Pseudonocardians A–C, New Diazaanthraquinone Derivatives from a Deap-Sea Actinomycete Pseudonocardia sp. SCSIO 01299
Abstract
:1. Introduction
2. Results and Discussion
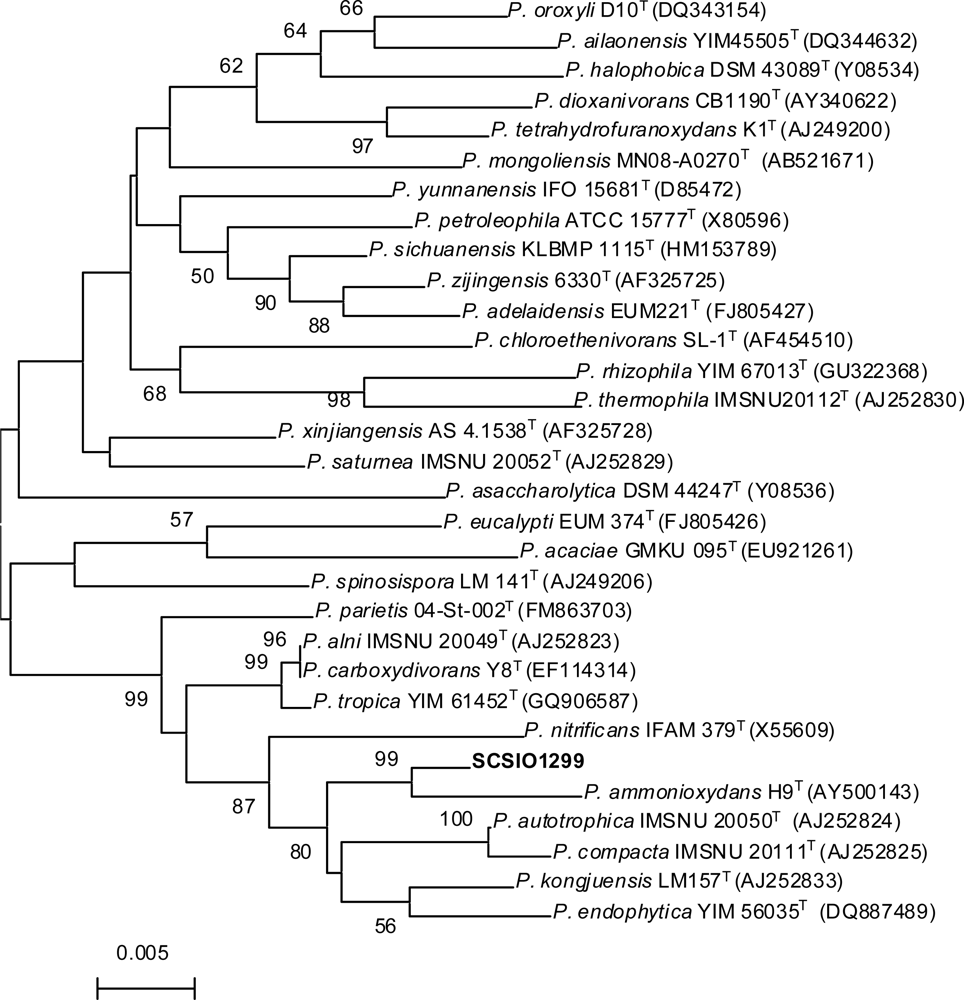
2.1. Taxonomy of the Producing Strain
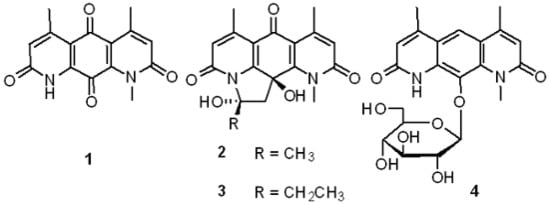
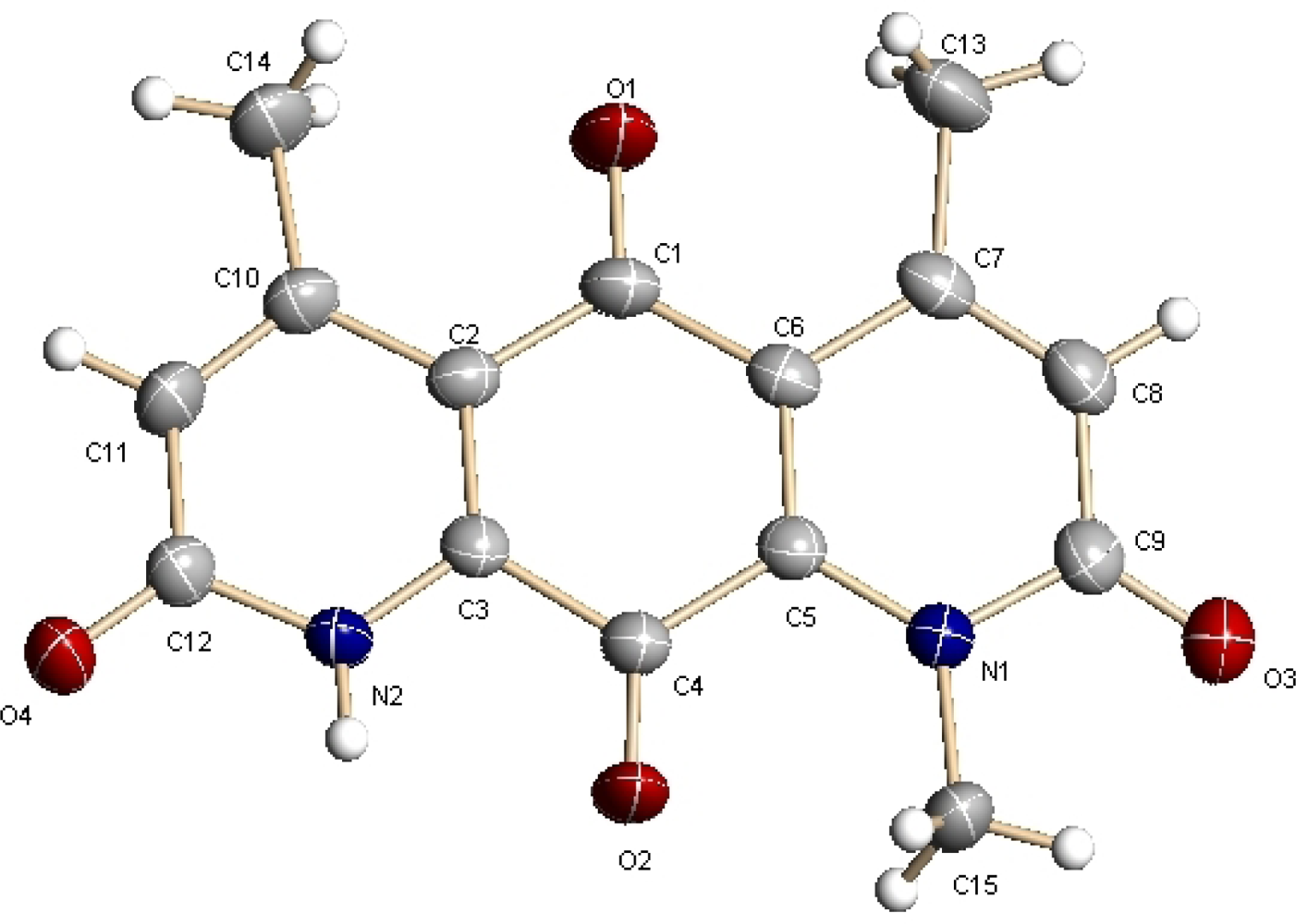
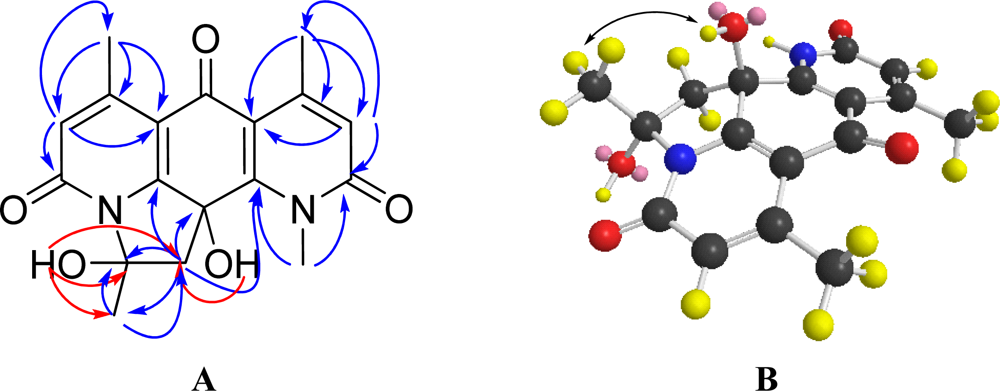
2.2. Structural Elucidation
2.3. Biological Activities
2.4. Discussion
3. Experimental Section
3.1. General Experimental Procedures
3.2. Microbiological Material
3.3. Extraction and Isolation
3.4. Characterization Data
3.5. Antibacterial and Cytotoxic Assay
4. Conclusions
Acknowledgments
References and Notes
- Blunt, JW; Copp, BR; Munro, MH; Northcote, PT; Prinsep, MR. Marine natural products. Nat. Prod. Rep. 2011, 28, 196–268. [Google Scholar]
- Bhatnagar, I; Kim, SK. Marine antitumor drugs: status, shortfalls and strategies. Mar. Drugs 2010, 8, 2702–2720. [Google Scholar]
- Olano, C; Mendez, C; Salas, JA. Antitumor compounds from marine actinomycetes. Mar. Drugs 2009, 7, 210–248. [Google Scholar]
- Rahman, H; Austin, B; Mitchell, WJ; Morris, PC; Jamieson, DJ; Adams, DR; Spragg, AM; Schweizer, M. Novel anti-infective compounds from marine bacteria. Mar. Drugs 2010, 8, 498–518. [Google Scholar]
- Fattorusso, E; Taglialatela-Scafati, O. Marine antimalarials. Mar. Drugs 2009, 7, 130–152. [Google Scholar]
- Tian, XP; Zhi, XY; Qiu, YQ; Zhang, YQ; Tang, SK; Xu, LH; Zhang, S; Li, WJ. Sciscionella marina gen. nov., sp. nov., a marine actinomycete isolated from a sediment in the northern South China Sea. Int. J. Syst. Evol. Microbiol. 2009, 59, 222–228. [Google Scholar]
- Tian, XP; Zhang, YQ; Li, QX; Zhi, XY; Tang, SK; Zhang, S; Li, WJ. Streptomyces nanshensis sp. nov., isolated from the Nansha Islands in the South China Sea. Int. J. Syst. Evol. Microbiol. 2009, 59, 745–749. [Google Scholar]
- Tian, XP; Tang, SK; Dong, JD; Zhang, YQ; Xu, LH; Zhang, S; Li, WJ. Marinactinospora thermotolerans gen. nov., sp. nov., a marine actinomycete isolated from a sediment in the northern South China Sea. Int. J. Syst. Evol. Microbiol. 2009, 59, 948–952. [Google Scholar]
- Dai, HQ; Wang, J; Xin, YH; Pei, G; Tang, SK; Ren, B; Ward, A; Ruan, JS; Li, WJ; Zhang, LX. Verrucosispora sediminis sp. nov., a cyclodipeptide-producing actinomycete from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 2010, 60, 1807–1812. [Google Scholar]
- Tian, XP; Long, LJ; Wang, FZ; Xu, Y; Li, J; Zhang, J; Zhang, CS; Zhang, S; Li, WJ. Streptomyces nanhaiensis sp. nov., a novel marine streptomycete isolated from a deep sea sediment in Southern China Sea. Int. J. Syst. Evol. Microbiol. 2011. [Google Scholar] [CrossRef]
- Niu, S; Li, S; Chen, Y; Tian, X; Zhang, H; Zhang, G; Zhang, W; Yang, X; Zhang, S; Ju, J; Zhang, C. Lobophorins E and F, new spirotetranate antibiotics from a South China Sea-derived Streptomyces sp. SCSIO 01127. J. Antibiot. 2011, in press.. [Google Scholar]
- Chu, M; Mierzwa, R; Xu, L; Yang, SW; He, L; Patel, M; Stafford, J; Macinga, D; Black, T; Chan, TM; Gullo, V. Structure elucidation of Sch 538415, a novel acyl carrier protein synthase inhibitor from a microorganism. Bioorg. Med. Chem. Lett. 2003, 13, 3827–3829. [Google Scholar]
- Lee, SD; Kim, ES; Hah, YC. Phylogenetic analysis of the genera Pseudonocardia and Actinobispora based on 16S ribosomal DNA sequences. FEMS Microbiol. Lett. 2000, 182, 125–129. [Google Scholar]
- Rinehart, KL; Renefroe, HB. The structure of nybomycin. J. Am. Chem. Soc. 1961, 83, 3729–3731. [Google Scholar]
- Bair, JS; Palchaudhuri, R; Hergenrother, PJ. Chemistry and biology of deoxynyboquinone, a potent inducer of cancer cell death. J. Am. Chem. Soc. 2010, 132, 5469–5478. [Google Scholar]
- Agrawal, PK. NMR spectroscopy in the structureal elucidation of oligosaccharides and glycosides. Phytochemistry 1992, 31, 3307–3330. [Google Scholar]
- Maskey, RP; Grun-Wollny, I; Laatsch, H. Isolation and structure elucidation of diazaquinomycin C from a terrestrial Streptomyces sp. and confirmation of the akashin structure. Nat. Prod. Res. 2005, 19, 137–142. [Google Scholar]
- Vicent, MJ; Manzanaro, S; de la Fuente, JA; Duncan, R. HPMA copolymer-1,5-diazaanthraquinone conjugates as novel anticancer therapeutics. J. Drug Target. 2004, 12, 503–515. [Google Scholar]
- Egawa, K; Yamori, T; Nosaka, C; Kunimoto, S; Takeuchi, T; Nos, K. Deoxynybomycin is a selective anti-tumor agent inducing apoptosis and inhibiting topoisomerase I. Biol. Pharm. Bull. 2000, 23, 1036–1040. [Google Scholar]
- Murata, M; Miyasaka, T; Tanaka, H; Omura, S. Diazaquinomycin A, a new antifolate antibiotic, inhibits thymidylate synthase. J. Antibiot. 1985, 38, 1025–1033. [Google Scholar]
- Nadzan, AM; Rinehart, KL, Jr; Sokolski, WT. Hydroxynybomycin: isolation, structure and bioactivity. J. Antibiot. 1977, 30, 523–524. [Google Scholar]
- Naganawa, H; Wakashiro, T; Yagi, A; Kondo, S; Takita, T. Deoxynybomycin from a Streptomyces. J. Antibiot. 1970, 23, 365–368. [Google Scholar]
- Manzanaro, S; Vicent, MJ; Martin, MJ; Salvador-Tormo, N; Perez, JM; del Mar Blanco, M; Avendano, C; Menendez, JC; de la Fuente, JA. Synthesis and biological evaluation of new 1,5-diazaanthraquinones with cytotoxic activity. Bioorg. Med. Chem. 2004, 12, 6505–6515. [Google Scholar]
- Avendano, C; Perez, JM; Blanco Mdel, M; de la Fuente, JA; Manzanaro, S; Vicent, MJ; Martin, MJ; Salvador-Tormo, N; Menendez, JC. Synthesis and structure-activity relationships of 1,5-diazaanthraquinones as antitumour compounds. Bioorg. Med. Chem. Lett. 2004, 14, 3929–3932. [Google Scholar]
- Lee, H; Lee, SI; Cho, J; Choi, SU; Yang, SI. Synthesis and in vitro evaluation of 1,8-diazaanthraquinones bearing 3-dialkylaminomethyl or 3-(N-alkyl- or N-aryl)carbamoyloxymethyl substituent. Eur. J. Med. Chem. 2003, 38, 695–702. [Google Scholar]
- Lee, H; Lee, SI; Yang, SI. Synthesis and in vitro cytotoxicity of 3-substituted-1, 8-diazaanthraquinones produced by Lewis-acid catalyzed hetero Diels-Alder reaction. Bioorg. Med. Chem. Lett. 1998, 8, 2991–2994. [Google Scholar]
- Ramos, MT; Diaz-Guerra, LM; Garcia-Copin, S; Avendano, C; Garcia-Gravalos, D; Garcia de Quesada, T. Synthesis and antitumour activity of fluorinated 1-aza and 1,8-diazaanthraquinones. Farmaco 1996, 51, 375–379. [Google Scholar]
- Nebois, P; do Nascimento, SC; Boitard, M; Bartoli, MH; Fillion, H. Synthesis and in vitro cytotoxic activity of aza- and diazaanthraquinone derivatives. Pharmazie 1994, 49, 819–821. [Google Scholar]
- Gesto, C; de la Cuesta, E; Avendano, C; Emling, F. Synthesis and biological activity of new 1,8-diaza-2,9,10-anthracenetrione derivatives. J. Pharm. Sci. 1992, 81, 815–816. [Google Scholar]
- Tsuzuki, K; Yokozuka, T; Murata, M; Tanaka, H; Omura, S. Synthesis and biological activity of analogues of diazaquinomycin A, a new thymidylate synthase inhibitor. J. Antibiot. 1989, 42, 727–737. [Google Scholar]
- Salas, JA; Hernandez, C; Mendez, C; Olano, C; Quiros, LM; Rodriguez, AM; Vilches, C. Intracellular glycosylation and active efflux as mechanisms for resistance to oleandomycin in Streptomyces antibioticus, the producer organism. Microbiologia 1994, 10, 37–48. [Google Scholar]
- Crystallographic data (excluding structure factors) for the structure of DNQ (1) in this paper have been deposited in the Cambridge Crystallographic Data Centre as supplementary publication numbers CCDC 834052. Copies of the data can be obtained, free of charge, on application to CCDC, 12 Union Road, Cambridge CB2 1EZ, UK. Fax: +44(0)-1223-336033 or E-Mail: deposit@ccdc.cam.ac.uk.
- Xiao, Y; Li, S; Niu, S; Ma, L; Zhang, G; Zhang, H; Zhang, G; Ju, J; Zhang, C. Characterization of tiacumicin B biosynthetic gene cluster affording diversified tiacumicin analogs and revealing a tailoring Di-halogenase. J. Am. Chem. Soc. 2011, 133, 1092–1105. [Google Scholar]
- Wu, ZC; Li, DL; Chen, YC; Zhang, WM. A new isofuranonaphthalenone and benzopyrans from the endophytic fungus Nodulisporium sp. A4 from Aquilaria sinensis. Helv. Chim. Acta 2010, 93, 920–924. [Google Scholar]
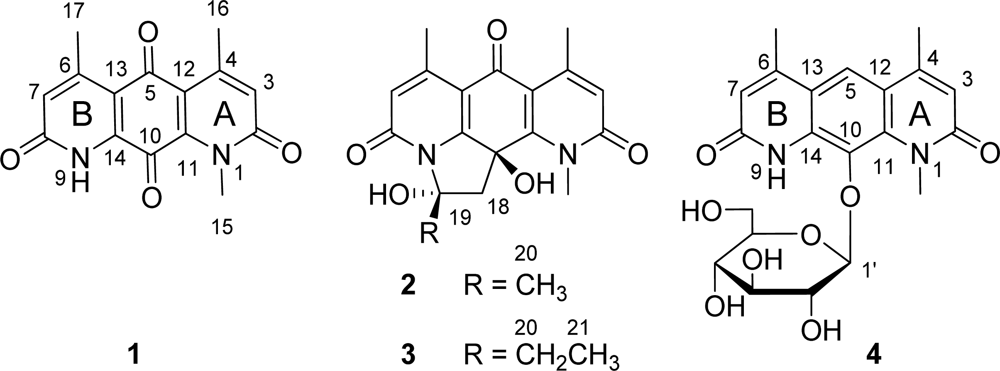
| No. | 1 a | 2 b | 2 c | 3 b | ||||
|---|---|---|---|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 2 | 161.4 s | 164.5 s | 161.0 s | 164.5 s | ||||
| 3 | 6.82 d (1.0) | 126.8 d | 6.48, s | 121.4 d | 6.39, d (1.0) | 119.8 d | 6.49, d (1.0) | 121.4 d |
| 4 | 149.0 s | 153.5s | 150.6 s | 153.5 s | ||||
| 5 | 182.4 s * | 183.5 s | 181.9 s | 183.6 s | ||||
| 6 | 150.5 s | 153.8 s | 150.1 s | 153.6 s | ||||
| 7 | 6.78 d (1.0) | 127.1 d | 6.32, s | 122.6 d | 6.24, d (1.0) | 121.2 d | 6.30, d (1.0) | 122.6 d |
| 8 | 162.4 s | 162.8 s | 159.3 s | 163.0 s | ||||
| 10 | 176.9 s * | 72.7 s | 70.8 s | 72.8 s | ||||
| 11 | 141.4 s | 151.8 s | 149.9 s | 152.0 s | ||||
| 12 | 118.2 s | 117.8 s | 114.7 s | 117.8 s | ||||
| 13 | 114.9 s | 110.8 s | 107.8 s | 111.0 s | ||||
| 14 | 141.7 s | 157.0 s | 155.8 s | 156.9 s | ||||
| 15 | 4.01, s | 33.9 q | 3.90, s | 34.1 q | 3.71, s | 32.6 q | 3.90, s | 34.2 q |
| 16 | 2.59, d (1.0) | 23.0 q | 2.57, s | 23.6 q | 2.44, d (1.0) | 22.7 q | 2.55, d, (1.0) | 23.5 q |
| 17 | 2.55, d (1.0) | 22.1 q | 2.58, s | 21.3 q | 2.45, d (1.0) | 20.5 q | 2.59, d (1.0) | 21.3 q |
| 18 | 2.71, d (13.5), | 52.4 t | 2.62, d (13.5), | 50.8 t | 2.51, d (13.8), | 47.9 t | ||
| 3.30, d (13.5) | 3.13, d (13.5) | 3.41, d (13.8) | ||||||
| 19 | 97.6 s | 95.5 s | 100.8 s | |||||
| 20 | 2.19, s | 26.5 q | 2.03, s | 25.9 q | 2.41, dd (7.5, 14.0), | 31.6 t | ||
| 2.84, dd (7.5, 14.0) | ||||||||
| 21 | 1.22, t (7.5) | 9.3 q | ||||||
| Compounds | MIC (μg mL–1) | IC50 (μM) | ||||
|---|---|---|---|---|---|---|
| B. thuringensis SCSIO BT01 | S. aureus ATCC 29213 | E. faecalis ATCC 29212 | SF-268 | MCF-7 | NCI-H460 | |
| 1 | 1 | 1 | 1 | 0.022 | 0.015 | 0.080 |
| 2 | 4 | 4 | 2 | 0.028 | 0.027 | 0.209 |
| 3 | 2 | 2 | 2 | 0.022 | 0.021 | 0.177 |
| 4 | >128 | >128 | >128 | 6.70 | 8.02 | 43.28 |
| Cisplatin | ND a | ND a | ND a | 3.99 | 9.24 | 1.53 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, S.; Tian, X.; Niu, S.; Zhang, W.; Chen, Y.; Zhang, H.; Yang, X.; Zhang, W.; Li, W.; Zhang, S.; et al. Pseudonocardians A–C, New Diazaanthraquinone Derivatives from a Deap-Sea Actinomycete Pseudonocardia sp. SCSIO 01299. Mar. Drugs 2011, 9, 1428-1439. https://doi.org/10.3390/md9081428
Li S, Tian X, Niu S, Zhang W, Chen Y, Zhang H, Yang X, Zhang W, Li W, Zhang S, et al. Pseudonocardians A–C, New Diazaanthraquinone Derivatives from a Deap-Sea Actinomycete Pseudonocardia sp. SCSIO 01299. Marine Drugs. 2011; 9(8):1428-1439. https://doi.org/10.3390/md9081428
Chicago/Turabian StyleLi, Sumei, Xinpeng Tian, Siwen Niu, Wenjun Zhang, Yuchan Chen, Haibo Zhang, Xianwen Yang, Weimin Zhang, Wenjun Li, Si Zhang, and et al. 2011. "Pseudonocardians A–C, New Diazaanthraquinone Derivatives from a Deap-Sea Actinomycete Pseudonocardia sp. SCSIO 01299" Marine Drugs 9, no. 8: 1428-1439. https://doi.org/10.3390/md9081428
APA StyleLi, S., Tian, X., Niu, S., Zhang, W., Chen, Y., Zhang, H., Yang, X., Zhang, W., Li, W., Zhang, S., Ju, J., & Zhang, C. (2011). Pseudonocardians A–C, New Diazaanthraquinone Derivatives from a Deap-Sea Actinomycete Pseudonocardia sp. SCSIO 01299. Marine Drugs, 9(8), 1428-1439. https://doi.org/10.3390/md9081428