Global Identification of HIF-1α Target Genes in Benzene Poisoning Mouse Bone Marrow Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Animals, Treatments, and Blood Routine Examination
2.3. Chromatin Immunoprecipitation (ChIP) Assay
2.4. Data Analysis
2.5. Real-Time PCR
2.6. Statistical Analysis
3. Results
3.1. Benzene Induced Hematopoietic Toxicity and Increased ROS in Mice
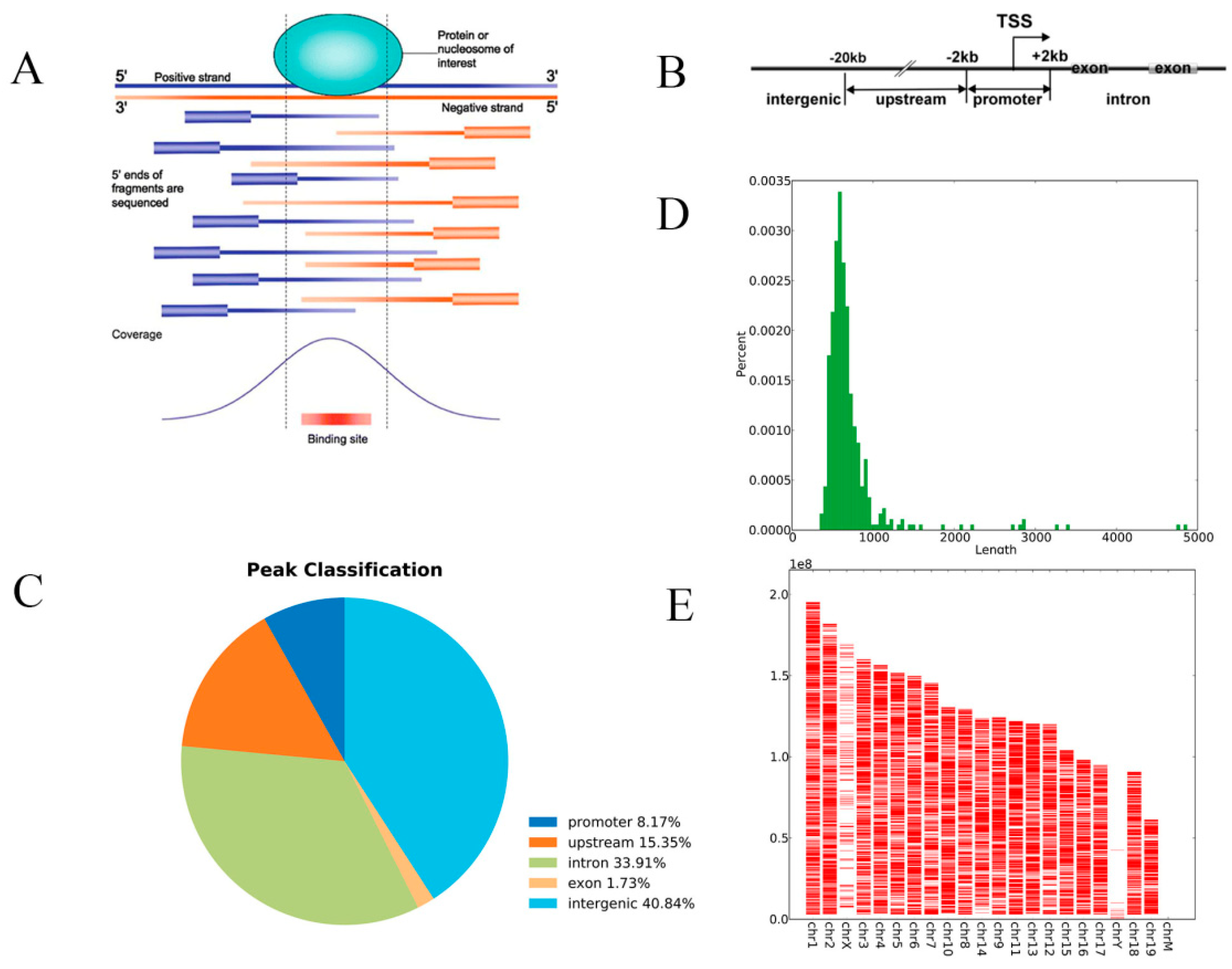
3.2. ChIP-Seq Analysis
3.3. Screening of HIF-1α Responsive Genes
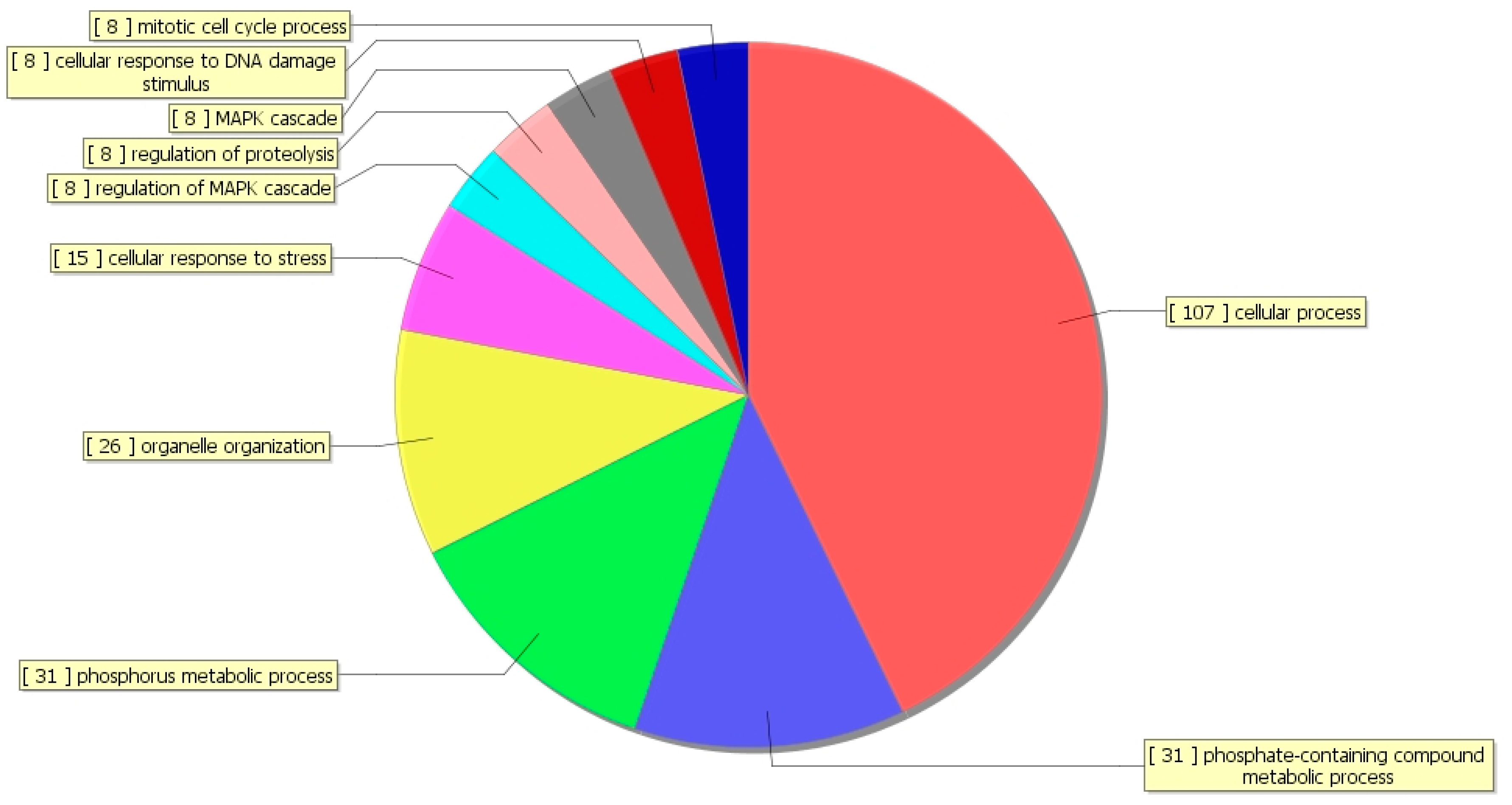
3.4. Gene Ontology (GO) Analysis
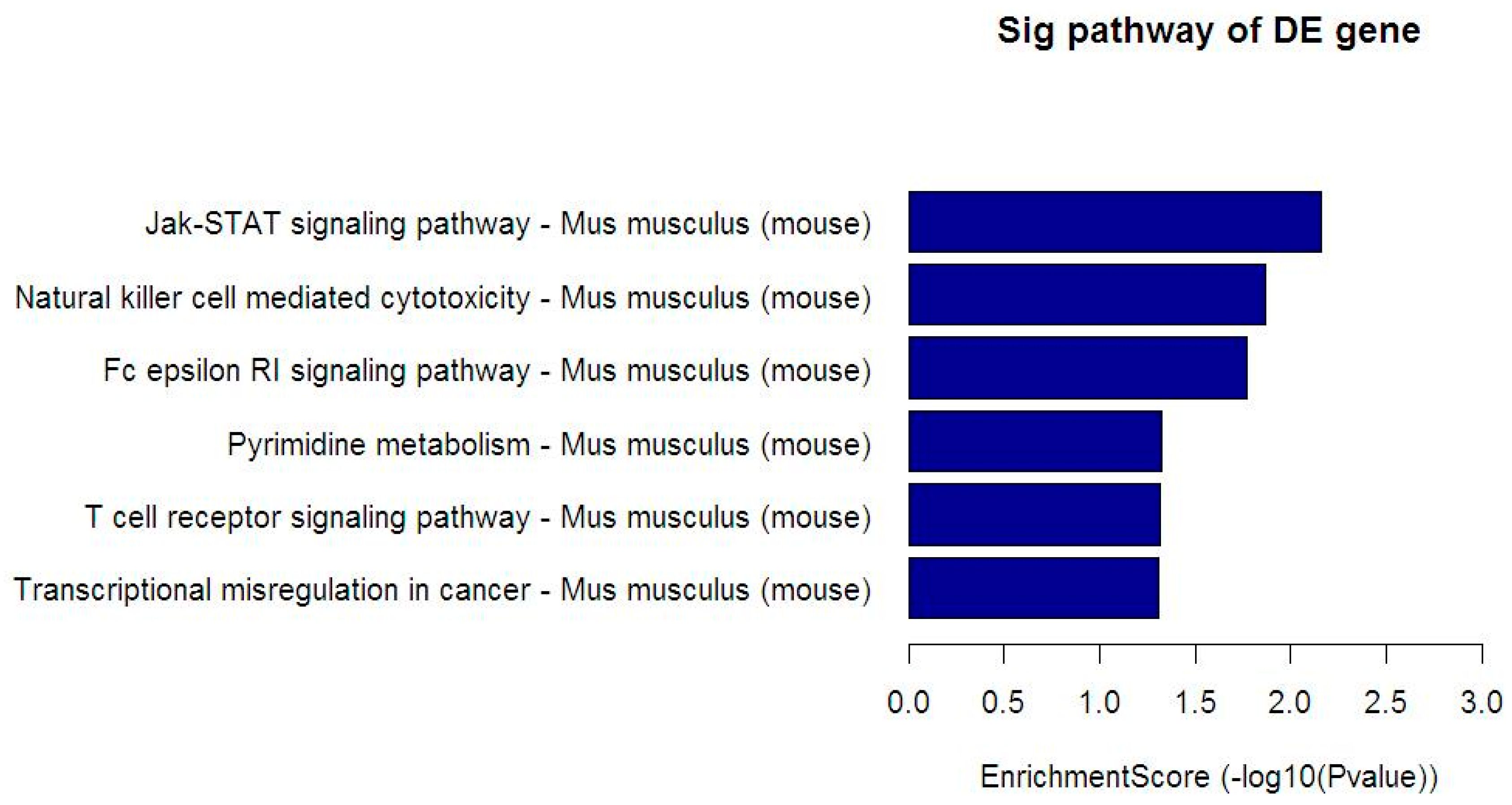
3.5. Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
3.6. Validation of HIF-1α Target Gene
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bird, M.G.; Greim, H.; Kaden, D.A.; Rice, J.M.; Snyder, R. Benzene 2009—Health effects and mechanisms of bone marrow toxicity: Implications for t-AML and the mode of action framework. Chem.-Biol. Interact. 2010, 184, 3–6. [Google Scholar] [CrossRef] [PubMed]
- McHale, C.M.; Zhang, L.; Smith, M.T. Current understanding of the mechanism of benzene-induced leukemia in humans: Implications for risk assessment. Carcinogenesis 2012, 33, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Pyatt, D.W.; Hays, S.M.; English, C.; Cushing, C.A. United States Voluntary Children’s Chemical Evaluation Program (VCCEP) risk assessment for children exposed to benzene. Toxicol. Mech. Methods 2012, 22, 81–104. [Google Scholar] [CrossRef] [PubMed]
- Santiago, F.; Alves, G.; Otero, U.B.; Tabalipa, M.M.; Scherrer, L.R.; Kosyakova, N.; Ornellas, M.H.; Liehr, T. Monitoring of gas station attendants exposure to benzene, toluene, xylene (BTX) using three-color chromosome painting. Mol. Cytogenet. 2014, 7, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Zhang, J.; Yin, L.; Pu, Y. Investigation into variation of endogenous metabolites in bone marrow cells and plasma in C3H/He mice exposed to benzene. Int. J. Mol. Sci. 2014, 15, 4994–5010. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Li, J.; Chen, L.; Xu, Z.; He, D.; Zhou, Y.; Zhu, Y.; Wei, F.; Li, J. Biomass fuels and coke plants are important sources of human exposure to polycyclic aromatic hydrocarbons, benzene and toluene. Environ. Res. 2014, 135, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Lai, Y.; Hu, K.; Wei, Q.; Liu, Y. Human CYP2E1-dependent and human sulfotransferase 1A1-modulated induction of micronuclei by benzene and its hydroxylated metabolites in Chinese hamster V79-derived cells. Mutat. Res. 2014, 770, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Ludin, A.; Gur-Cohen, S.; Golan, K.; Kaufmann, K.B.; Itkin, T.; Medaglia, C.; Lu, X.J.; Ledergor, G.; Kollet, O.; Lapidot, T. Reactive oxygen species regulate hematopoietic stem cell self-renewal, migration and development, as well as their bone marrow microenvironment. Antioxid. Redox Signal. 2014, 21, 1605–1619. [Google Scholar] [CrossRef] [PubMed]
- Richardson, C.; Yan, S.; Vestal, C.G. Oxidative stress, bone marrow failure, and genome instability in hematopoietic stem cells. Int. J. Mol. Sci. 2015, 16, 2366–2385. [Google Scholar] [CrossRef] [PubMed]
- Suda, T.; Takubo, K.; Semenza, G.L. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 2011, 9, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Urao, N.; Ushio-Fukai, M. Redox regulation of stem/progenitor cells and bone marrow niche. Free Rad. Biol. Med. 2013, 54, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Snyder, R. The bone marrow niche, stem cells, and leukemia: Impact of drugs, chemicals, and the environment. Ann. N.Y. Acad. Sci. 2014, 1310, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Labbaye, C.; Castelli, G.; Pelosi, E. Oxidative stress and hypoxia in normal and leukemic stem cells. Exp. Hematol. 2016, 44, 540–560. [Google Scholar] [CrossRef] [PubMed]
- Danet, G.H.; Pan, Y.; Luongo, J.L.; Bonnet, D.A.; Simon, M.C. Expansion of human SCID-repopulating cells under hypoxic conditions. J. Clin. Investig. 2003, 112, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Forristal, C.E.; Nowlan, B.; Jacobsen, R.N.; Barbier, V.; Walkinshaw, G.; Walkley, C.R.; Winkler, I.G.; Levesque, J.P. HIF-1alpha is required for hematopoietic stem cell mobilization and 4-prolyl hydroxylase inhibitors enhance mobilization by stabilizing HIF-1alpha. Leukemia 2015, 29, 1366–1378. [Google Scholar] [CrossRef] [PubMed]
- Regan, J.N.; Lim, J.; Shi, Y.; Joeng, K.S.; Arbeit, J.M.; Shohet, R.V.; Long, F. Up-regulation of glycolytic metabolism is required for HIF1alpha-driven bone formation. Proc. Natl. Acad. Sci. USA 2014, 111, 8673–8678. [Google Scholar] [CrossRef] [PubMed]
- Takubo, K.; Nagamatsu, G.; Kobayashi, C.I.; Nakamura-Ishizu, A.; Kobayashi, H.; Ikeda, E.; Goda, N.; Rahimi, Y.; Johnson, R.S.; Soga, T.; et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 2013, 12, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, C.; D’Aprile, A.; Ripoli, M.; Scrima, R.; Lecce, L.; Boffoli, D.; Tabilio, A.; Capitanio, N. Bone-marrow derived hematopoietic stem/progenitor cells express multiple isoforms of NADPH oxidase and produce constitutively reactive oxygen species. Biochem. Biophys. Res. Commun. 2007, 353, 965–972. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, C.; Ria, R.; Scrima, R.; Cela, O.; D’Aprile, A.; Boffoli, D.; Falzetti, F.; Tabilio, A.; Capitanio, N. Characterization of mitochondrial and extra-mitochondrial oxygen consuming reactions in human hematopoietic stem cells. Novel evidence of the occurrence of NAD(P)H oxidase activity. J. Biol. Chem. 2005, 280, 26467–26476. [Google Scholar] [CrossRef] [PubMed]
- Urao, N.; McKinney, R.D.; Fukai, T.; Ushio-Fukai, M. NADPH oxidase 2 regulates bone marrow microenvironment following hindlimb ischemia: Role in reparative mobilization of progenitor cells. Stem Cells 2012, 30, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Hirao, A.; Arai, F.; Takubo, K.; Matsuoka, S.; Miyamoto, K.; Ohmura, M.; Naka, K.; Hosokawa, K.; Ikeda, Y.; et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat. Med. 2006, 12, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.Y.; Sharkis, S.J. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood 2007, 110, 3056–3063. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, J.; Yin, L.; Pu, Y. Involvement of hypoxia-inducible factor-1 alpha (HIF-1alpha) in inhibition of benzene on mouse hematopoietic system. J. Toxicol. Environ. Health A 2016, 79, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Ciurea, A.V.; Palade, C.; Voinescu, D.; Nica, D.A. Subarachnoid hemorrhage and cerebral vasospasm—Literature review. J. Med. Life 2013, 6, 120–125. [Google Scholar] [PubMed]
- Pugh, C.W.; Ratcliffe, P.J. Regulation of angiogenesis by hypoxia role of the HIF system. Nat. Med. 2003, 9, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Jaakkola, P.; Mole, D.R.; Tian, Y.-M.; Wilson, M.I.; Gielbert, J.; Gaskell, S.J.; Kriegsheim, A.V.; Hebestreit, H.F.; Mukherji, M.; Schofield, C.J.; et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001, 292, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, N.R.; Semenza, G.L. Oxygen Sensing and Homeostasis. Physiology 2015, 30, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Sahin, E.; Colla, S.; Liesa, M.; Moslehi, J.; Muller, F.L.; Guo, M.; Cooper, M.; Kotton, D.; Fabian, A.J.; Walkey, C.; et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 2011, 470, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr.; Ratcliffe, P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Keswani, S.C.; Bosch-Marce, M.; Reed, N.; Fischer, A.; Semenza, G.L.; Hoke, A. Nitric oxide prevents axonal degeneration by inducing HIF-1-dependent expression of erythropoietin. Proc. Natl. Acad. Sci. USA 2011, 108, 4986–4990. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.; Ponka, P.; Prchal, J.T. Hypoxia. 5. Hypoxia and hematopoiesis. Am. J. Physiol. Cell Physiol. 2011, 300, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.-G.; Han, Z.-T.; He, S.-H.; Wu, X.-D.; Chen, T.-R.; Shao, C.-H.; Chen, D.-L.; Su, N.; Chen, Y.-M.; Wang, T.; et al. HIF1/2α mediates hypoxia-induced LDHA expression in human pancreatic cancer cells. Oncotarget 2017, 8, 24840–24852. [Google Scholar] [PubMed]
- Ren, H.; Accili, D.; Duan, C. Hypoxia converts the myogenic action of insulin-like growth factors into mitogenic action by differentially regulating multiple signaling pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 5857–5862. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Xia, L.; Ning, X.; Liu, L.; Sun, W.; Huang, C.; Wang, H.; Sun, S. Hypoxia-induced Bmi1 promotes renal tubular epithelial cell-mesenchymal transition and renal fibrosis via PI3K/Akt signal. Mol. Biol. Cell 2014, 25, 2650–2659. [Google Scholar] [CrossRef] [PubMed]
- Park, I.-K.; Qian, D.; Kiel, M.; Becker, M.W.; Pihalja, M.; Weissman, I.L.; Morrison, S.J.; Clarke, M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 2003, 423, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, A.; Esplugues, A.; Estarlich, M.; Llop, S.; Cases, A.; Mantilla, E.; Ballester, F.; Iniguez, C. Infants’ indoor and outdoor residential exposure to benzene and respiratory health in a Spanish cohort. Environ. Pollut. 2017, 222, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Simsek, T.; Kocabas, F.; Zheng, J.; Deberardinis, R.J.; Mahmoud, A.I.; Olson, E.N.; Schneider, J.W.; Zhang, C.C.; Sadek, H.A. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 2010, 7, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Hu, H.; Chang, R.; Zhong, J.; Knabel, M.; O’Meally, R.; Cole, R.N.; Pandey, A.; Semenza, G.L. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 2011, 145, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Nishida, C.; Kusubata, K.; Tashiro, Y.; Gritli, I.; Sato, A.; Ohki-Koizumi, M.; Morita, Y.; Nagano, M.; Sakamoto, T.; Koshikawa, N.; et al. MT1-MMP plays a critical role in hematopoiesis by regulating HIF-mediated chemokine/cytokine gene transcription within niche cells. Blood 2012, 119, 5405–5416. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin. Cancer Biol. 2009, 19, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Chachami, G.; Paraskeva, E.; Mingot, J.M.; Braliou, G.G.; Gorlich, D.; Simos, G. Transport of hypoxia-inducible factor HIF-1alpha into the nucleus involves importins 4 and 7. Biochem. Biophys. Res. Commun. 2009, 390, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Russell, L.; Naora, H.; Naora, H. Down-regulated RPS3a/nbl Expression during Retinoid-induced Differentiation of HL-60 Cell: A Close Association with Diminished Susceptibility to Actinomycin D-stimulated Apoptosis. Cell Struct. Funct. 2000, 25, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Penna, I.; Du, H.; Ferriani, R.; Taylor, H.S. Calpain5 expression is decreased in endometriosis and regulated by HOXA10 in human endometrial cells. Mol. Hum. Reprod. 2008, 14, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Chiusolo, V.; Jacquemin, G.; Yonca Bassoy, E.; Vinet, L.; Liguori, L.; Walch, M.; Kozjak-Pavlovic, V.; Martinvalet, D. Granzyme B enters the mitochondria in a Sam50-, Tim22- and mtHsp70-dependent manner to induce apoptosis. Cell Death Differ. 2017, 24, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Radke, I.; Gotte, M.; Smollich, M.; Scharle, N.; Kiesel, L.; Wulfing, P. Expression of PRL-3 regulates proliferation and invasion of breast cancer cells in vitro. Arch. Gynecol. Obstet. 2017, 296, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Moreira, S.; Moreno, S.G.; Ghinatti, G.; Lewandowski, D.; Hoffschir, F.; Ferri, F.; Gallouet, A.S.; Gay, D.; Motohashi, H.; Yamamoto, M.; et al. Low-Dose Irradiation Promotes Persistent Oxidative Stress and Decreases Self-Renewal in Hematopoietic Stem Cells. Cell Rep. 2017, 20, 3199–3211. [Google Scholar] [CrossRef] [PubMed]
- Bakkar, N.; Kousari, A.; Kovalik, T.; Li, Y.; Bowser, R. RBM45 Modulates the Antioxidant Response in Amyotrophic Lateral Sclerosis through Interactions with KEAP1. Mol. Cell. Biol. 2015, 35, 2385–2399. [Google Scholar] [CrossRef] [PubMed]
| Reagent | Volume |
|---|---|
| DNase/RNase-Free Water | 6 μL |
| SYBR Green PCR Master mix (Toyobo) | 10 μL |
| Forward /Reverse primer (100 μM) | 0.8 μL |
| ROX Reference Dye | 0.4 μL |
| cDNA | 2 μL |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| β-actin | CTATGCTCTCCCTCACGCCA | TCACGCACGATTTCCCTCTC |
| Ptp4a3 | CCTGTAAGGCAGCCCCAACTA | GTGTCTTAGCCAGGGTTTTATG |
| Samd4 | CAGACGAGGAAGAGTAGAGGG | ACAGACGCATTACTATCACCAA |
| Ifitm3 | GAGGACCAAGGTGCTGATGTT | TAGCCTATGCCTACTCCGTGAA |
| Gzmb | GCCAGTCTTTGCAGTCCTTTA | CTCTGATTACCCATCGTCCCT |
| Acta1 | CCTTCTGACCCATACCTACCAT | AAGCCTCACTTCCTACCCTCG |
| Rbm45 | TTTAGGTTCAGCCAAGAGTGC | CGGGAGAAGTTCAAGGTGTAT |
| Capn5 | TGATTCCTCTTAGCCTCGTCA | GTGGATTTCACAGGTGGTGTT |
| Rps3a1 | AGCAAGGCTCACTTCAAACAC | TTAGGAACATCGGGAAGACAC |
| Ipo4 | AGCCACTCCTCCATGTCTTCC | CATCTTTGGGTTGGGCGTACT |
| Asb15 | GAGCCTCAGCATAATCTCATC | TATACTTCGCCGTCTCCAATA |
| Rabgap1l | AGAGGCGGCTTAGTTGTTTGG | GCGGTCTACCTGTTGATTGCC |
| Name | Raw Reads | Mapped to Reference Genome | Mapped Percentage |
|---|---|---|---|
| C-Input | 13,294,498 | 13,049,446 | 98.16% |
| C1-IP | 16,759,284 | 16,585,336 | 98.96% |
| C2-IP | 15,330,270 | 14,798,086 | 96.53% |
| L-Input | 14,177,435 | 13,932,833 | 98.27% |
| L1-IP | 17,959,618 | 17,666,744 | 98.37% |
| L2-IP | 16,528,262 | 15,512,603 | 93.86% |
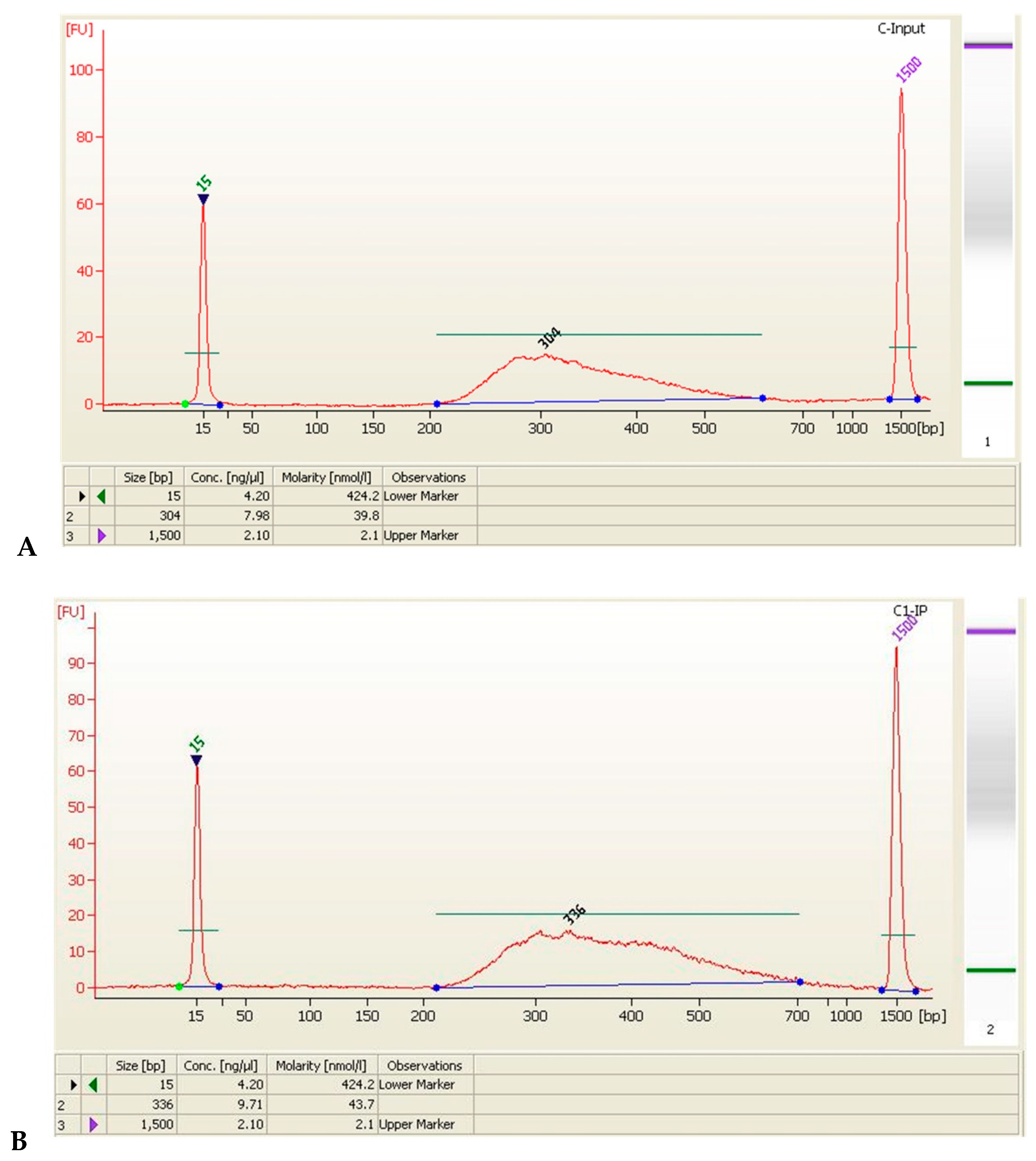
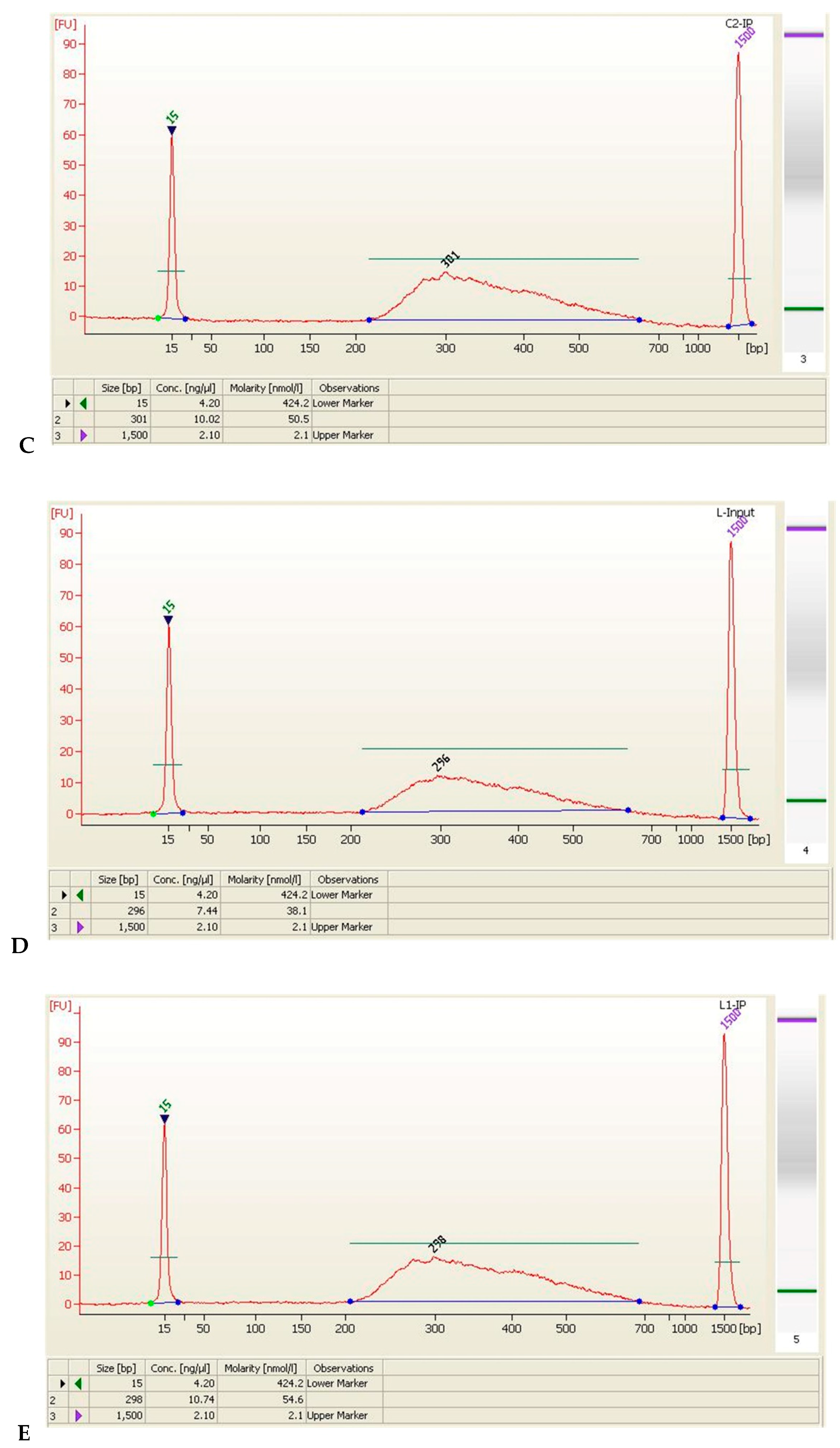
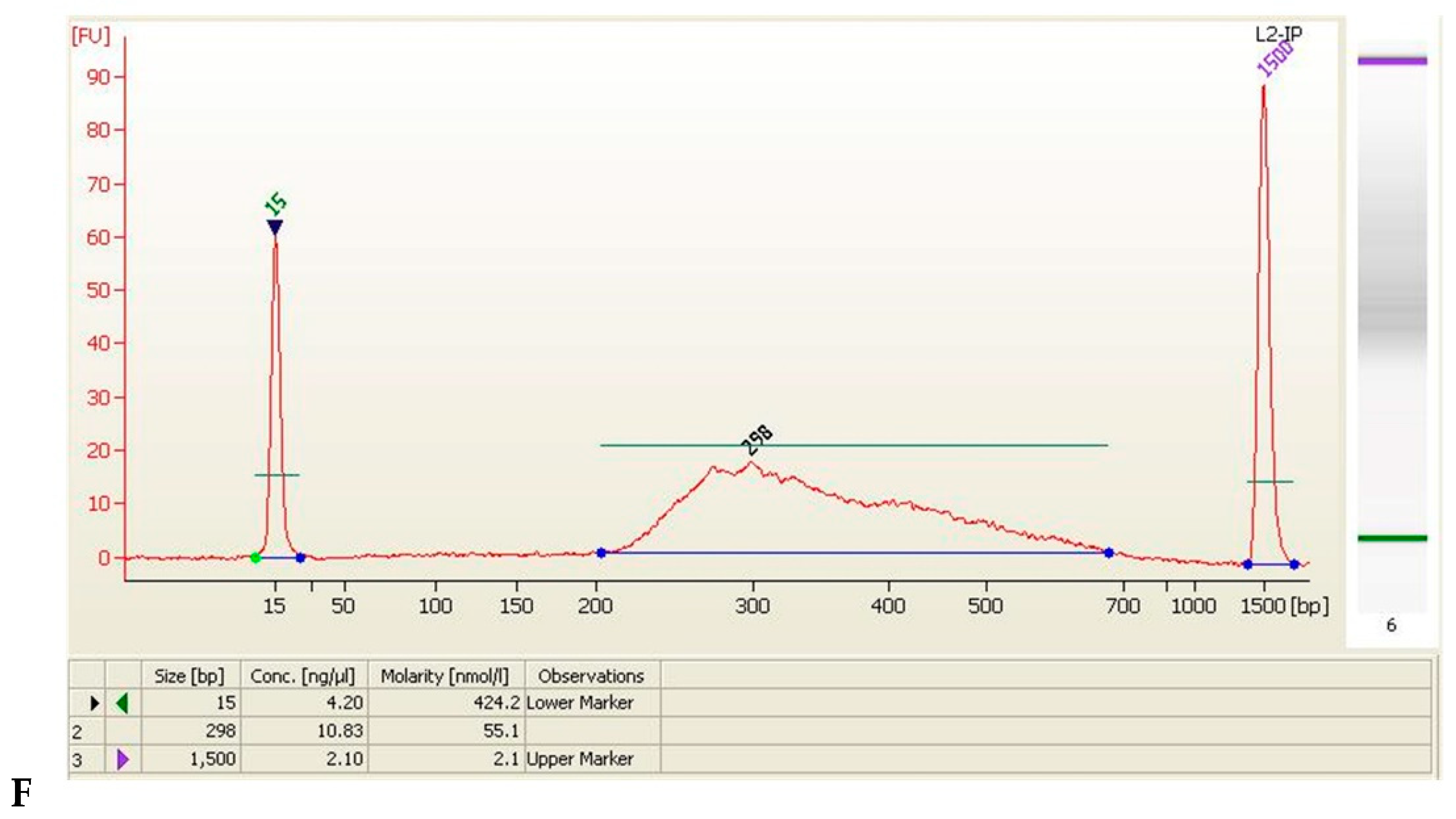
| Sample Name | Size (bp) | Concentration (ng/μL) | Concentration (nmol/L) | Volume (μL) | Total Amount (ng) |
|---|---|---|---|---|---|
| C-Input | 304 | 7.98 | 39.8 | 20 | 159.6 |
| C1-IP | 336 | 9.71 | 43.7 | 20 | 194.2 |
| C2-IP | 301 | 10.02 | 50.5 | 20 | 200.4 |
| L-Input | 296 | 7.44 | 38.1 | 20 | 148.8 |
| L1-IP | 298 | 10.74 | 54.6 | 20 | 214.8 |
| L2-IP | 298 | 10.83 | 55.1 | 20 | 216.6 |
| Down-Regulated Gene Name | Fold Change | FDR | Up-Regulated Gene Name | Fold Change | FDR |
|---|---|---|---|---|---|
| Olfr1120 | −138.5 | 0.000 | Fpgt | 128.8 | 0.005 |
| Hilpda | −116.3 | 0.003 | Lrriq3 | 128.8 | 0.005 |
| Ebag9 | −116.3 | 0.003 | Pitpnm2 | 113.6 | 0.007 |
| Ptp4a3 | −105.4 | 0.002 | Rspry1 | 111 | 0.006 |
| Rgs1 | −105.4 | 0.002 | Fam192a | 111 | 0.006 |
| Tmc1 | −99.9 | 0.005 | Otud1 | 106 | 0.007 |
| Snx33 | −94.4 | 0.007 | Rab5c | 104.3 | 0.010 |
| Spred3 | −94.3 | 0.007 | Edn3 | 102.3 | 0.004 |
| Snhg17 | −94.3 | 0.007 | Ccdc88a | 97.3 | 0.007 |
| Olfr1113 | −94.3 | 0.007 | Sod2 | 96.7 | 0.012 |
| Commd9 | −89 | 0.005 | Sept9 | 96.7 | 0.012 |
| Rhno1 | −89 | 0.005 | C330013E15Rik | 96.1 | 0.012 |
| Foxm1 | −89 | 0.005 | Dynll1 | 95.5 | 0.012 |
| Pgm5 | −88.9 | 0.005 | Gm13830 | 95.5 | 0.012 |
| Mug1 | −88.8 | 0.009 | Asnsd1 | 94.1 | 0.007 |
| Samd4 | −88.8 | 0.009 | 4930486L24Rik | 90.4 | 0.012 |
| Pathway | Total | Hits | Target Gene Name |
|---|---|---|---|
| Jak-STAT signaling pathway | 155 | 5 | CSF2RA, GRB2, PIK3CA, SPRED2, SPRED3 |
| Natural killer cell mediated cytotoxicity | 119 | 4 | GRB2, GZMB, LCP2, PIK3CA |
| Fc epsilon RI signaling pathway | 70 | 3 | GRB2, LCP2, PIK3CA |
| Pyrimidine metabolism | 104 | 3 | CTPS, NT5C3B, TXNRD1 |
| T cell receptor signaling pathway | 105 | 3 | GRB2, LCP2, PIK3CA |
| Transcriptional misregulation in cancer | 178 | 4 | BCL2A1C, GZMB, JMJD1C, LYL1 |
| Gene | 2−ΔΔC(t) | |
|---|---|---|
| LB/C | HB/C | |
| Ptp4a3 | 0.11985 *** | 0.07554 *** |
| Samd4 | 0.10351 *** | 0.04879 *** |
| Ifitm3 | 0.39707 * | 0.53172 |
| Gzmb | 0.41548 | 0.35731 * |
| Acta1 | 0.06175 ** | 0.11185 * |
| Rbm45 | 0.70175 | 0.49878 ** |
| Capn5 | 0.08935 *** | 0.06172 *** |
| Rps3a1 | 0.48881 *** | 0.57754 ** |
| Ipo4 | 0.39123 ** | 0.31920 ** |
| Asb15 | 0.09352 ** | 0.25600 |
| Rabgap1l | 0.23461 *** | 0.41362 ** |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Man, Z.; Meng, X.; Sun, F.; Pu, Y.; Xu, K.; Sun, R.; Zhang, J.; Yin, L.; Pu, Y. Global Identification of HIF-1α Target Genes in Benzene Poisoning Mouse Bone Marrow Cells. Int. J. Environ. Res. Public Health 2018, 15, 2531. https://doi.org/10.3390/ijerph15112531
Man Z, Meng X, Sun F, Pu Y, Xu K, Sun R, Zhang J, Yin L, Pu Y. Global Identification of HIF-1α Target Genes in Benzene Poisoning Mouse Bone Marrow Cells. International Journal of Environmental Research and Public Health. 2018; 15(11):2531. https://doi.org/10.3390/ijerph15112531
Chicago/Turabian StyleMan, Zhaodi, Xing Meng, Fengxia Sun, Yunqiu Pu, Kai Xu, Rongli Sun, Juan Zhang, Lihong Yin, and Yuepu Pu. 2018. "Global Identification of HIF-1α Target Genes in Benzene Poisoning Mouse Bone Marrow Cells" International Journal of Environmental Research and Public Health 15, no. 11: 2531. https://doi.org/10.3390/ijerph15112531
APA StyleMan, Z., Meng, X., Sun, F., Pu, Y., Xu, K., Sun, R., Zhang, J., Yin, L., & Pu, Y. (2018). Global Identification of HIF-1α Target Genes in Benzene Poisoning Mouse Bone Marrow Cells. International Journal of Environmental Research and Public Health, 15(11), 2531. https://doi.org/10.3390/ijerph15112531





