Characteristics of Inorganic Phosphate-Solubilizing Bacteria from the Sediments of a Eutrophic Lake
Abstract
1. Introduction
2. Materials and Methods
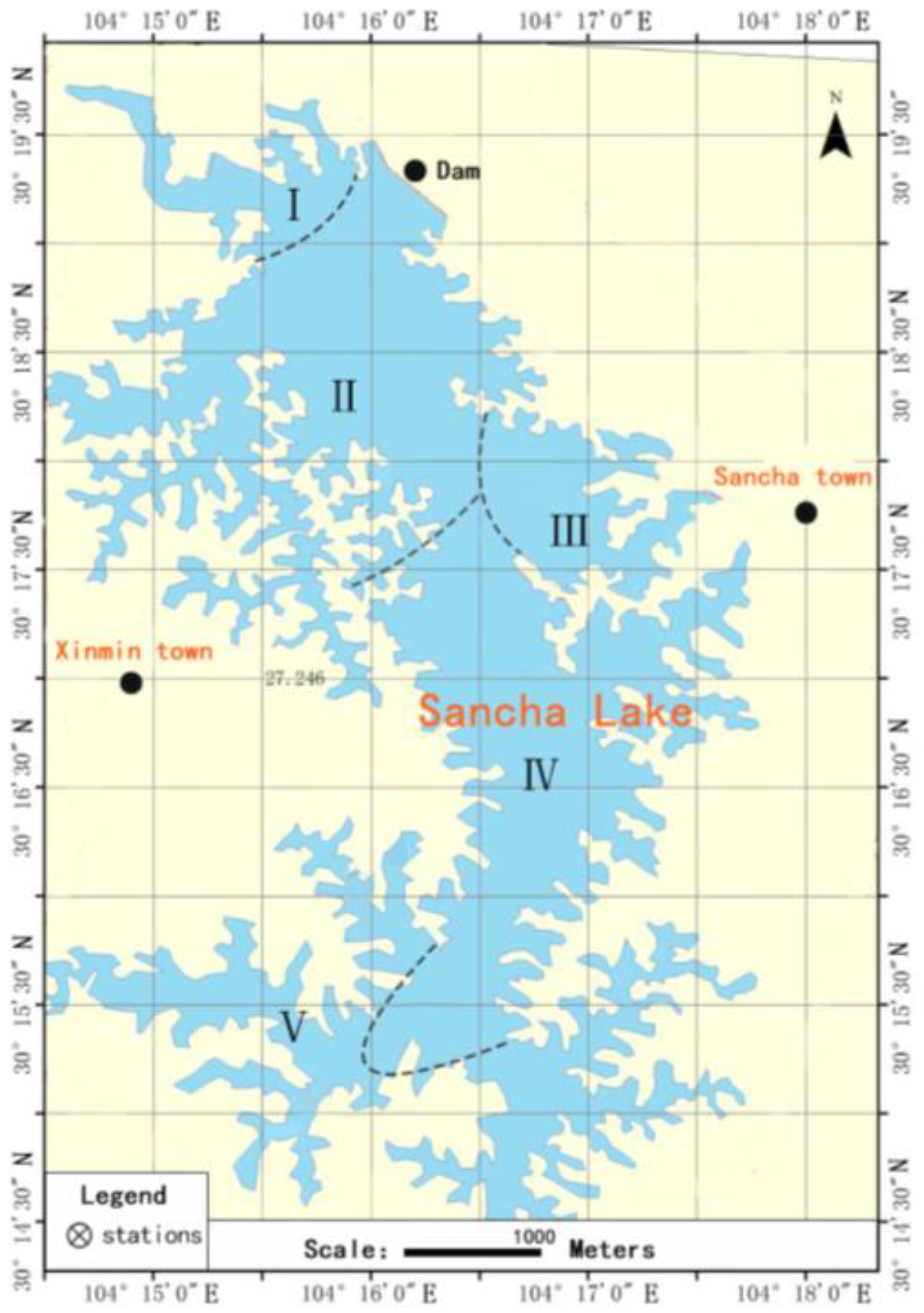
2.1. Overview of Sancha Lake and Collection of Sediments
2.2. Screening and Isolation of IPB
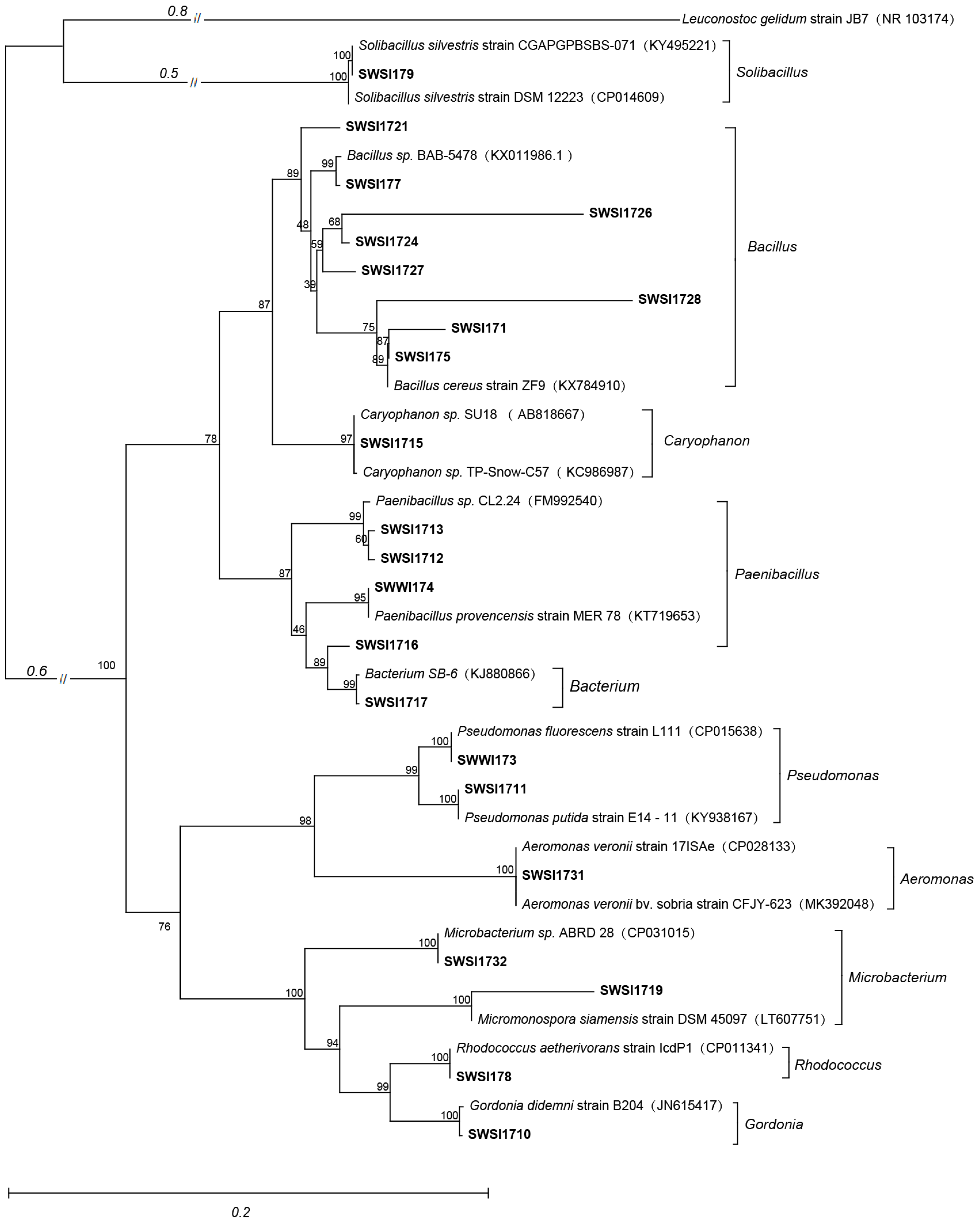
2.3. 16S rRNA Gene Amplification, Sequencing, and Phylogenetic Analysis
2.4. Evaluation of Phosphate-Solubilizing Ability of the IPB Strains
2.4.1. NBRIP Solid Culture
2.4.2. PVK Liquid Culture
2.4.3. Microcosm Setup and P Release by IPB Strains
2.4.4. Statistical Analysis
3. Results and Discussion
3.1. Results of IPB Screening and Identification
3.2. P-Solubilizing Ability
3.2.1. P-Solubilizing Activity of Screened IPB Strains
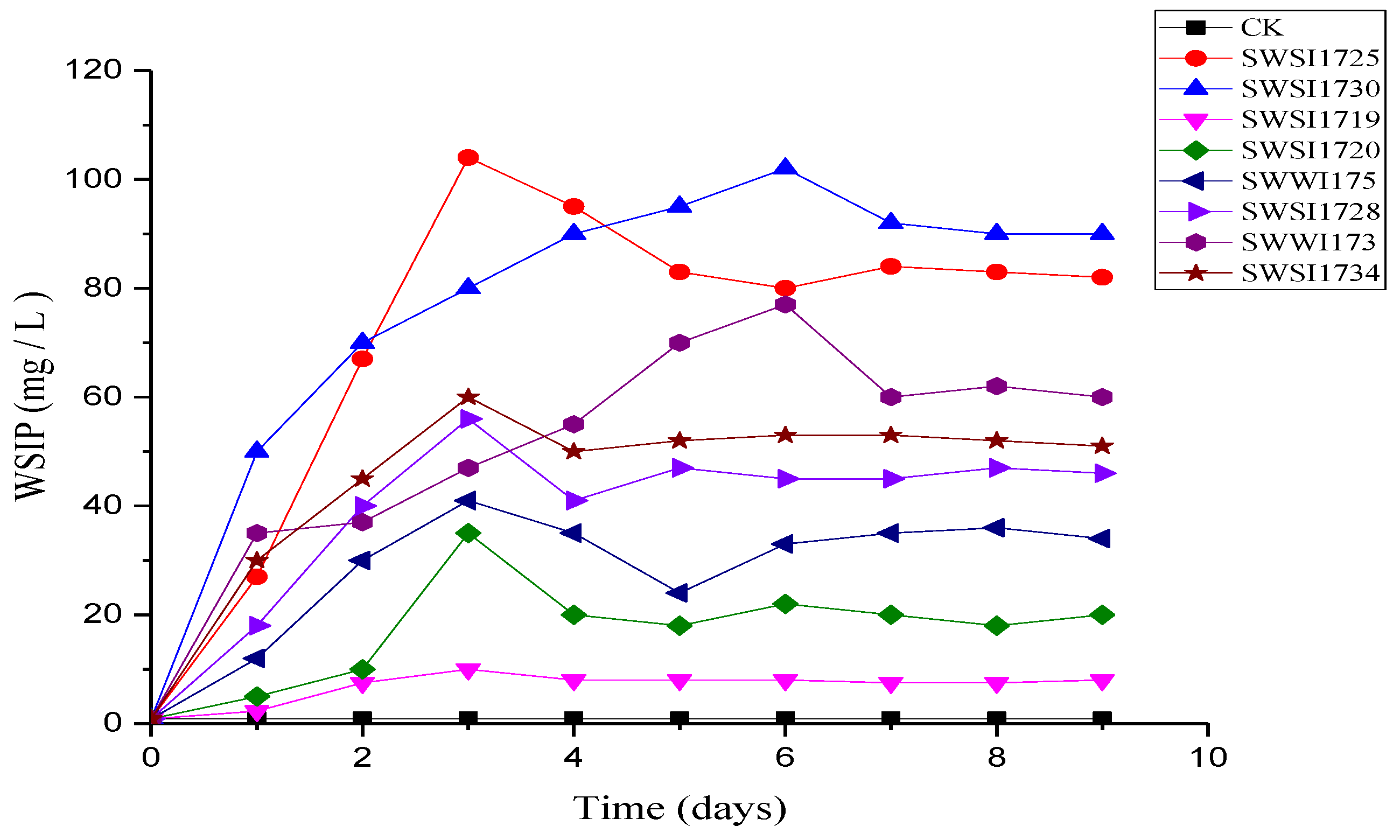
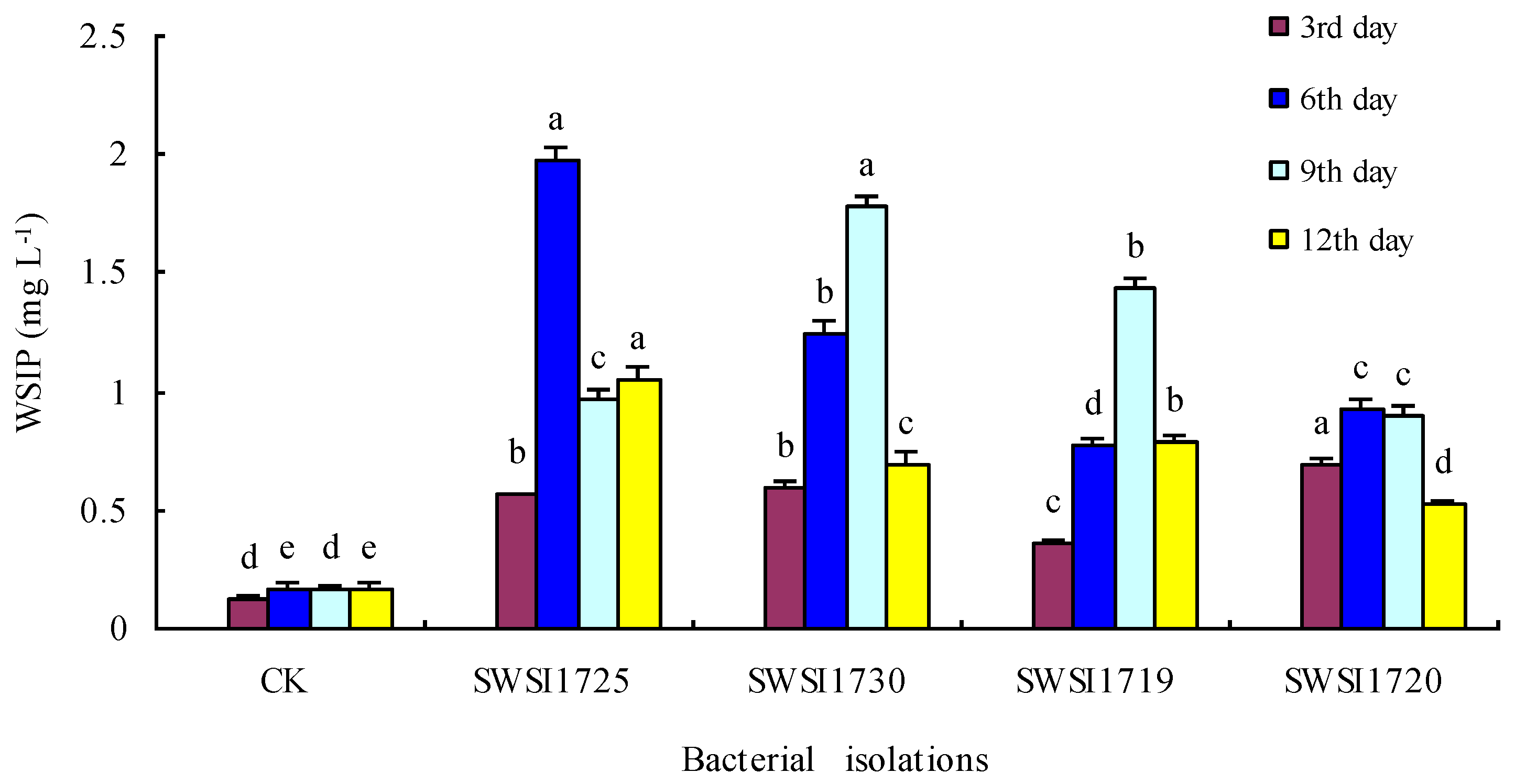
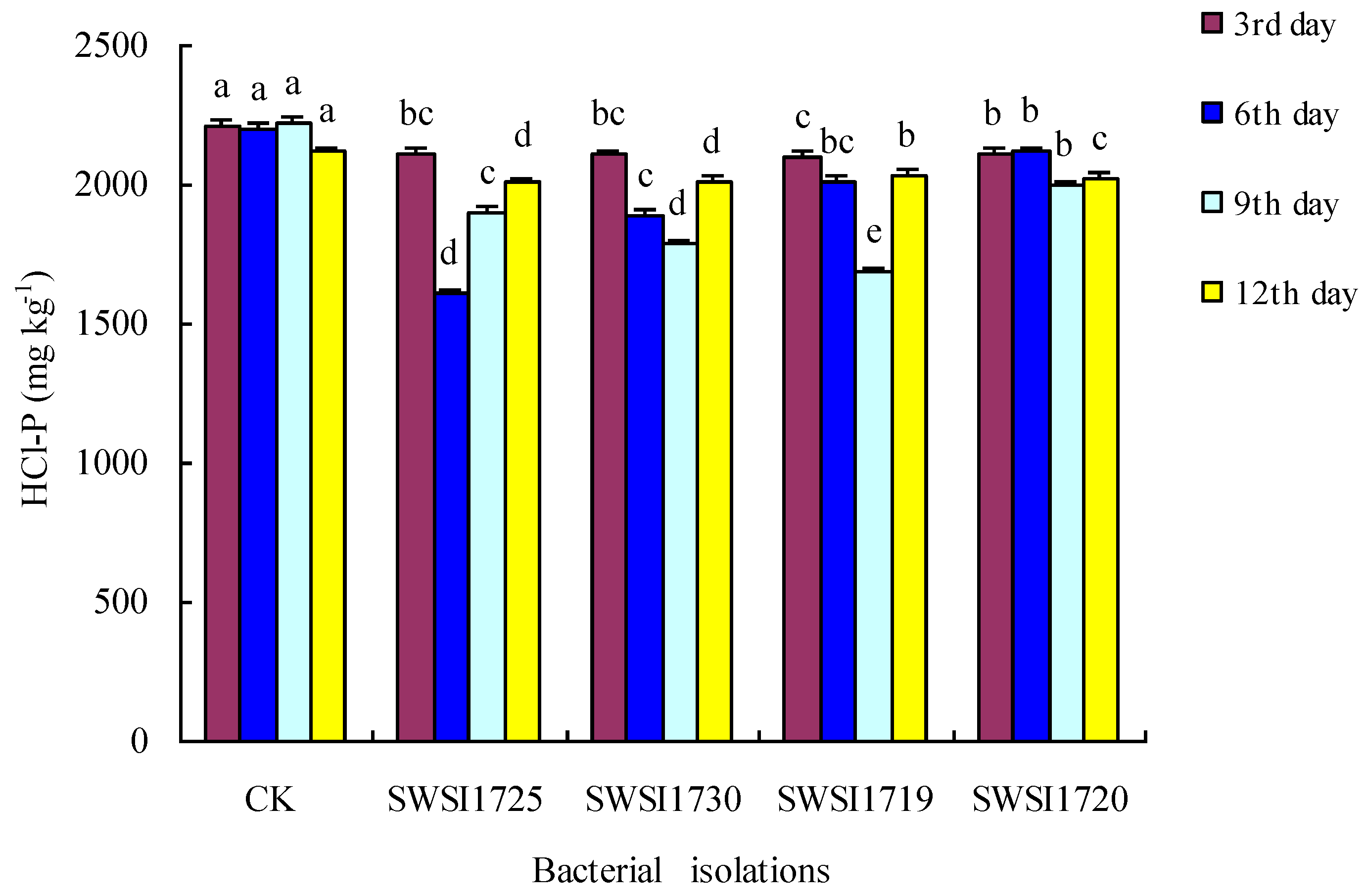
3.2.2. P Release by IPB Strains in Sediments
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xie, L.Q.; Xie, P.; Tang, H.J. Enhancement of dissolved phosphorus release from sediment to lake water by Microcystis blooms-an enclosure experiment in a hyper-eutrophic, subtropical Chinese lake. Environ. Pollut. 2003, 122, 391–399. [Google Scholar] [CrossRef]
- Ruban, V.; Brigault, S.; Demare, D. An investigation of the origin and mobility of phosphorus in freshwater sediments from Bort-Les-Orgues Reservoir, France. J. Environ. Monitor. 1999, 1, 403–407. [Google Scholar] [CrossRef]
- Wu, Q.H.; Zhang, R.D.; Huang, S. Efects of bacteria on nitrogen and phosphorus release from river sediment. J. Environ. Sci. 2008, 20, 404–412. [Google Scholar]
- Nilanjan, M.; Sanjib, K.M.; Srikanta, S. Ecological significance and phosphorus release potential of phosphate solubilizing bacteria in freshwater ecosystems. Hydrobiologia 2015, 745, 69–83. [Google Scholar]
- Mander, C.; Wakelin, S.; Young, S. Incidence and diversity of phosphate-solubilising bacteria are linked to phosphorus status in grassland soils. Soil Biol. Biochem. 2012, 44, 93–101. [Google Scholar] [CrossRef]
- Behera, B.C.; Singdevsachan, S.K.; Mishra, R.R. Diversity mechanism and biotechnology of phosphate solubilising microorganism in mangrove—A review. Biocataly. Agric. Biotechnol. 2014, 3, 97–110. [Google Scholar]
- Srinivasan, R.; Alagawadi, A.R.; Yandigeri, M.S. Characterization of phosphate-solubilizing microorganisms from salt-affected soils of India and their effect on growth of sorghum plants [Sorghum bicolor (L.) Moench]. Ann. Microbiol. 2012, 62, 93–105. [Google Scholar] [CrossRef]
- Mudryk, Z.J. Decomposition of organic and solubilization of inorganic phosphorus compounds by bacteria isolated from a marine sandy beach. Mar. Biol. 2004, 145, 1227–1234. [Google Scholar] [CrossRef]
- Qian, Y.C.; Shi, J.Y.; Chen, Y.X. Characterization of phosphate solubilizing bacteria from the sediments of shallow eutrophic lake and wetland: Isolation, molecular identification and phosphorus release ability determination. Molecules 2010, 15, 8518–8533. [Google Scholar] [CrossRef]
- Zhou, C.C.; Song, D.; Huang, Y. Isolation and characterization of organic phosphorus-mineralizing bacteria in sediment of a Chinese large shallow eutrophic lake (Lake Taihu). Geomicrobiol. J. 2011, 28, 660–666. [Google Scholar] [CrossRef]
- Del Giorgio, P.A.; Newell, R. Phosphorusand DOC vailability influence the partitioning between Bacterioplankton production and respiration in tidal marsh ecosystems. Environ. Microbiol. 2012, 14, 1296–1307. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.Y.; Tang, Y.; Fu, W.L. Relationship among sediment characteristics, eutrophication process and human activities in the Sancha Lake. China Environ. Sci. 2013, 33, 1638–1644. [Google Scholar]
- Li, Y.; Zhang, J.Q.; Gong, Z.L. Gcd gene diversity of quinoprotein glucose dehydrogenase in the sediment of Sancha lake and its response to the environment. Int. J. Environ. Res. Public Health 2019, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing bacteria. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A. 16S ribosomal DNA amplification for phylogenetic studies. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Studier, J.A.; Keppler, K.J. A note on the neighbor-joining algorithm of Saitou and Nei. Mol. Biol. Evol. 1988, 5, 729–731. [Google Scholar]
- Qiao, C.C.; Wang, T.T.; Wang, R.F. Screening phosphate solubilizing bacterial strains from maize rhizosphere and research on their plant growth promotion effect. J. Nanjing Agric. Univ. 2017, 40, 664–670. [Google Scholar]
- Zhou, Z.Y.; Luo, Y.; Ma, W.Y. Identification and determination of growth curve of four bacterium isolated from Taihu lake. J. Lake Sci. 1998, 10, 60–62. [Google Scholar]
- Pikovskaya, R.I. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Mikrobiologiya 1948, 17, 362–370. [Google Scholar]
- Huang, Q.H.; Wang, D.H.; Wang, C.X. Relationship between phosphorus morphology in sediment and lake eutrophication. China Environ. Sci. 2003, 23, 583–586. [Google Scholar]
- Ruban, V.; López-Sánchez, J.F.; Pardo, P. Harmonized Protocol and certified reference material for the determination of extractable contents of phosphorus in fresh water sediments—A synthesis of recent works. Fresen J. Anal. Chem. 2001, 370, 224–228. [Google Scholar] [CrossRef]
- Wei, G.F.; Pan, I.; Du, H. ERIC-PCR fingerprinting based community DNA hybridization to pinpoint genome specific fragments as molecular markers to identify and track populations common to healthy human guts. J. Microbiol. Methods 2004, 59, 91–108. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; He, C.G.; Yu, F.Q. Bacterial diversity cultured from shiros of tricholoma matsutake. Chin. J. Ecol. 2015, 34, 150–156. [Google Scholar]
- Li, M.; Teng, Z.D.; Zhu, J.; Song, M.Y. Research advances in heavy metal contaminated soil remediation by phosphate solubilizing microorganisms. Acta Ecol. Sin. 2018, 38, 3393–3402. [Google Scholar]
- Liu, Z.G.; Li, Y.C.; Zhang, S.A. Characterization of phosphate-solubilizing bacteria isolated from calcareous soils. Appl. Soil Ecol. 2015, 96, 217–224. [Google Scholar] [CrossRef]
- Lin, Q.M.; Zhao, X.R.; Sun, Y.X. Community characters of soil phosphobacteria in four ecosystems. Soil Environ. Sci. 2000, 9, 34–37. [Google Scholar]
- Asea, P.E.A.; Kucey, R.M.; Stewart, J.W.B. Inorganic phosphate solubilization by two Penicillum species in solution culture and soil. Soil Biol. Biochem. 1988, 20, 459–464. [Google Scholar] [CrossRef]
- Bianco, C.; Defez, R. Improvement of phosphate solubilization and medicago plant yield by an indole-3-acetic acid-overproducing strain of Sinorhizobium meliloti. Appli. Environ Microbiol. 2010, 76, 4626–4632. [Google Scholar] [CrossRef]
- Bagyaraj, D.J.; Krishnaraj, P.U.; Khanuja, S.P.S. Mineral phosphate solubilization: Agronomic implications, mechanism and molecular genetics. Proc. Indian Natl. Sci. Acad. 2000, B66, 69–82. [Google Scholar]
- You, X.J.; Su, Y.P.; Zhang, X.C. Research on effect of microbe on phosphorus releasing from sediment in dutang Reservoir. J. Fujian Norm. Univ. 2012, 28, 88–93. [Google Scholar]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Halder, A.K.; Mishra, A.K.; Bhattacharyya, P. Solubilization of rock phosphate by Rhizobium and Bradyrhizobimn. J. Gen. Appl. Microbiol. 1990, 36, 81–92. [Google Scholar] [CrossRef]
- Huang, T.L.; Chai, B.B.; Qiu, E.S. Microbial Effects on Phosphorus Release from Sediments on the Multi-phase Interface of Water-Sediment-Biofacies. J. Basic Sci. Eng. 2010, 18, 61–70. [Google Scholar]
- Kozerski, H.P.; Kleeberg, A. The sediments and the benthic pelagic exchange in the shallow lake Muggelsee. Int. Rev. Hydrobiol. 1998, 83, 77–112. [Google Scholar] [CrossRef]
- Yoo, J.K.; Ro, H.M.; Choi, W.J. Phosphorus adsorption and removal by sediments of a constructed marsh in Korea. Ecol. Eng. 2006, 27, 109–117. [Google Scholar] [CrossRef]
- Fankem, H.D.; Nwaga, A.; Deubel, L. Occurrence and functioning of phosphate solubilizing microorganisms from oil palm tree (Elaeis guineensis) rhizosphere in Cameroon. Afr. J. Biotechnol. 2006, 5, 2450–2460. [Google Scholar]

| Bacteria Strain | Nearest Phylogenetic Neighbor (Accession Number) a | Gene Identity (%) b | Taxonomical Assignment | Accession Number c |
|---|---|---|---|---|
| SWSI171 | Bacillus cereus strain ATCC 14579 (AE016877) | 98.3% | Bacillus cereus | MK569699 |
| SWSI172 | Bacillus indicus strain LMG 22858 (JGVU01000003) | 99.01% | Bacillus indicus | MK569700 |
| SWSI173 | Bacillus idriensis strain SMC 4352-2 (AY904033) | 100% | Bacillus idriensis | MK569701 |
| SWSI174 | Bacillus cibi strain DSM 16189 (JNVC01000024) | 98.61% | Bacillus idriensis | MK569702 |
| SWSI175 | Bacillus proteolyticus strain TD42 (MACH01000033) | 100% | Bacillus proteolyticus | MK569703 |
| SWSI177 | Bacillus aryabhattai strain B8W22 (EF114313) | 100% | Bacillus aryabhattai | MK569705 |
| SWSI1714 | Bacillus proteolyticus strain TD42 (MACH01000033) | 100% | Bacillus proteolyticus | MK569712 |
| SWSI1718 | Bacillus idriensis strain SMC 4352-2 (AY904033) | 100% | Bacillus idriensis | MK569716 |
| SWSI1721 | Bacillus firmus strain NBRC 15306 (BCUY01000205) | 99.6% | Bacillus firmus | MK569719 |
| SWSI1722 | Bacillus idriensis strain SMC 4352-2 (AY904033) | 100% | Bacillus idriensis | MK569720 |
| SWSI1723 | Bacillus flexus strain NBRC 15715 (BCVD01000224) | 99% | Bacillus flexus | MK569721 |
| SWSI1724 | Bacillus idriensis strain SMC 4352-2 (AY904033) | 99% | Bacillus idriensis | MK569722 |
| SWSI1725 | Bacillus idriensis strain SMC 4352-2 (AY904033) | 100% | Bacillus idriensis | MK569723 |
| SWSI1726 | Bacillus indicus strain LMG 22858 (JGVU01000003) | 98.61% | Bacillus indicus | MK569724 |
| SWSI1727 | Bacillus pumilus Strain FJAT-21963 (LJIY01000004) | 100% | Bacillus pumilus | MK569725 |
| SWSI1728 | Bacillus paramycoides strain NH24A2 (MAOI01000012) | 96.2% | Bacillus sancha sp.nov. | MK569726 |
| SWSI1729 | Bacillus idriensis SMC 4352-2 (AY904033) | 100% | Bacillus idriensis | MK569727 |
| SWSI1734 | Bacillus idriensis strain SMC 4352-2 (AY904033) | 95.9% | Bacillus jianyang sp.nov. | MK559751 |
| SWWI171 | Bacillus firmus strain NBRC 15306 (BCUY01000205) | 100% | Bacillus firmus | MK569731 |
| SWWI172 | Bacillus idriensis strain SMC 4352-2 (AY904033) | 99% | Bacillus idriensis | MK569732 |
| SWWI175 | Bacillus idriensis strain SMC 4352-2 (AY904033) | 99% | Bacillus idriensis | MK569735 |
| SWWI177 | Bacillus altitudinis strain 41KF2b (ASJC01000029) | 100% | Bacillus altitudinis | MK569737 |
| SWWI179 | Bacillus idriensis SMC 4352-2 (AY904033) | 99% | Bacillus idriensis | MK569739 |
| SWSI1716 | Paenibacillus populi strain LAM0705 (KJ000069) | 99% | Paenibacillus populi | MK569714 |
| SWSI1712 | Paenibacillus sp. strain FSL A5-0031 (MRTD01000002) | 99.3% | Paenibacillus sp. | MK569710 |
| SWSI1713 | Paenibacillus sp. strain FSL A5-0031 (MRTD01000002) | 99% | Paenibacillus sp. | MK569711 |
| SWSI1720 | Paenibacillus agaridevorans strain DSM 1355 (AJ345023) | 99% | Paenibacillus agaridevorans | MK569718 |
| SWWI174 | Paenibacillus provencensis strain 4401170 (EF212893) | 100% | Paenibacillus provencensis | MK569734 |
| SWWI178 | Paenibacillus provencensis strain 4401170 (EF212893) | 100% | Paenibacillus provencensis | MK569738 |
| SWSI1717 | Paenibacillus sp. strain AZH4 (EU592044) | 99% | Paenibacillus sp. | MK569715 |
| SWSI1711 | Pseudomonas asiatica strain RYU5 (MH517510) | 100% | Pseudomonas asiatica | MK569709 |
| SWSI1730 | Pseudomonas sp. Strain TKP (CP006852) | 100% | Pseudomonas sp. | MK569728 |
| SWWI173 | Pseudomonas sp. Strain TKP (CP006852) | 98.3% | Pseudomonas sp. | MK569733 |
| SWSI176 | Solibacillus silvestris strain DSM 12223 (CP014609) | 99% | Solibacillus silvestris | MK569704 |
| SWSI179 | Solibacillus isronensis strain B3W22 (AMCK01000046) | 99% | Solibacillus isronensis | MK569707 |
| SWSI1710 | Gordonia terrae strain NBRC 100016 (BAFD01000032) | 100% | Gordonia terrae | MK569708 |
| SWWI176 | Gordonia hongkongensis strain HKU50 (LC072670) | 100% | Gordonia hongkongensis | MK569736 |
| SWSI1731 | Aeromonas ichthiosmia strain DSM 6393(X71120) | 100% | Aeromonas ichthiosmia | MK569729 |
| SWSI1733 | Aeromonas veronii strain CECT 4257 (CDDK01000015) | 99% | Aeromonas veronii | MK562701 |
| SWSI1719 | Micromonospora halophytica DSM 43171 (jgi.1058864) | 94.2% | Micromonosporaceae jiansan sp.nov. | MK569717 |
| SWSI1732 | Microbacterium invictum strain DC-200 (AM949677) | 98.74% | Microbacterium invictum | MK569730 |
| SWSI178 | Rhodococcus aetherivorans strain 10bc312 (AF447391) | 99% | Rhodococcus aetherivorans | MK569706 |
| SWSI1715 | Caryophanon tenue strain DSM 14152(T) (MASJ01000025) | 97.39% | Caryophanon tenue | MK569713 |
| Bacteria Strain | Phosphate-Solubilizing Halo (HD/CD) a | P-Liberated (mgL−1) b | Time of Growth (h) c | pH d |
|---|---|---|---|---|
| CK | 0.87 ± 0.09 | 7.03 ± 0.06 a | ||
| SWSI171 | 1.2 ± 0.1 h | 4.37 ± 0.34 j | 72 | 4.1 ± 0.1 f |
| SWSI172 | 1.9 ± 0.2 fg | 5.41 ± 0.42 j | 72 | 6.2 ± 0.1 b |
| SWSI173 | 1.5 ± 0.2 gh | 4.02 ± 0.37 j | 72 | 5.1 ± 0.2 d |
| SWSI174 | 1.8 ± 0.2 fg | 5.09 ± 0.44 j | 72 | 3.9 ± 0.1 f |
| SWSI175 | 2.7 ± 0.3 e | 17.02 ± 1.34 gh | 72 | 3.6 ± 0.0 g |
| SWSI177 | 2.1 ± 0.3 ef | 10.703 ± 1.03 i | 72 | 4.6 ± 0.2 e |
| SWSI1714 | 1.4 ± 0.2 gh | 4.092 ± 0.44 j | 72 | 5.0 ± 0.1 d |
| SWSI1718 | 1.9 ± 0.3 fg | 4.86 ± 0.39 j | 72 | 5.3 ± 0.2 d |
| SWSI1721 | 1.2 ± 0.1 h | 4.65 ± 0.49 j | 72 | 5.1 ± 0.3 d |
| SWSI1722 | 2.9 ± 0.3 de | 19.72 ± 2.49 g | 72 | 3.7 ± 0.2 g |
| SWSI1723 | 2.5 ± 0.3 ef | 15.72 ± 1.43 h | 72 | 3.6 ± 0.1 g |
| SWSI1724 | 3.5 ± 0.4 cd | 21.15 ± 1.76 g | 72 | 3.9 ± 0.2 f |
| SWSI1725 | 4.3 ± 0.5 ab | 102.77 ± 4.06 a | 72 | 3.1 ± 0.1 h |
| SWSI1726 | 2.9 ± 0.3 de | 21.25 ± 2.70 g | 72 | 3.9 ± 0.2 f |
| SWSI1727 | 2.3 ± 0.1 ef | 13.84 ± 1.00 hi | 72 | 3.8 ± 0.3 f |
| SWSI1728 | 2.8 ± 0.3 e | 55.27 ± 3.72 d | 72 | 3.4 ± 0.1 g |
| SWSI1729 | 1.8 ± 0.2 fg | 17.51 ± 1.02 gh | 72 | 3.9 ± 0.2f |
| SWSI1734 | 4.1 ± 0.3 bc | 59.32 ± 2.71 d | 72 | 3.5 ± 0.4 g |
| SWWI171 | 2.6 ± 0.3 ef | 15.71 ± 1.70 h | 72 | 3.7 ± 0.1 g |
| SWWI172 | 4.7 ± 0.5 a | 82.43 ± 0.70 c | 72 | 3.2 ± 0.1 h |
| SWWI175 | 3.0 ± 0.4 de | 41.10 ± 0.42 e | 72 | 3.4 ± 0.1 g |
| SWWI177 | 2.1 ± 0.1 ef | 12.31 ± 0.82 i | 72 | 3.7 ± 0.3 g |
| SWWI179 | 1.2 ± 0.1 h | 2.95 ± 0.34 k | 144 | 6.0 ± 0.2 b |
| SWSI1716 | 1.9 ± 0.3 fg | 6.22 ± 0.71 j | 72 | 5.9 ± 0.0 c |
| SWSI1712 | 1.6 ± 0.2 fg | 5.08 ± 0.41 j | 144 | 6.1 ± 0.1 b |
| SWSI1713 | 1.1 ± 0.1 h | 4.85 ± 0.54 j | 144 | 6.2 ± 0.2 b |
| SWSI1720 | 2.5 ± 0.2 ef | 34.23 ± 1.91 f | 72 | 4.9 ± 0.2 e |
| SWWI174 | 2.1 ± 0.3 ef | 5.51 ± 0.56 j | 144 | 6.1 ± 0.2 b |
| SWWI178 | 1.5 ± 0.1 gh | 7.90 ± 0.65 j | 144 | 5.1 ± 0.1 d |
| SWSI1711 | 2.2 ± 0.2 ef | 12.27 ± 0.85 i | 144 | 4.0 ± 0.3 f |
| SWSI1730 | 3.5 ± 0.4 cd | 101.71 ± 5.25 a | 144 | 3.2 ± 0.1 h |
| SWWI173 | 3.0 ± 0.3 de | 76.91 ± 2.26 cd | 144 | 4.4 ± 0.2 e |
| SWSI176 | 1.6 ± 0.2 fg | 2.95 ± 0.23 k | 144 | 5.8 ± 0.1 c |
| SWSI179 | 1.7 ± 0.1 fg | 4.93 ± 0.36 j | 48 | 6.0 ± 0.1 b |
| SWSI1710 | ND | 2.29 ± 0.16 k | 48 | 6.5 ± 0.1 b |
| SWWI176 | ND | 3.22 ± 0.45 j | 48 | 5.2 ± 0.1 d |
| SWSI1731 | 3.5 ± 0.3 cd | 71.71 ± 3.15 cd | 72 | 4.0 ± 0.1 f |
| SWSI1733 | 3.4 ± 0.4 cd | 96.21 ± 7.05 b | 72 | 4.0 ± 0.11 f |
| SWSI1719 | 2.3 ± 0.3 ef | 8.37 ± 0.85 i | 72 | 6.0 ± 0.1 b |
| SWSI1732 | 1.1 ± 0.2 h | 3.35 ± 0.45 j | 72 | 6.0 ± 0.3 b |
| SWSI178 | ND | 3.04 ± 0.49 j | 72 | 6.7 ± 0.3 a |
| SWSI1715 | ND | 3.96 ± 0.42 j | 72 | 6.4 ± 0.2 b |
| SWSI1717 | ND | 1.79 ± 0.36 k | 72 | 6.4 ± 0.1 b |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhang, J.; Zhang, J.; Xu, W.; Mou, Z. Characteristics of Inorganic Phosphate-Solubilizing Bacteria from the Sediments of a Eutrophic Lake. Int. J. Environ. Res. Public Health 2019, 16, 2141. https://doi.org/10.3390/ijerph16122141
Li Y, Zhang J, Zhang J, Xu W, Mou Z. Characteristics of Inorganic Phosphate-Solubilizing Bacteria from the Sediments of a Eutrophic Lake. International Journal of Environmental Research and Public Health. 2019; 16(12):2141. https://doi.org/10.3390/ijerph16122141
Chicago/Turabian StyleLi, Yong, Jiejie Zhang, Jianqiang Zhang, Wenlai Xu, and Zishen Mou. 2019. "Characteristics of Inorganic Phosphate-Solubilizing Bacteria from the Sediments of a Eutrophic Lake" International Journal of Environmental Research and Public Health 16, no. 12: 2141. https://doi.org/10.3390/ijerph16122141
APA StyleLi, Y., Zhang, J., Zhang, J., Xu, W., & Mou, Z. (2019). Characteristics of Inorganic Phosphate-Solubilizing Bacteria from the Sediments of a Eutrophic Lake. International Journal of Environmental Research and Public Health, 16(12), 2141. https://doi.org/10.3390/ijerph16122141




