Spatial and Seasonal Variations in the Abundance of Nitrogen-Transforming Genes and the Microbial Community Structure in Freshwater Lakes with Different Trophic Statuses
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site and Procedure
2.2. Physicochemical Analysis
2.3. PCR Amplification, Clone Libraries Construction, and Phylogenetic Analysis
2.4. Real-Time Quantitative PCR
2.5. Statistical Analysis
3. Results
3.1. Physicochemical Properties of the Lake Regions
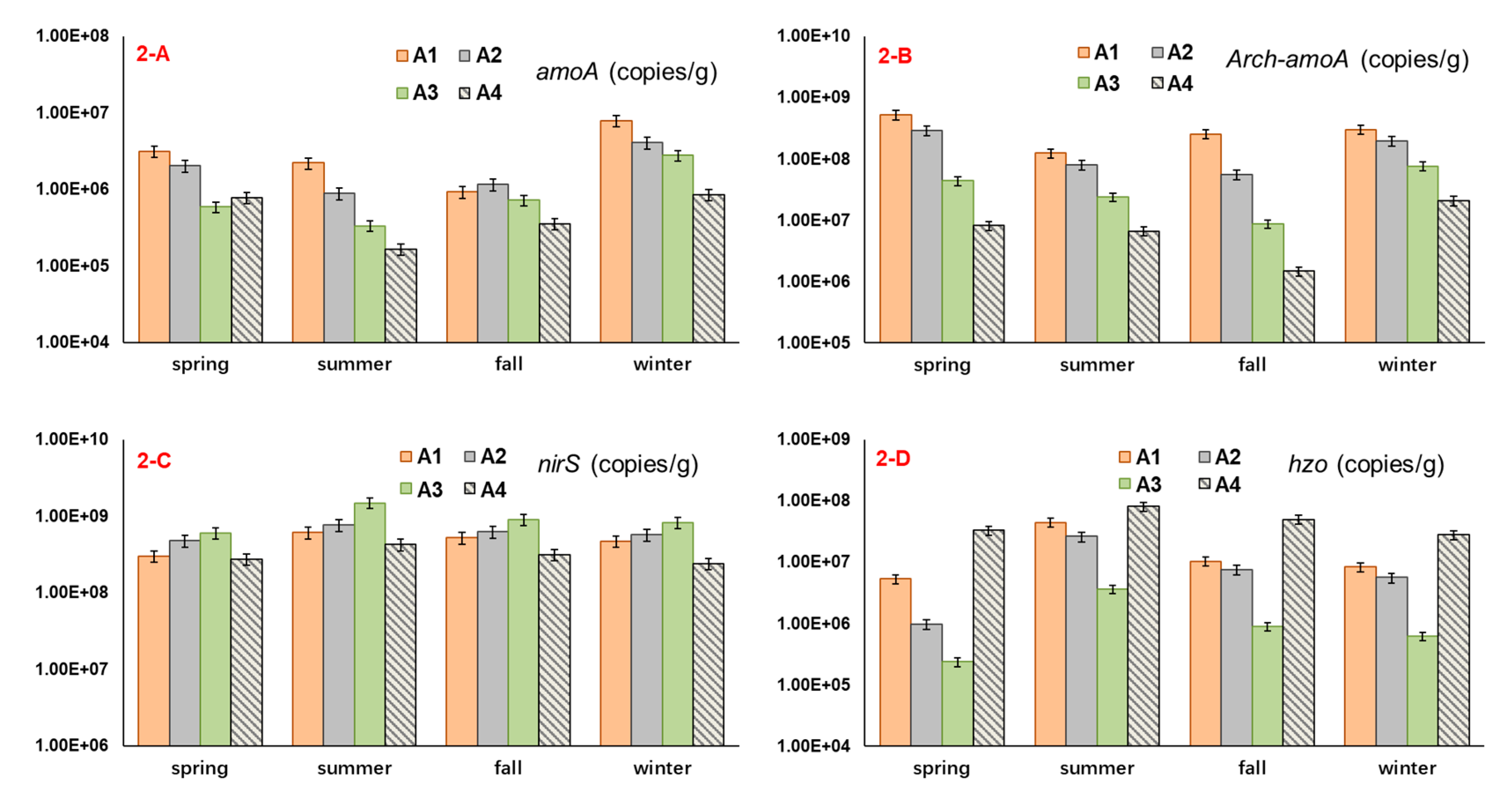
3.2. The Abundance of Microbial Nitrogen-Transforming Genes
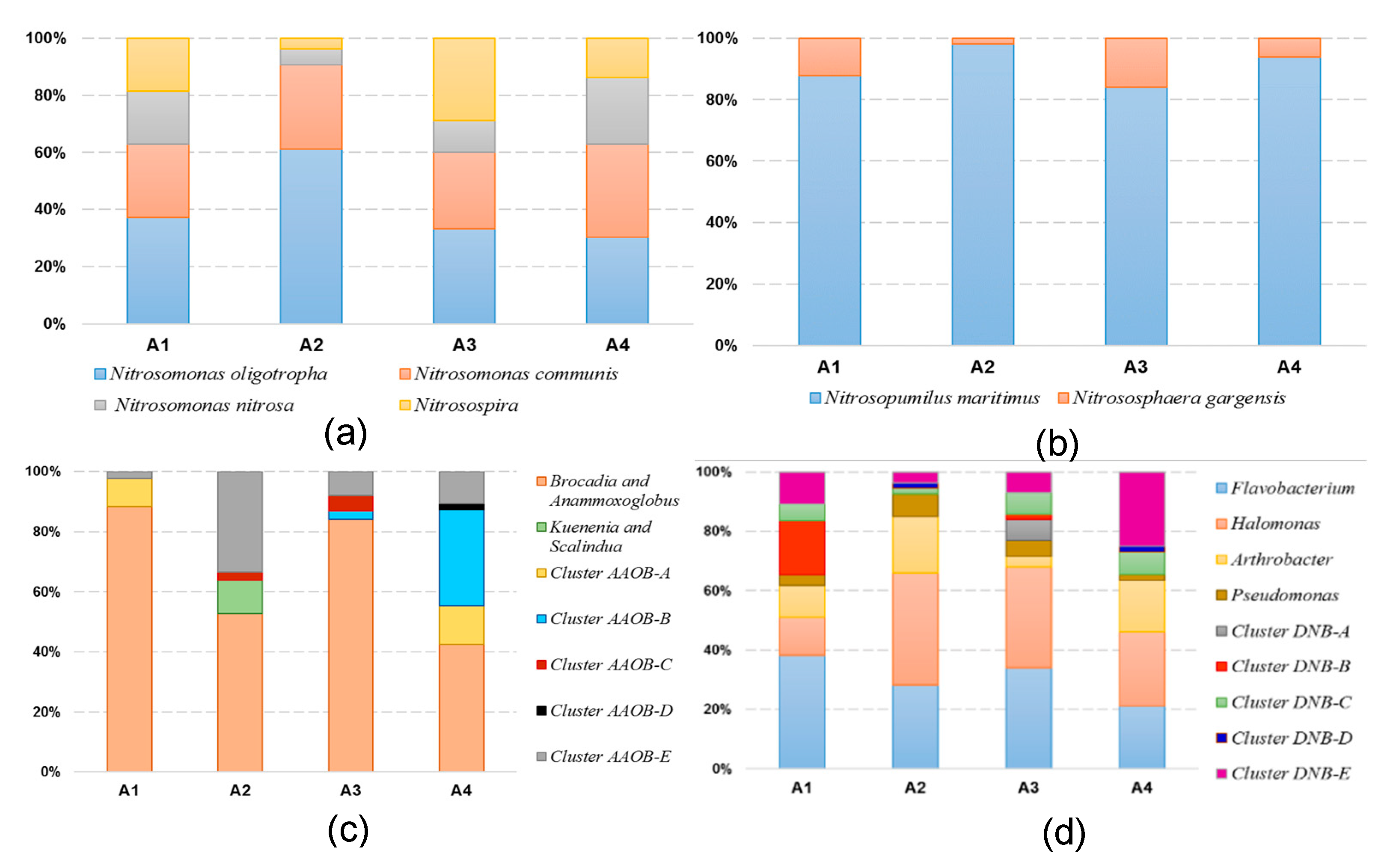
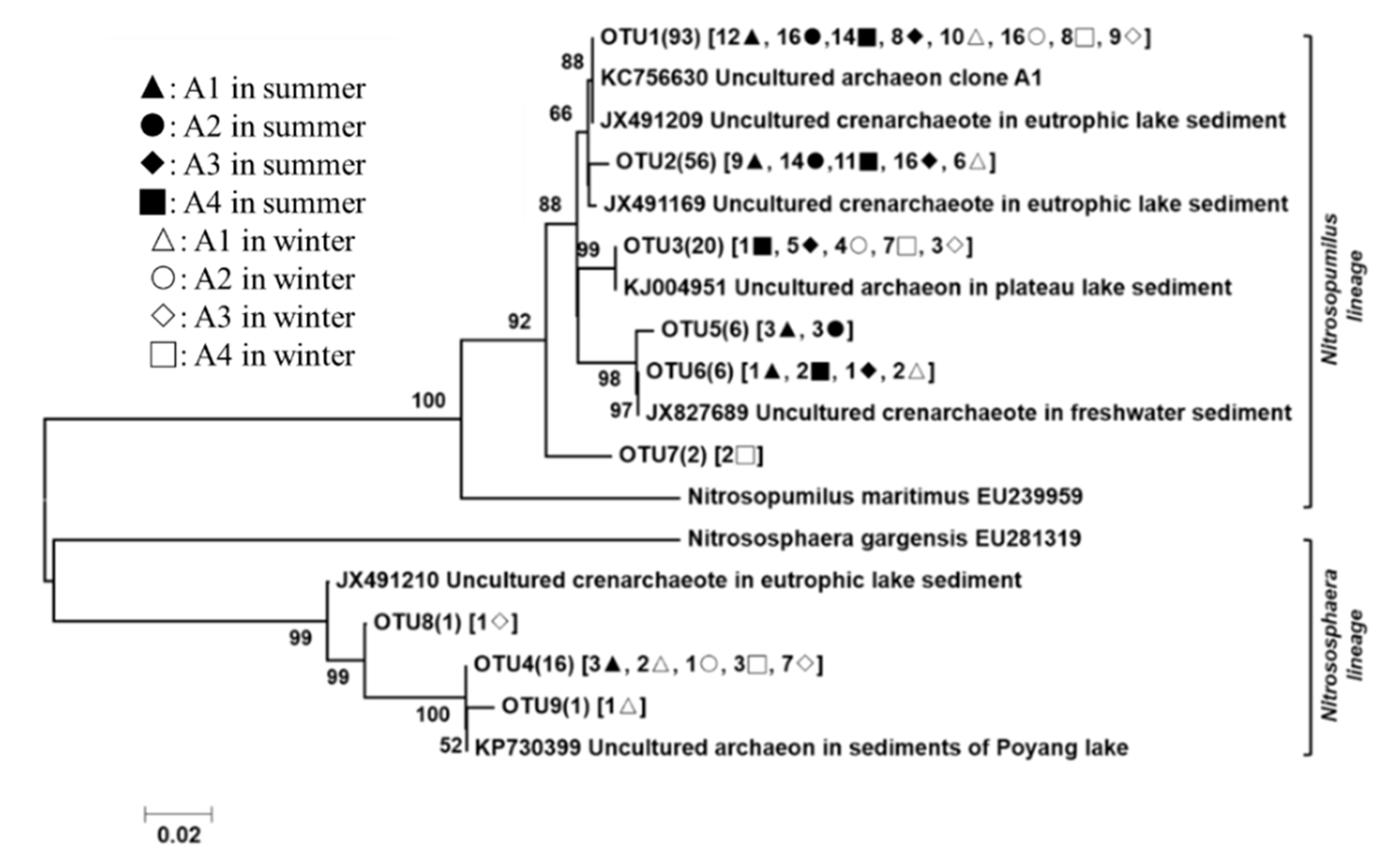
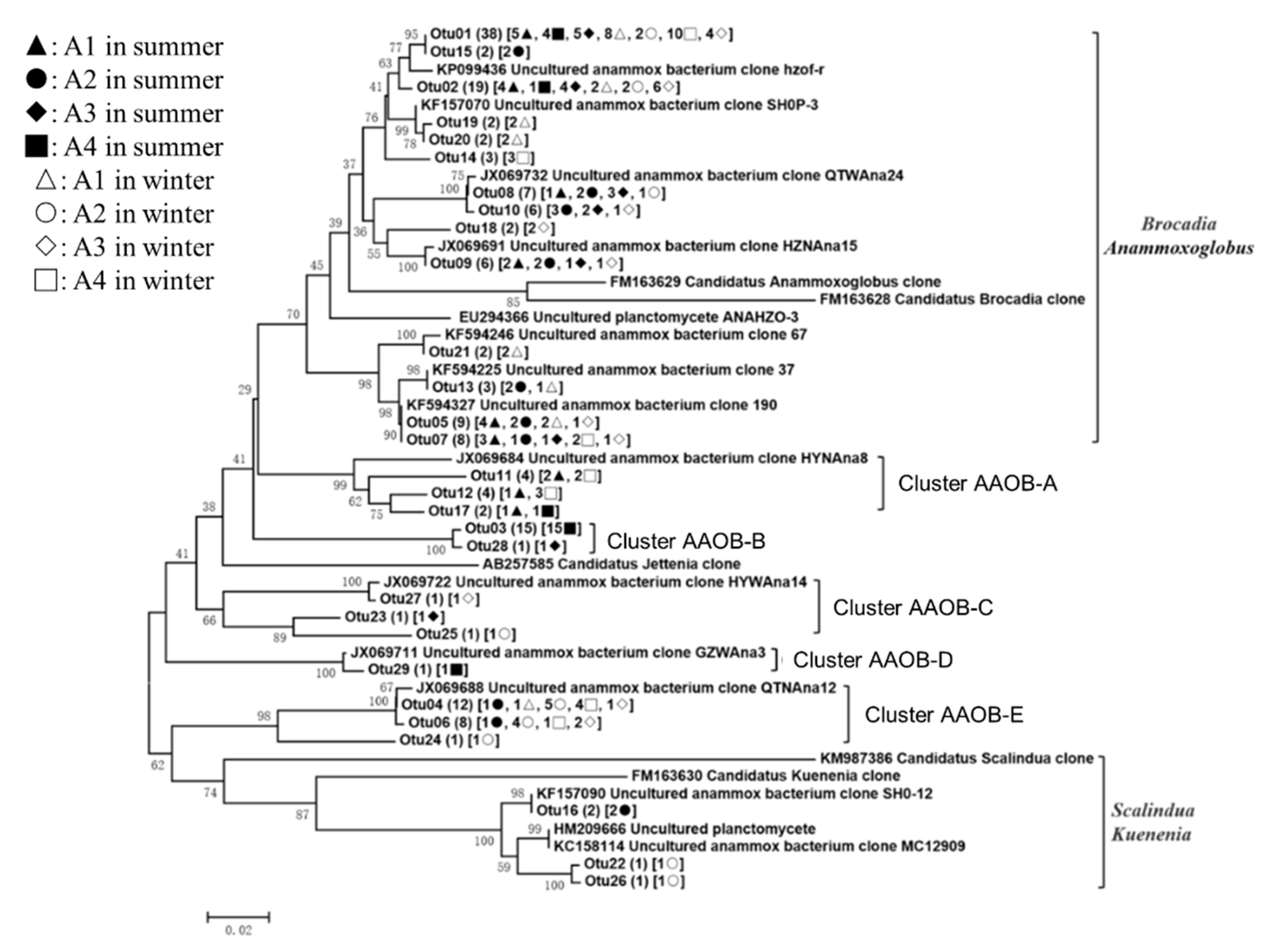
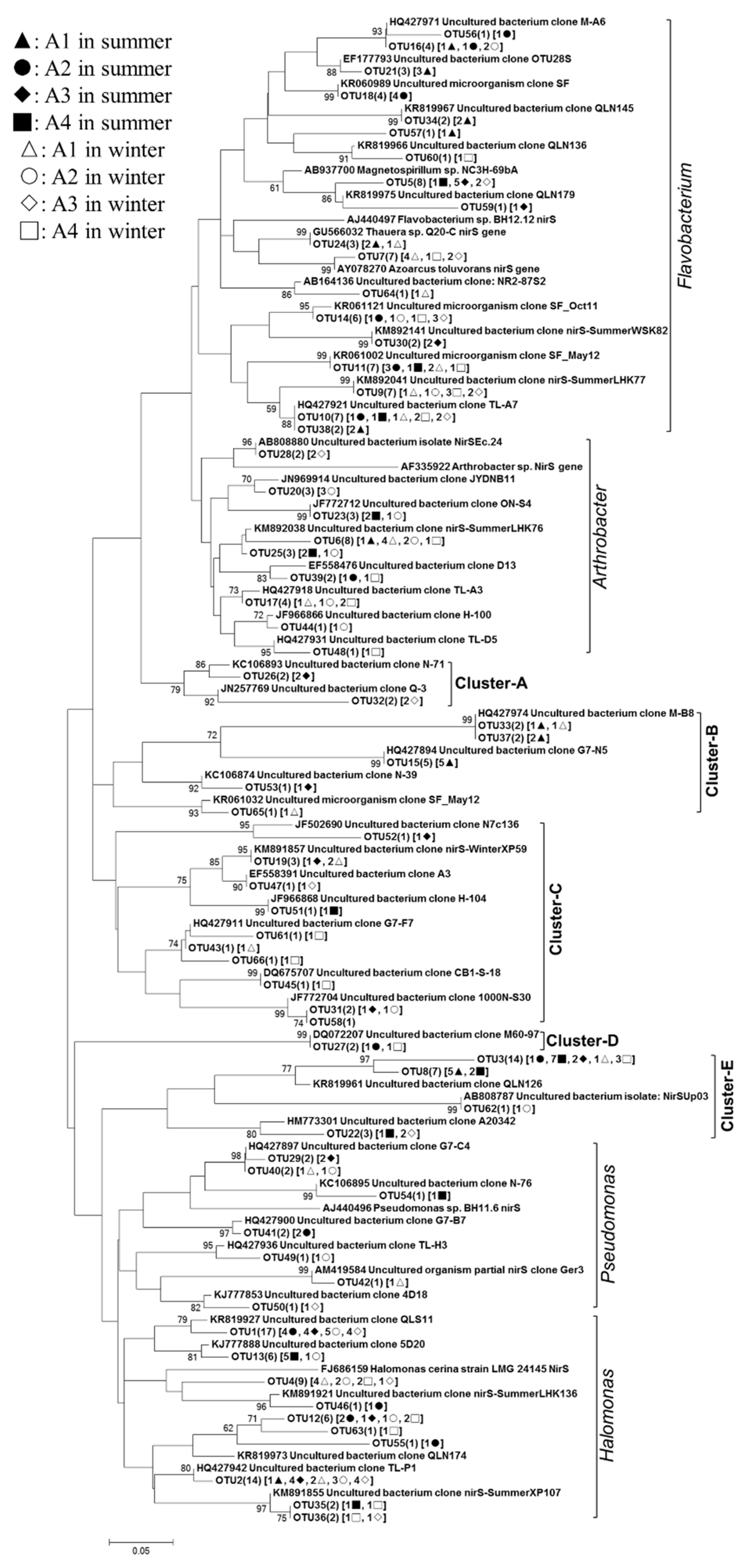
3.3. Phylogenetic Analyses of the Nitrogen-Transforming Microbes
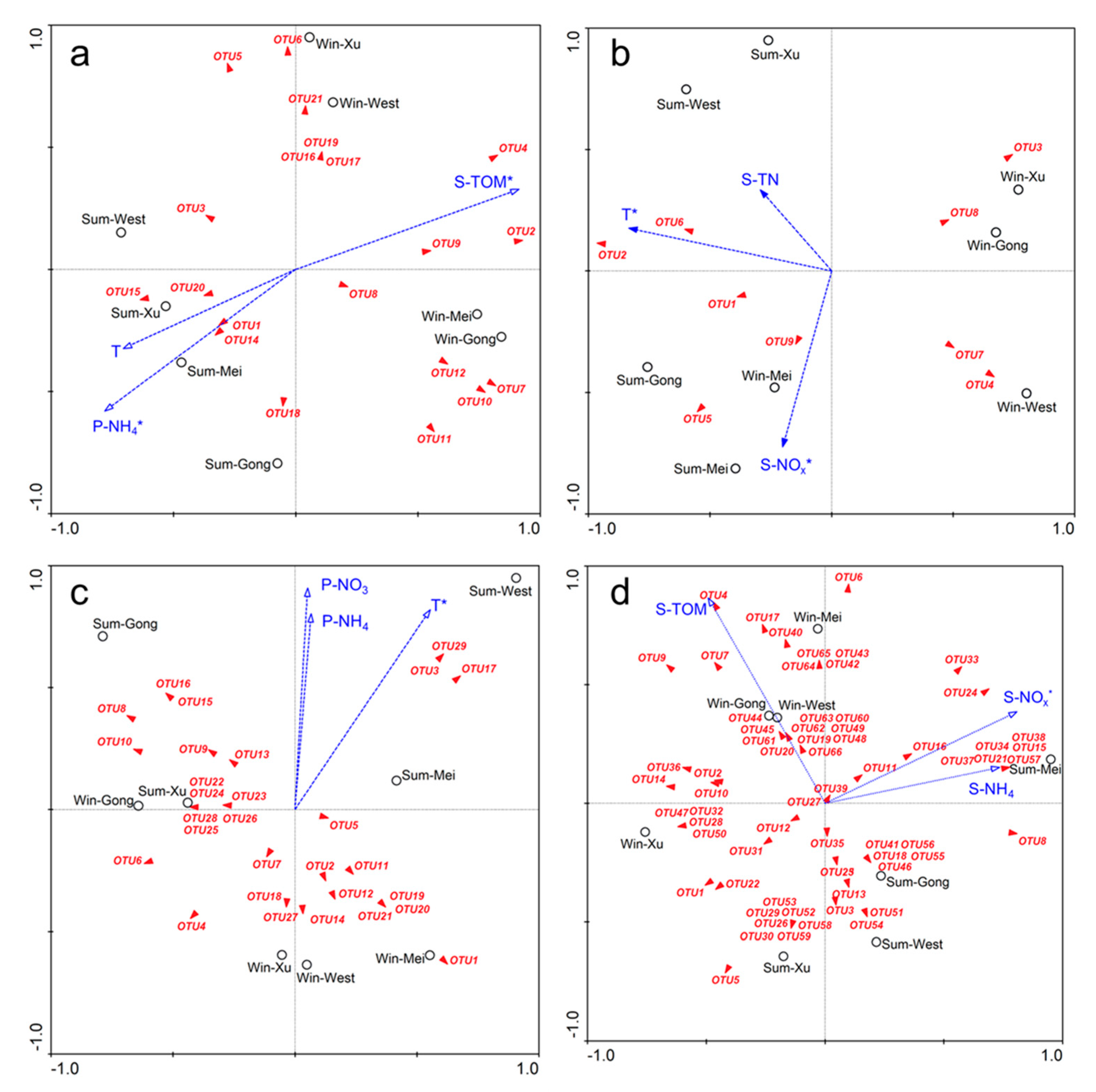
3.4. Relationships between Microbial Community Composition and Environmental Variables
4. Discussion
4.1. Distribution Characteristics of Nitrogen Occurrence Forms
4.2. The Relation between Functional Gene Abundance and Nitrogen Occurrence Characteristic
4.3. Effect of Trophic Status on the Microbial Community Distribution Characteristics
4.4. Factors Affecting Nitrogen-Transforming Microbial Community Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wan, Y.; Ruan, X.; Zhang, Y.; Li, R. Illumina sequencing-based analysis of sediment bacteria community in different trophic status freshwater lakes. MicrobiologyOpen 2017, 6, e00450. [Google Scholar] [CrossRef]
- Barnard, R.; Leadley, P.W.; Lensi, R.; Barthes, L. Plant, soil microbial and soil inorganic nitrogen responses to elevated CO2: A study in microcosms of Holcus lanatus. Acta Oecologica 2005, 27, 171–178. [Google Scholar] [CrossRef]
- Henry, S.; Baudoin, E.; López-Gutiérrez, J.C.; Martin-Laurent, F.; Brauman, A.; Philippot, L. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 2004, 59, 327–335. [Google Scholar] [CrossRef]
- Nogales, B.; Timmis, K.N.; Nedwell, D.B.; Osborn, A.M. Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Appl. Environ. Microbiol. 2002, 68, 5017–5025. [Google Scholar] [CrossRef]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997, 63, 4704–4712. Available online: https://aem.asm.org/content/63/12/4704.short (accessed on 27 June 2017).
- Venter, J.C.; Remington, K.; Heidelberg, J.F.; Halpern, A.L.; Rusch, D.; Eisen, J.A.; Fouts, D.E. Environmental genome shotgun sequencing of the Sargasso Sea. Science 2004, 304, 66–74. [Google Scholar] [CrossRef]
- Humbert, S.; Tarnawski, S.; Fromin, N.; Mallet, M.P.; Aragno, M.; Zopfi, J. Molecular detection of anammox bacteria in terrestrial ecosystems: Distribution and diversity. ISME J. 2010, 4, 450. [Google Scholar] [CrossRef]
- Wang, J.; Shen, J.; Wu, Y.; Tu, C.; Soininen, J.; Stegen, J.C.; Zhang, E. Phylogenetic beta diversity in bacterial assemblages across ecosystems: Deterministic versus stochastic processes. ISME J. 2013, 7, 1310–1321. [Google Scholar] [CrossRef]
- Mosier, A.C.; Francis, C.A. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 2008, 10, 3002–3016. [Google Scholar] [CrossRef]
- Caffrey, J.M.; Bano, N.; Kalanetra, K.; Hollibaugh, J.T. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 2007, 1, 660. [Google Scholar] [CrossRef]
- Li, M.; Cao, H.; Hong, Y.G.; Gu, J.D. Seasonal dynamics of anammox bacteria in estuarial sediment of the Mai Po Nature Reserve revealed by analyzing the 16S rRNA and hydrazine oxidoreductase (hzo) genes. Microbes. Environ. 2011, 26, 15–22. [Google Scholar] [CrossRef]
- Sun, W.; Xu, M.Y.; Wu, W.M.; Guo, J.; Xia, C.Y.; Sun, G.P.; Wang, A.J. Molecular diversity and distribution of anammox community in sediments of the Dongjiang River, a drinking water source of Hong Kong. J. Appl. Microbiol. 2014, 116, 464–476. [Google Scholar] [CrossRef]
- Lisa, J.A.; Song, B.; Tobias, C.R.; Hines, D.E. Genetic and biogeochemical investigation of sedimentary nitrogen cycling communities responding to tidal and seasonal dynamics in Cape Fear River Estuary. Estuar. Coast. Shelf Sci. 2015, 167, A313–A323. [Google Scholar] [CrossRef]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 2006, 72, 5181–5189. [Google Scholar] [CrossRef]
- Wu, Y.; Xiang, Y.; Wang, J.; Wu, Q.L. Molecular detection of novel anammox bacterial clusters in the sediments of the shallow freshwater Lake Taihu. Geomicrobiol. J. 2012, 29, 852–859. [Google Scholar] [CrossRef]
- Dai, J.; Gao, G.; Chen, D.; Tang, X.; Shao, K.; Cai, X. Effects of trophic status and temperature on communities of sedimentary ammonia oxidizers in Lake Taihu. Geomicrobiol. J. 2013, 30, 886–896. [Google Scholar] [CrossRef]
- Wu, Y.; Xiang, Y.; Wang, J.; Zhong, J.; He, J.; Wu, Q.L. Heterogeneity of archaeal and bacterial ammonia-oxidizing communities in Lake Taihu, China. Environ. Microbiol. Rep. 2010, 2, 569–576. [Google Scholar] [CrossRef]
- Guo, L.; Hu, Z.; Fang, F.; Liu, T.; Chuai, X.; Yang, L. Trophic status determines the nirS-denitrifier community in shallow freshwater lakes. Ann. Microbiol. 2014, 64, 999–1006. [Google Scholar] [CrossRef]
- Taihu Basin Authority of Ministry of Water Resources. The Health Status Report of Lake Taihu. 2014. Available online: http://www.tba.gov.cn/contents/45/14729.html (accessed on 27 June 2017). (In Chinese)
- Lu, R.K. Analytical Methods of Soil Agrochemistry; China Agricultural Science and Technology Press: Beijing, China, 1999. (In Chinese) [Google Scholar]
- Wei, F.S. Determination Methods for Examination of Water and Wastewater; China Environmental Science Press: Beijing, China, 2002. (In Chinese) [Google Scholar]
- Braker, G.; Fesefeldt, A.; Witzel, K.P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 1998, 64, 3769–3775. Available online: https://aem.asm.org/content/64/10/3769.short (accessed on 27 June 2017).
- Francis, C.A.; Roberts, K.J.; Beman, J.M.; Santoro, A.E.; Oakley, B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 2005, 102, 14683–14688. [Google Scholar] [CrossRef]
- Li, H.; Chen, S.; Mu, B.Z.; Gu, J.D. Molecular detection of anaerobic ammonium-oxidizing (anammox) bacteria in high-temperature petroleum reservoirs. Microb. Ecol. 2010, 60, 771–783. [Google Scholar] [CrossRef]
- Pratscher, J.; Dumont, M.G.; Conrad, R. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc. Natl. Acad. Sci. USA 2011, 108, 4170–4175. [Google Scholar] [CrossRef]
- Mullins, T.D.; Britschgi, T.B.; Krest, R.L.; Giovannoni, S.J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 1995, 40, 148–158. [Google Scholar] [CrossRef]
- Wan, Y.; Bai, Y.; He, J.; Zhang, Y.; Li, R.; Ruan, X. Temporal and spatial variations of aquatic environmental characteristics and sediment bacterial community in five regions of Lake Taihu. Aquat. Ecol. 2017, 51, 343–358. [Google Scholar] [CrossRef]
- Xia, X.; Yang, Z.; Zhang, X. Effect of suspended-sediment concentration on nitrification in river water: Importance of suspended sediment - water interface. Enviro. Sci. Technol. 2009, 43, 3681–3687. [Google Scholar] [CrossRef]
- Wu, Q.; Ruan, X.; Wu, C.; Li, R.; Wang, C.; Wan, Y. Analysis of sources and transformation of nitrogen as a contaminant in the river and lake water in the western region of the Taihu Lake basin. Acta Sci. Circumstantiae 2015, 35, 3883–3889, (In Chinese with English abstract). [Google Scholar]
- Hu, B.; Shen, L.; Du, P.; Zheng, P.; Xu, X.; Zeng, J. The influence of intense chemical pollution on the community composition, diversity and abundance of anammox bacteria in the Jiaojiang Estuary (China). PLoS ONE 2012, 7, e33826. [Google Scholar] [CrossRef]
- Lam, P.; Lavik, G.; Jensen, M.M.; van de Vossenberg, J.; Schmid, M.; Woebken, D.; Kuypers, M.M. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc. Natl. Acad. Sci. USA 2009, 106, 4752–4757. [Google Scholar] [CrossRef]
- Liu, S.; Shen, L.; Lou, L.; Tian, G.; Zheng, P.; Hu, B. Spatial distribution and factors shaping the niche segregation of ammonia-oxidizing microorganisms in the Qiantang River, China. Appl. Environ. Microbiol. 2013, 79, 4065–4071. [Google Scholar] [CrossRef]
- Jin, T.; Zhang, T.; Ye, L.; Lee, O.O.; Wong, Y.H.; Qian, P.Y. Diversity and quantity of ammonia-oxidizing Archaea and Bacteria in sediment of the Pearl River Estuary, China. Appl. Microbiol. Biotechnol. 2011, 90, 1137–1145. [Google Scholar] [CrossRef]
- Zheng, Y.; Hou, L.; Liu, M.; Lu, M.; Zhao, H.; Yin, G.; Zhou, J. Diversity, abundance, and activity of ammonia-oxidizing bacteria and archaea in Chongming eastern intertidal sediments. Appl. Microbiol. Biotechnol. 2013, 97, 8351–8363. [Google Scholar] [CrossRef]
- Sims, A.; Horton, J.; Gajaraj, S.; McIntosh, S.; Miles, R.J.; Mueller, R.; Hu, Z. Temporal and spatial distributions of ammonia-oxidizing archaea and bacteria and their ratio as an indicator of oligotrophic conditions in natural wetlands. Water Res. 2012, 46, 4121–4129. [Google Scholar] [CrossRef]
- Hugoni, M.; Etien, S.; Bourges, A.; Lepère, C.; Domaizon, I.; Mallet, C.; Mary, I. Dynamics of ammonia-oxidizing Archaea and Bacteria in contrasted freshwater ecosystems. Res. Microbiol. 2013, 164, 360–370. [Google Scholar] [CrossRef]
- Stehr, G.; Böttcher, B.; Dittberner, P.; Rath, G.; Koops, H.P. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol. Ecol. 1995, 17, 177–186. [Google Scholar] [CrossRef]
- de Bie, M.J.; Speksnijder, A.G.; Kowalchuk, G.A.; Schuurman, T.; Zwart, G.; Stephen, J.R.; Laanbroek, H.J. Shifts in the dominant populations of ammonia-oxidizing b-subclass Proteobacteria along the eutrophic Schelde estuary. Aquat. Microb. Ecol. 2001, 23, 225–236. [Google Scholar] [CrossRef]
- Bollmann, A.; Bär-Gilissen, M.J.; Laanbroek, H.J. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 2002, 68, 4751–4757. [Google Scholar] [CrossRef]
- Geets, J.; Boon, N.; Verstraete, W. Strategies of aerobic ammonia-oxidizing bacteria for coping with nutrient and oxygen fluctuations. FEMS Microbiol. Ecol. 2006, 58, 1–13. [Google Scholar] [CrossRef]
- Wei, B.; Yu, X.; Zhang, S.; Gu, L. Comparison of the community structures of ammonia-oxidizing bacteria and archaea in rhizoplanes of floating aquatic macrophytes. Microbiol. Res. 2011, 166, 468–474. [Google Scholar] [CrossRef]
- Schramm, A.; de Beer, D.; van den Heuvel, J.C.; Ottengraf, S.; Amann, R. Microscale Distribution of Populations and Activities of Nitrosospira and Nitrospira spp. along a Macroscale Gradient in a Nitrifying Bioreactor: Quantification by In Situ Hybridization and the Use of Microsensors. Appl. Environ. Microbiol. 1999, 65, 3690–3696. Available online: https://aem.asm.org/content/65/8/3690.short (accessed on 27 June 2017).
- Zeng, J.; Zhao, D.; Yu, Z.; Huang, R.; Wu, Q.L. Temperature responses of ammonia-oxidizing prokaryotes in freshwater sediment microcosms. PLoS ONE 2014, 9, e100653. [Google Scholar] [CrossRef]
- Zhang, Y.; Ruan, X.H.; Op den Camp, H.J.; Smits, T.J.; Jetten, M.S.; Schmid, M.C. Diversity and abundance of aerobic and anaerobic ammonium-oxidizing bacteria in freshwater sediments of the Xinyi River (China). Environ. Microbiol. 2007, 9, 2375–2382. [Google Scholar] [CrossRef]
- Kartal, B.; Rattray, J.; van Niftrik, L.A.; van de Vossenberg, J.; Schmid, M.C.; Webb, R.I.; Strous, M. Candidatus “Anammoxoglobus propionicus” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 2007, 30, 39–49. [Google Scholar] [CrossRef]
- Hirsch, M.D.; Long, Z.T.; Song, B. Anammox bacterial diversity in various aquatic ecosystems based on the detection of hydrazine oxidase genes (hzoA/hzoB). Microb. Ecol. 2011, 61, 264–276. [Google Scholar] [CrossRef]
- Dang, H.; Wang, C.; Li, J.; Li, T.; Tian, F.; Jin, W.; Zhang, Z. Diversity and distribution of sediment nirS-encoding bacterial assemblages in response to environmental gradients in the eutrophied Jiaozhou Bay, China. Microb. Ecol. 2009, 58, 161–169. [Google Scholar] [CrossRef]
- Gaboyer, F.; Vandenabeele-Trambouze, O.; Cao, J.; Ciobanu, M.C.; Jebbar, M.; Le Romancer, M.; Alain, K. Physiological features of Halomonas lionensis sp. nov., a novel bacterium isolated from a Mediterranean Sea sediment. Res. Microbiol. 2014, 165, 490–500. [Google Scholar] [CrossRef]
- Berendes, F.; Gottschalk, G.; Heine-Dobbernack, E.; Moore, E.R.B.; Tindall, B.J. Halomonas desiderata sp. nov, a new alkaliphilic, halotolerant and denitrifying bacterium isolated from a municipal sewage works. Syst. Appl. Microbiol. 1996, 19, 158–167. [Google Scholar] [CrossRef]
- Mergaert, J.; Swings, J. Biodiversity of microorganisms that degrade bacterial and synthetic polyesters. J. Ind. Microbiol. Biotechnol. 1996, 17, 463–469. [Google Scholar] [CrossRef]
- Qiang, K.; Zongming, R.; Rongshu, F. Identification of aerobic denitrifier isolated from Beiyunhe River sediment and denitrification. Water Technol. 2010, 3, 3, (In Chinese with English abstract). [Google Scholar]
- He, T.; Li, Z.; Sun, Q.; Xu, Y.; Ye, Q. Heterotrophic nitrification and aerobic denitrification by Pseudomonas tolaasii Y-11 without nitrite accumulation during nitrogen conversion. Bioresour. Technol. 2016, 200, 493–499. [Google Scholar] [CrossRef]
- Avrahami, S.; Bohannan, B.J. Response of Nitrosospira sp. AF-like ammonia-oxidizing bacteria to changes in temperature, soil moisture and fertilizer concentration. Appl. Environ. Microbiol. 2006, 73, 1166–1173. [Google Scholar] [CrossRef]
- Yuan, F.; Ran, W.; Shen, Q.; Wang, D. Characterization of nitrifying bacteria communities of soils from different ecological regions of China by molecular and conventional methods. Biol. Fertil. Soils 2005, 41, 22–27. [Google Scholar] [CrossRef]
- Urakawa, H.; Tajima, Y.; Numata, Y.; Tsuneda, S. Low temperature decreases the phylogenetic diversity of ammonia-oxidizing archaea and bacteria in aquarium biofiltration systems. Appl. Environ. Microbiol. 2008, 74, 894–900. [Google Scholar] [CrossRef]
- Han, P.; Gu, J.D. More refined diversity of anammox bacteria recovered and distribution in different ecosystems. Appl. Microbiol. Biotechnol. 2013, 97, 3653–3663. [Google Scholar] [CrossRef]
- Yuan, Q.; Liu, P.; Lu, Y. Differential responses of nirK-and nirS-carrying bacteria to denitrifying conditions in the anoxic rice field soil. Environ. Microbiol. Rep. 2012, 4, 113–122. [Google Scholar] [CrossRef]
- Saggar, S.; Jha, N.; Deslippe, J.; Bolan, N.S.; Luo, J.; Giltrap, D.L.; Tillman, R.W. Denitrification and N2O: N2 production in temperate grasslands: Processes, measurements, modelling and mitigating negative impacts. Sci. Total Environ. 2013, 465, 173–195. [Google Scholar] [CrossRef]


| Genes | Lake Region | Summer | Winter | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | A4 | A1 | A2 | A3 | A4 | ||
| nirS | The number of clones | 26 | 24 | 28 | 25 | 29 | 29 | 29 | 28 |
| The number of OTUs | 12 | 14 | 14 | 12 | 17 | 18 | 14 | 19 | |
| Shannon | 3.32 | 3.55 | 3.54 | 3.15 | 3.82 | 3.92 | 3.66 | 4.18 | |
| Coverage (%) | 80.8 | 62.5 | 75.0 | 72.0 | 62.1 | 58.6 | 86.2 | 53.6 | |
| amoA | The number of clones | 13 | 24 | 14 | 14 | 30 | 30 | 31 | 29 |
| The number of OTUs | 8 | 9 | 4 | 5 | 10 | 8 | 7 | 11 | |
| Shannon | 2.82 | 2.43 | 1.57 | 2.07 | 2.98 | 2.55 | 2.66 | 2.99 | |
| Coverage (%) | 61.5 | 79.2 | 85.7 | 85.7 | 86.7 | 93.3 | 96.8 | 75.9 | |
| Arch-amoA | The number of clones | 28 | 33 | 30 | 28 | 21 | 21 | 20 | 20 |
| The number of OTUs | 5 | 3 | 4 | 4 | 5 | 3 | 4 | 4 | |
| Shannon | 1.91 | 1.35 | 1.59 | 1.47 | 1.88 | 1.03 | 1.68 | 1.80 | |
| Coverage (%) | 96.4 | 97.0 | 96.7 | 96.4 | 95.2 | 95.2 | 95.0 | 95.0 | |
| hzo | The number of clones | 23 | 18 | 18 | 22 | 20 | 18 | 20 | 25 |
| The number of OTUs | 9 | 10 | 8 | 5 | 8 | 9 | 10 | 7 | |
| Shannon | 2.94 | 3.24 | 2.71 | 1.43 | 2.62 | 2.86 | 2.95 | 2.45 | |
| Coverage (%) | 87.0 | 83.3 | 77.8 | 86.4 | 90.0 | 72.2 | 70.0 | 96.0 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, Y.; Ruan, X.; Wang, J.; Shi, X. Spatial and Seasonal Variations in the Abundance of Nitrogen-Transforming Genes and the Microbial Community Structure in Freshwater Lakes with Different Trophic Statuses. Int. J. Environ. Res. Public Health 2019, 16, 2298. https://doi.org/10.3390/ijerph16132298
Wan Y, Ruan X, Wang J, Shi X. Spatial and Seasonal Variations in the Abundance of Nitrogen-Transforming Genes and the Microbial Community Structure in Freshwater Lakes with Different Trophic Statuses. International Journal of Environmental Research and Public Health. 2019; 16(13):2298. https://doi.org/10.3390/ijerph16132298
Chicago/Turabian StyleWan, Yu, Xiaohong Ruan, Jie Wang, and Xiaojun Shi. 2019. "Spatial and Seasonal Variations in the Abundance of Nitrogen-Transforming Genes and the Microbial Community Structure in Freshwater Lakes with Different Trophic Statuses" International Journal of Environmental Research and Public Health 16, no. 13: 2298. https://doi.org/10.3390/ijerph16132298
APA StyleWan, Y., Ruan, X., Wang, J., & Shi, X. (2019). Spatial and Seasonal Variations in the Abundance of Nitrogen-Transforming Genes and the Microbial Community Structure in Freshwater Lakes with Different Trophic Statuses. International Journal of Environmental Research and Public Health, 16(13), 2298. https://doi.org/10.3390/ijerph16132298





