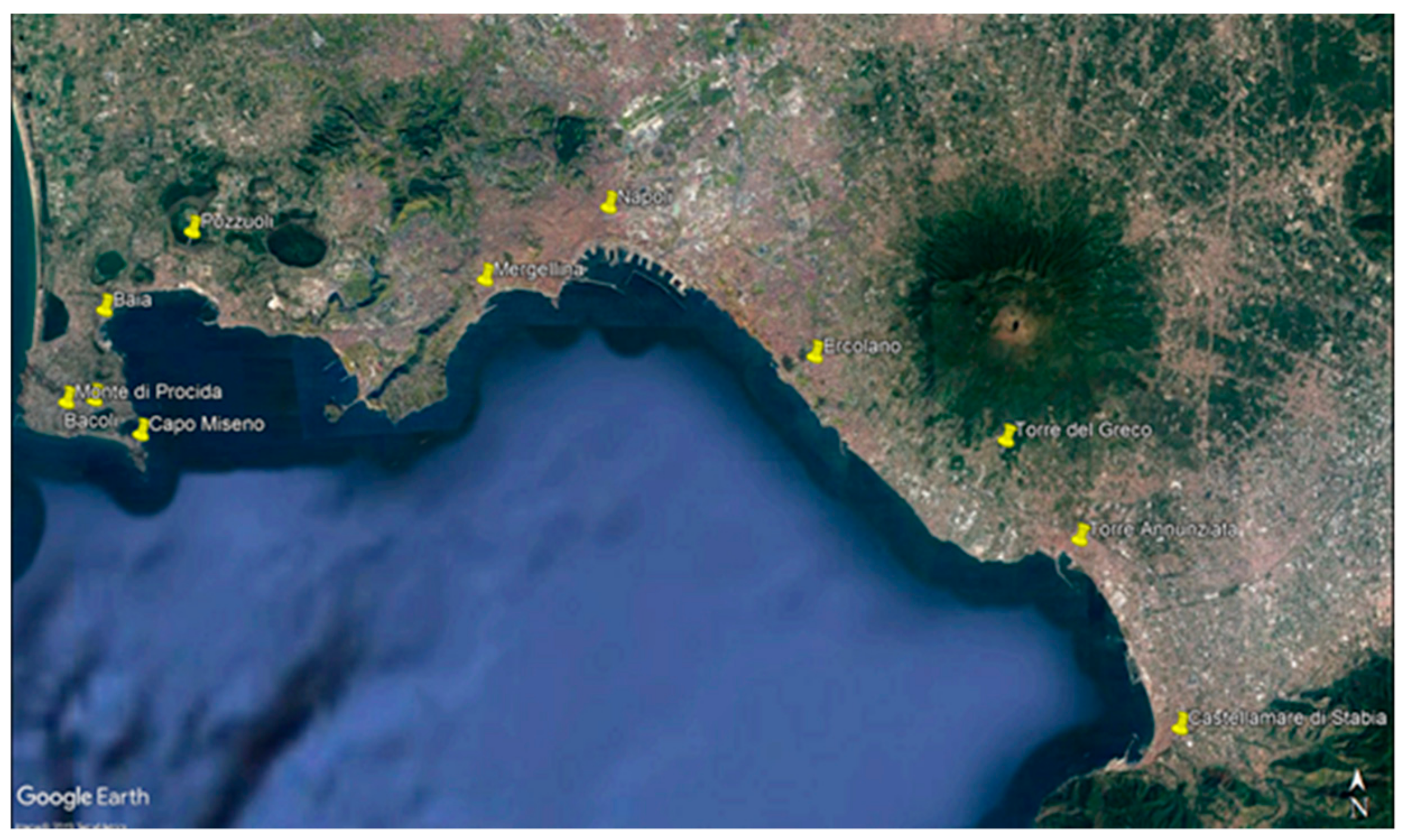
2.1. Sampling
2.2. Nucleic Acid Extraction and Detection of Viruses by Quantitative Real-Time PCR
2.3. Sequencing and Phylogenetic Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fusco, G.; Aprea, G.; Galiero, G.; Guarino, A.; Viscardi, M. Escherichia coli, Salmonella spp., hepatitis A virus and norovirus in bivalve molluscs in Southern Italy. Vet. Ital. 2013, 49, 55–58. [Google Scholar] [PubMed]
- Fusco, G.; Di Bartolo, I.; Cioffi, B.; Ianiro, G.; Palermo, P.; Monini, M.; Amoroso, M.G. Prevalence of foodborne viruses in mussels in Southern Italy. Food Environ. Virol. 2017, 9, 187–194. [Google Scholar] [CrossRef]
- SEIEVA 2015 Annual Rates/100,000 Inhabitants by Type of Hepatitis, Age, Sex, and Geographical Area of Acute Viral Hepatitis. Available online: http://www.epicentro.iss.it (accessed on 15 July 2018).
- SEIEVA 2016 Annual Rates/100,000 Inhabitants by Type of Hepatitis, Age, Sex, and Geographical Area of Acute Viral Hepatitis. Available online: http://www.epicentro.iss.it (accessed on 20 July 2018).
- Tosone, G.; Mascolo, S.; Bruni, R.; Taffon, S.; Equestre, M.; Tosti, M.E.; Ciccaglione, A.R.; Martucci, F.; Liberti, A.; Iannece, M.D.; et al. A family cluster of hepatitis A virus due to an uncommon IA strain circulating in Campania (southern Italy), not associated with raw shellfish or berries: A wake-up call to implement vaccination against hepatitis A. Infez. Med. 2016, 24, 30–33. [Google Scholar]
- Chiapponi, C.; Pavoni, E.; Bertasi, B.; Baioni, L.; Scaltriti, E.; Chiesa, E.; Cianti, L.; Losio, M.N.; Pongolini, S. Isolation and genomic sequence of hepatitis A virus from mixed frozen berries in Italy. Food Environ. Virol. 2014, 6, 202–206. [Google Scholar] [CrossRef][Green Version]
- SEIEVA 2017 Annual Rates/100,000 Inhabitants by Type of Hepatitis, Age, Sex, and Geographical Area of Acute Viral Hepatitis. Available online: http://www.epicentro.iss.it (accessed on 15 July 2018).
- Comelli, A.; Izzo, I.; Casari, S.; Spinetti, A.; Bergamasco, A.; Castelli, F. Hepatitis A outbreak in men who have sex with men (MSM) in Brescia (Northern Italy), July 2016–July 2017. Infez. Med. 2018, 26, 46–51. [Google Scholar] [PubMed]
- Araud, E.; Di Caprio, E.; Ma, Y.; Lou, F.; Gao, Y.; Kingsley, D.H.; Hughes, J.H.; Li, J. Thermal inactivation of enteric viruses and bioaccumulation of enteric foodborne viruses in live oysters (Crassostrea virginica). Appl. Environ. Microbiol. 2016, 82, 86–99. [Google Scholar] [CrossRef]
- Le Guyader, F.S.; Le Saux, J.C.; Ambert-Balay, K.; Krol, J.; Serais, O.; Parnaudeau, S.; Giraudon, H.; Delmas, G.; Pommepuy, M.; Pothier, P.; et al. Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster-related gastroenteritis outbreak. J. Clin. Microbiol. 2008, 46, 4011–4017. [Google Scholar] [CrossRef]
- Nakagawa-Okamoto, R.; Arita-Nishida, T.; Toda, S.; Kato, H.; Iwata, H.M.; Akiyama, O.; Nishio, H.; Kimura, M.; Noda, N.; Takeda, N.; et al. Detection of multiple sapovirus genotypes and genogroups in oyster-associated outbreaks. Jpn. J. Infect. Dis. 2009, 62, 63–66. [Google Scholar]
- Pina, S.; Puig, M.; Lucena, F.; Jofre, J.; Girones, R. Viral pollution in the environment and in shellfish: Human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 1998, 64, 3376–3382. [Google Scholar]
- Iritani, N.; Kaida, A.; Abe, N.; Kubo, H.; Sekiguchi, J.; Yamamoto, S.P.; Goto, K.; Tanaka, T.; Noda, M. Detection and genetic characterization of human enteric viruses in oyster-associated gastroenteritis outbreaks between 2001 and 2012 in Osaka City, Japan. J. Med. Virol. 2014, 86, 2019–2025. [Google Scholar] [CrossRef]
- Béji-Hamza, A.; Hassine-Zaafrane, M.; Khélifi-Gharbi, H.; Della Libera, S.; Iaconelli, M.; Muscillo, M.; Petricca, S.; Ciccaglione, A.R.; Bruni, R.; Taffon, S.; et al. Hepatitis E virus genotypes 1 and 3 in wastewater samples in Tunisia. Arch. Virol. 2015, 160, 183–189. [Google Scholar] [CrossRef]
- La Rosa, G.; Della Libera, S.; Brambilla, M.; Bisaglia, C.; Pisani, G.; Ciccaglione, A.R.; Bruni, R.; Taffon, S.; Equestre, M.; Iaconelli, M. Hepatitis E virus (genotype 3) in slurry samples from swine farming activities in Italy. Food Environ. Virol. 2017, 9, 219–229. [Google Scholar] [CrossRef]
- Lazaro, D.R.; Cook, N.; Ruggeri, F.M.; Sellwood, J.; Nasser, A.; Nascimento, M.S.; D’Agostino, M.; Santos, R.; Saiz, J.C.; Rzezutka, A.; et al. Virus hazards from food, water and other contaminated environments. FEMS Microbiol. Rev. 2012, 36, 786–814. [Google Scholar] [CrossRef]
- Reg.CE854/2004. Specific Rules for the Organization of Official Controls on Products of Animal Origin Intended for Human Consumption–All II, Chapter II–Classification of Production and Relaying Areas. Available online: https://aeaconsulenze.files.wordpress.com/2015/07/reg_ce_854–2004.pdf (accessed on 21 May 2018).
- Reg CE 2285/2015. Reg. (UE) 2015/2285 of the COMMISSION of 8 December 2015 Amending Annex II to Regulation (EC) No 854/2004 of the European Parliament and of the Council Laying down Rules for the Organization of Official Controls on Products of Animal Origin Intended for all Live Bivalve Molluscs, Echinoderms, Tunicates and Marine Gastropods, as well as Annex I of the Regulation (EC) n. 2073/2005 on the Microbiological Criteria Principle of Food Products; EUR-Lex: Brussels, Belgium, 2015. [Google Scholar]
- Reg CE 2073/2008. Microbiological Criteria Applicable to Food Products. Available online: http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2005:338:0001:0026:EN:PDF (accessed on 15 July 2019).
- UNI CEN ISO/TS 15216–2:2013. Microbiology of Food and Animal Feed—Horizontal Method for Determination of Hepatitis a Virus and Norovirus in Food Using Real-Time RT-PCR—Part 2: Method for Quantitative Detection; ISO: Geneva, Switzerland, 2013. [Google Scholar]
- Kitajima, M.; Gerba, C.P. Aichi virus 1: Environmental occurrence and behavior. Pathogens 2015, 4, 256–268. [Google Scholar] [CrossRef]
- Zeng, M.; Chen, J.; Gong, S.T.; Xu, X.H.; Zhu, C.M.; Zhu, Q.R. Epidemiological surveillance of norovirus and rotavirus diarrhea among outpatient children in five metropolitan cities. Jpn. Infect. Dis. 2008, 64, 3376–3382. [Google Scholar]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef]
- Varela, M.F.; Hooper, A.S.; Rivadulla, E.; Romalde, J.L. Human sapovirus in mussels from Ría do Burgo, A Coruña (Spain). Food Environ. Virol. 2016, 8, 187–193. [Google Scholar] [CrossRef]
- Hernroth, B.E.; Conden-Hansson, A.C.; Rehnstam-Holm, A.S.; Girones, R.; Allard, A.K. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: The first Scandinavian report. Appl. Environ. Microbiol. 2002, 68, 4523–4533. [Google Scholar] [CrossRef]
- Le Cann, P.; Ranarijaona, S.; Monpoeho, S.; Le Guyader, F.; Ferré, V. Quantification of human astroviruses in sewage using real-time RT-PCR. Res. Microbiol. 2004, 155, 11–15. [Google Scholar] [CrossRef]
- Taffon, S.; Bidini, G.; Vichi, F.; Corti, G.; Genovese, D.; Kondili, L.A.; Bindi, R.; Armellini, F.; Leoncini, F.; Bartoloni, A.; et al. A unique HAV strain circulated in patients with acute HAV infection with different risk exposures in Tuscany Italy. J. Clin. Virol. 2011, 50, 142–147. [Google Scholar] [CrossRef]
- Allard, A.; Albinsson, B.; Wadell, G. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 2001, 39, 498–505. [Google Scholar] [CrossRef]
- Bruni, R.; Costantino, A.; Equestre, M.; Garbuglia, A.R.; Pacenti, M.; Morea, A.; Madonna, E.; Chionne, P.; Alfonsi, V.; Rizzo, C.; et al. Ongoing European Outbreak of Hepatitis a Among MSM: Four Strains Observed in Italy, August 2016 to February 2017; Submitted (23-MAR-2017); Biomedical Sciences and Human Oncology–Hygiene Section, University of Bari Aldo Moro, Policlinico PiazzaG: Cesare, Bari, Italy, 2017. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KY886891 (accessed on 15 July 2019).
- Garbuglia, A.R.; Minosse, C.; Scognamiglio, P.; Capobianchi, M.R. Surveillance Study on Hepatitis a Virus in LAZIO; Submitted (05-DEC-2016); National Institute for Infectious Diseases’L: Spallanzani, Portuense, Rome, Italy, 2016. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KY292290 (accessed on 15 July 2019).
- Franchin, E.; Pacenti, M.; Barzon, L.; Lavezzo, E.; Palu, G. HAV in Veneto Region, Italy, 2017; Submitted (17-MAR-2017); Department of Molecular Medicine, University of Padua: Gabelli, Padua, PD, Italy, 2017. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KY782330 (accessed on 15 July 2019).
- Costantino, A.; Coppola, N.; Spada, E.; Bruni, R.; Taffon, S.; Equestre, M.; Marcantonio, C.; Sagnelli, C.; Dell’Isola, C.; Tosone, G.; et al. Hepatitis A virus strains circulating during 1997–2015 in Campania, a Southern Italy region with periodic outbreaks. J. Med. Virol. 2017, 89, 1931–1936. [Google Scholar] [CrossRef]
- Polo, D.; Varela, M.F.; Romalde, J.L. Detection and quantification of hepatitis A virus and norovirus in Spanish authorized shellfish harvesting areas. Int. J. Food Microbiol. 2015, 193, 43–50. [Google Scholar] [CrossRef]
- Siebenga, J.J.; Vennema, H.; Duizer, E.; Koopmans, M.P.G. Gastroenteritis caused by norovirus GGII.4 in the Netherlands, 1994–2005. Emerg. Infect. Dis. 2007, 13, 144–146. [Google Scholar] [CrossRef]
- Atmar, R.L.; Opekum, A.R.; Gilger, M.A.; Estes, M.K.; Crawford, S.E.; Neill, F.H.; Graham, D.Y. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 2008, 14, 1553–1557. [Google Scholar] [CrossRef]
- Chan, M.M.C.W.; Sung, J.J.Y.; Lam, R.K.Y.; Chan, P.K.S.; Lee, N.L.S.; Lai, R.W.M.; Leung, W.K. Fecal viral load and norovirus associated gastroenteritis. Emerg. Infect. Dis. 2006, 12, 1278–1280. [Google Scholar] [CrossRef]
- Rivadulla, E.; Varela, M.F.; Romalde, J.L. Low prevalence of Aichi virus in molluscan shellfish samples from Galicia (NW Spain). J. Appl. Microbiol. 2017, 122, 516–521. [Google Scholar] [CrossRef]
- Vilariño, M.L.; Le Guyader, F.S.; Polo, D.; Schaeffer, J.; Kröl, J.; Romalde, J.L. Assessment of human enteric viruses in cultured and wild bivalve molluscs. Int. Microbiol. 2009, 12, 145–151. [Google Scholar]
- Aprea, G.; Amoroso, M.G.; Di Bartolo, I.; D’Alessio, N.; Di Sabatino, D.; Boni, A.; Cioffi, B.; D’Angelantonio, D.; Scattolini, S.; De Sabato, L.; et al. Molecular detection and phylogenetic analysis of hepatitis E virus strains circulating in wild boars in south-central Italy. Transbound. Emerg. Dis. 2018, 65, e25–e31. [Google Scholar] [CrossRef]
| Collection Area | Number of Samples Collected | Percentage of Samples Collected (%) |
|---|---|---|
| ASL Na 1 Centre | 27 | 9.4 |
| ASL Na 2 North | 201 | 70.0 |
| ASL Na 3 South | 56 | 19.9 |
| Collection area not indicated | 3 | 1.0 |
| Classification | Number of Samples Analyzed | Percentage of Samples Analyzed (%) |
|---|---|---|
| Class A 1 | 46 | 16.0 |
| Class B 2 | 237 | 82.1 |
| Class C 3 | 1 | 0.3 |
| Natural banks 4 | 5 | 1.7 |
| Target | Primers/Probe Name | Sequence | Reference |
|---|---|---|---|
| HAV Primer forward | HAV 68 | 5’TCACCGCCGTTTGCCTAG3’ | |
| Primer reverse | HAV 240 | 5’GGAGAGCCCTGGAAGAAAG3’ | [20] |
| Probe | HAV 150 FAM | 5’CCTGAACCTGCAGGAATTAA-3’-MGB/NFQ | |
| NOROVIRUS GI | |||
| Primer forward | NGI QNIF4 | 5’-CGCTGGATGCGNTTCCAT-3’ | |
| Primer reverse | NGI NV1LCR | 5’CCTTAGACGCCATCATCATTTAC3’ | [20] |
| Probe | NVGG1P-FAM | 5’TGGACAGGAGAYCGCRATCT3’TAMRA | |
| NOROVIRUS GII | |||
| Primer forward | NGII QNIF2 | 5’ATGTTCAGRTGGATGAGRTTCTCWGA-3’ | |
| Primer reverse | NGII COG2R | 5’-TCGACGCCATCTTCATTCACA-3’ | [20] |
| Probe | NGII QNIF FAM | 5’-AGCACGTGGGAGGGCGATCG-3’TAMRA | |
| HEV | |||
| Primer forward | JVHEV1 | 5’-GGTGGTTTCTGGGGTGAC-3’ | |
| Primer reverse | JVHEV2 | 5’-AGGGGTTGGTTGGATGAA-3’ | [23] |
| Probe | JVHEV P-FAM | 5’-TGATTCTCAGCCCTTCGC-3’TAMRA | |
| AICHIVIRUS | |||
| Primer forward | AiV-AB-F | 5’-GTCTCCACHGACACYAAYTGGAC-3’ | |
| Primer reverse | AiV-AB-R | 5’-GTTGTACATRGCAGCCCAGG-3’ | [21] |
| Probe | AiV-AB-TP FAM | 5’-TTYTCCTTYGTGCGTGC- 3’NFQ (MGB) | |
| ROTAVIRUS | |||
| Primer forward | NSP3F | 5’ACCATCTWCACRTRACCCTCTATGAG-3’ | |
| Primer reverse | NSP3R | 5’-GGTCACATAACGCCCCTATAGC-3’ | [22] |
| Probe | NSP3P-FAM | 5’-AGTTAAAAGCTAACACTGTCAAA3’(MGB) | |
| SAPOVIRUS | |||
| Primer forward | SAV124F | 5’-GAYCASGCTCTCGCYACCTAC-3’ | |
| Primer reverse | SAV1245R | 5’-CCCTCCATYTCAAACACTA-3’ | [24] |
| Probe | SAV124TPFAM | 5’- CCCCTATRAACCA-3’NFQ (MGB) | |
| ADENOVIRUS | |||
| Primer forward | AdV1 | 5’-CWTACATGCACATCKCSGG-3’ | |
| Primer reverse | AdV2 | 5’-CRCGGGCRAAYTGCACCAG-3’ | [25] |
| Probe | Advs-FAM | 5’CCGGGCTCAGGTACTCCGAGGCGTCCT-3’ | |
| ASTROVIRUS | |||
| Primer forward | AsV1 | 5’-CCGAGTAGGATCGAGGGT-3’ | |
| Primer reverse | AsV2 | 5’-GCTTCTGATTAAATCAATTTTAA-3’ | [26] |
| Probe | AsV FAM | 5’CTTTTCTGTCTCTGTTTAGATTATTTTAATCACC-3’ TAMRA |
| Viruses Detected | 2015 Number of Positive Samples (%) | 2016 Number of Positive Samples (%) | 2017 Number of Positive Samples (%) | Total Positivity over Three Years (%) |
|---|---|---|---|---|
| HAV | 7 (4.7) | 4 (5.1) | 15 (22.7) | 26 (8.9) |
| NoVGI | 6 (4.1) | 7 (9.0) | 18 (28.1) | 31 (10.8) |
| NoVGII | 31 (21.2) | 53 (68.8) | 30 (46.8) | 114 (39.7) |
| RV | 18 (12.3) | 0 (0) | 8 (12.5) | 26 (9.0) |
| SaV | 16 (10.9) | 20 (25.9) | 17 (26.5) | 54 (18.8) |
| AsV | 30 (20.5) | 11 (14.2) | 19 (29.0) | 60 (20.8) |
| AiV | 9 (6.1) | 2 (2.6) | 5 (7.8) | 16 (5.5) |
| AdV | 4 (2.7) | 6 (7.7) | 6 (9.3) | 16 (5.5) |
| HEV | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Total samples analyzed | 146 | 77 | 66 | 289 |
| Positivity of Samples | Number of Positive Samples | Percentage of Samples out of the Total Positive (%) |
|---|---|---|
| Positivity for one target | 60 | 37.7 |
| Positivity for at least two targets | 99 | 62.2 |
| Total positivity | 159 | 55.0 |
| Month | Virus Positivity Number of Positive Samples/Number of Samples Analyzed (%) | HAV % | NoVGI % | NoVGII % | RV % | SaV % | AsV % | AiV % | AdV % |
|---|---|---|---|---|---|---|---|---|---|
| Jan. | 23/28 (82.1) | 25 | 12.9 | 20.1 | 0 | 12.9 | 10 | 25 | 3.7 |
| Feb. | 22/23 (95.6) | 3.8 | 29.0 | 17.5 | 7.6 | 1.8 | 26.6 | 18.7 | 18.7 |
| Mar. | 18/26 (69.2) | 11.5 | 9.6 | 8.7 | 0 | 12.9 | 20 | 12.5 | 6.2 |
| Apr. | 33/44 (75) | 23.0 | 19.3 | 21.9 | 15.3 | 11.1 | 30 | 18.7 | 6.2 |
| May | 14/24 (58.3) | 11.5 | 9.6 | 8.7 | 15.3 | 5.5 | 10 | 6.2 | 0 |
| Jun. | 3/4 (75) | 0 | 0 | 1 | 0 | 0 | 1.6 | 0 | 0 |
| Jul. | 8/33 (24.2) | 0 | 6.4 | 1.7 | 7.6 | 5.5 | 1.6 | 0 | 0 |
| Aug. | 0/13 (0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sept. | 9/27 (33.3) | 0 | 6.4 | 0 | 26.9 | 5.5 | 0 | 0 | 6.2 |
| Oct. | 0/18 (0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Nov. | 8/18 (44.4) | 0 | 9.6 | 2.6 | 11.5 | 3.7 | 0 | 0 | 0 |
| Dec. | 22/28 (78.5) | 15.3 | 64.5 | 17.5 | 15.3 | 11.1 | 5 | 25 | 25 |
| Classification | Number of Samples Analyzed | Number of Positive Samples | Virus Identified |
|---|---|---|---|
| Class A | 46 | 20 | HAV, NoVGI, NoVGII, RV, AsV, SaV, Adv. |
| Class B | 237 | 136 | HAV, NoVGI, NoVGII, RV, AsV, SaV, Adv, AiV |
| Class C | 1 | 1 | HAV, NoVGI, NoVGII, RV, AsV, SaV, Adv, AiV |
| Natural banks | 5 | 2 | HAV, NoVGI, NoVGII, RV, AsV, SaV, Adv, AiV |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fusco, G.; Anastasio, A.; Kingsley, D.H.; Amoroso, M.G.; Pepe, T.; Fratamico, P.M.; Cioffi, B.; Rossi, R.; La Rosa, G.; Boccia, F. Detection of Hepatitis A Virus and Other Enteric Viruses in Shellfish Collected in the Gulf of Naples, Italy. Int. J. Environ. Res. Public Health 2019, 16, 2588. https://doi.org/10.3390/ijerph16142588
Fusco G, Anastasio A, Kingsley DH, Amoroso MG, Pepe T, Fratamico PM, Cioffi B, Rossi R, La Rosa G, Boccia F. Detection of Hepatitis A Virus and Other Enteric Viruses in Shellfish Collected in the Gulf of Naples, Italy. International Journal of Environmental Research and Public Health. 2019; 16(14):2588. https://doi.org/10.3390/ijerph16142588
Chicago/Turabian StyleFusco, Giovanna, Aniello Anastasio, David H. Kingsley, Maria Grazia Amoroso, Tiziana Pepe, Pina M. Fratamico, Barbara Cioffi, Rachele Rossi, Giuseppina La Rosa, and Federica Boccia. 2019. "Detection of Hepatitis A Virus and Other Enteric Viruses in Shellfish Collected in the Gulf of Naples, Italy" International Journal of Environmental Research and Public Health 16, no. 14: 2588. https://doi.org/10.3390/ijerph16142588
APA StyleFusco, G., Anastasio, A., Kingsley, D. H., Amoroso, M. G., Pepe, T., Fratamico, P. M., Cioffi, B., Rossi, R., La Rosa, G., & Boccia, F. (2019). Detection of Hepatitis A Virus and Other Enteric Viruses in Shellfish Collected in the Gulf of Naples, Italy. International Journal of Environmental Research and Public Health, 16(14), 2588. https://doi.org/10.3390/ijerph16142588








