Toward the Synthesis and Biological Screening of a Cyclotetrapeptide from Marine Bacteria
Abstract
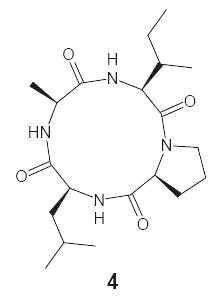
:1. Introduction
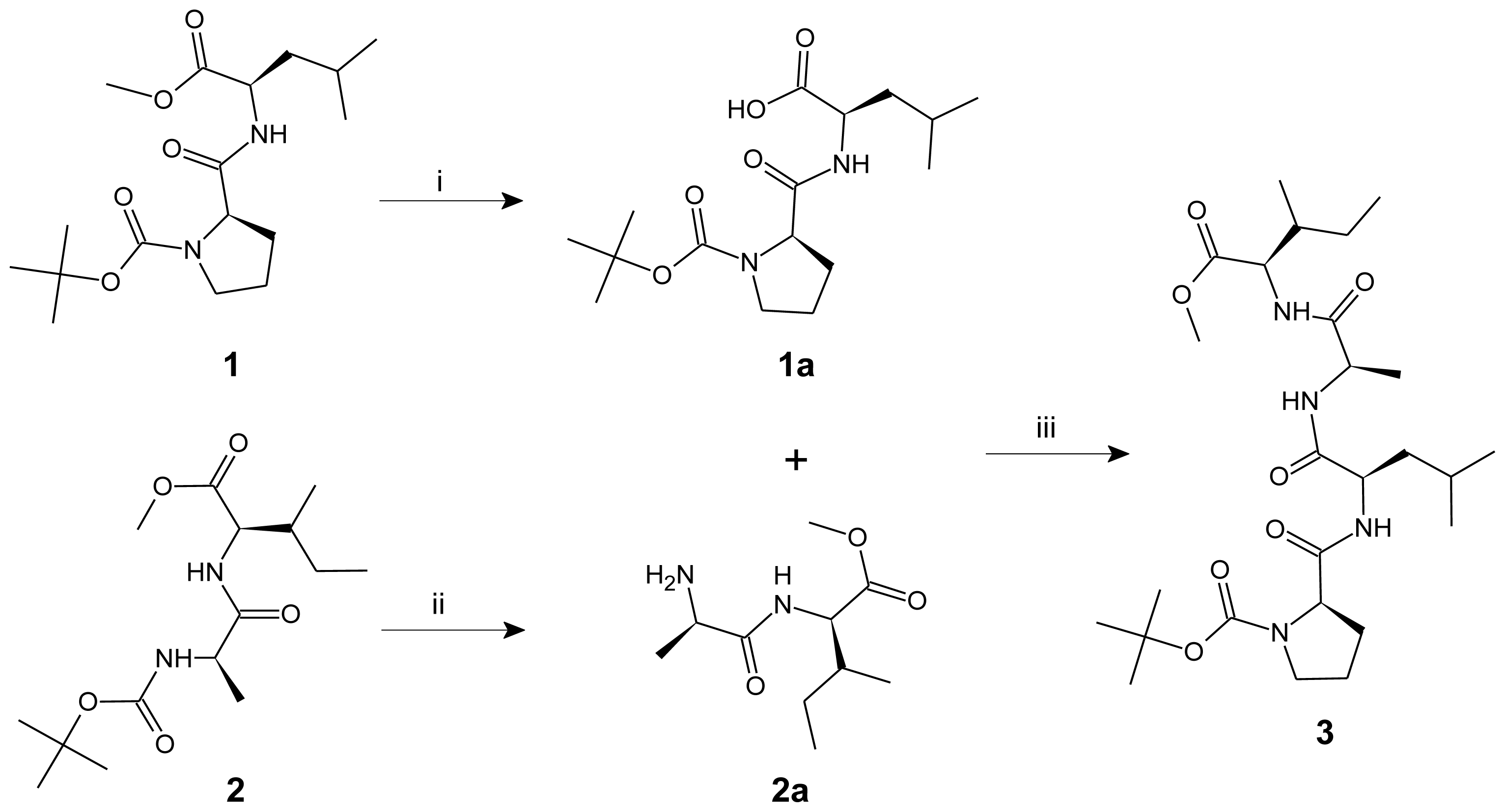
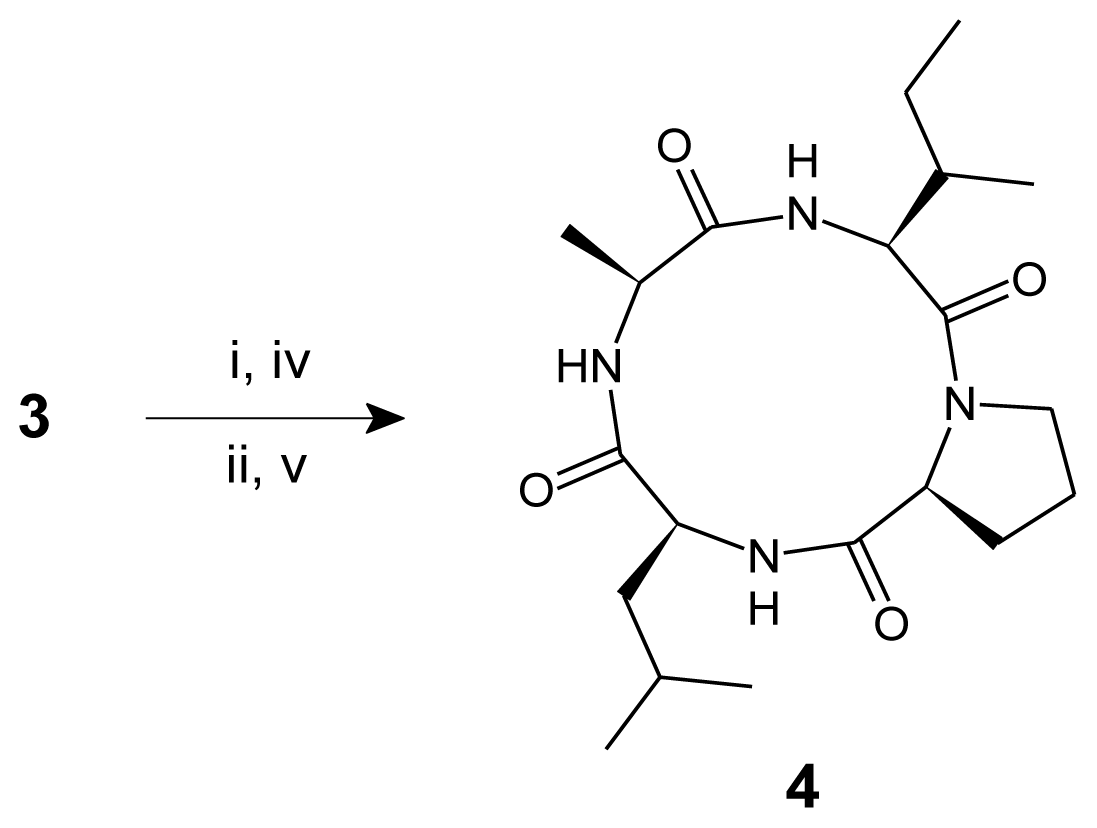
2. Results and Discussion
3. Experimental Section
3.1. Extraction and Isolation of Natural Cyclic Tetrapeptide
3.2. General Procedure for the Preparation of Linear Dipeptide Fragments
3.3. Procedure for the Synthesis of Linear Tetrapeptide Unit and Its Cyclization
3.4. Biological Activity Studies
3.4.1. Antihelmintic Screening
3.4.2. Antimicrobial Screening
4. Conclusions
Acknowledgements
- Samples Availability: Available from the authors.
References
- Fenical, W. Chemical Studies of Marine Bacteria: Developing a New Resource. Chem Rev 1993, 93, 1673–1683. [Google Scholar]
- De Rosa, S; Mitova, M; Tommonaro, G. Marine Bacteria associated with Sponge as Source of Cyclic Peptides. Biomol Eng 2003, 20, 311–316. [Google Scholar]
- Mitova, M; Tommonaro, G; De Rosa, S. A Novel Cyclopeptide from a Bacterium associated with the Marine Sponge Ircinia muscarum. Z Naturforsch C 2003, 58, 740–745. [Google Scholar]
- Shin, J; Seo, Y; Lee, HS; Rho, JR; Mo, SJ. A New Cyclic Peptide from a Marine-derived Bacterium of the Genus Nocardiopsis. J Nat Prod 2003, 66, 883–884. [Google Scholar]
- Faulkner, DJ. Marine Natural Products. Nat Prod Rep 1998, 15, 113–158. [Google Scholar]
- Mitova, M; Popov, S; De Rosa, S. Cyclic Peptides from a Ruegeria strain of Bacteria associated with the Sponge Suberites domuncula. J Nat Prod 2004, 67, 1178–1181. [Google Scholar]
- Cueto, M; Jensen, PR; Fenical, W. N-Methylsansalvamide, a Cytotoxic Cyclic Depsipeptide from a Marine Fungus of the Genus Fusarium. Phytochemistry 2000, 55, 223–226. [Google Scholar]
- Tan, LT; Cheng, XC; Jensen, PR; Fenical, W. Scytalidamides A and B, New Cytotoxic Cyclic Heptapeptides from a Marine Fungus of the Genus Scytalidium. J Org Chem 2003, 68, 8767–8773. [Google Scholar]
- Adachi, K; Kanoh, K; Wisespongp, P; Nishijima, M; Shizuri, Y. Clonostachysins A and B, New Anti-dinoflagellate Cyclic Peptides from a Marine-derived Fungus. J Antibiot 2005, 58, 145–150. [Google Scholar]
- Lee, HS; Shin, HJ; Jang, KH; Kim, TS; Oh, KB; Shin, J. Cyclic Peptides of the Nocardamine Class from a Marine-derived Bacterium of the Genus Streptomyces. J Nat Prod 2005, 68, 623–625. [Google Scholar]
- Rungprom, W; Siwu, ERO; Lambert, LK; Dechsakulwatana, C; Barden, MC; Kokpol, U; Blanchfield, JT; Kita, M; Garson, MJ. Cyclic Tetrapeptides from Marine Bacteria associated with the Seaweed Diginea sp. and the Sponge Halisarca ectofibrosa. Tetrahedron 2008, 64, 3147–3152. [Google Scholar]
- Dahiya, R; Pathak, D. Cyclic Peptides: New Hope for Antifungal Therapy. Egypt. Pharm. J. (NRC) 2006, 5, 189–199. [Google Scholar]
- Pathak, D; Dahiya, R. Cyclic Peptides as Novel Antineoplastic Agents: A Review. J Sci Pharm 2003, 4, 125–131. [Google Scholar]
- Dahiya, R; Pathak, D; Himaja, M; Bhatt, S. First Total Synthesis and Biological Screening of Hymenamide E. Acta Pharm 2006, 56, 399–415. [Google Scholar]
- Dahiya, R. Synthetic and Pharmacological Studies on Longicalycinin A. Pak J Pharm Sci 2007, 20, 317–323. [Google Scholar]
- Dahiya, R. Synthetic Studies on a Cyclic Hexapeptide from Dianthus superbus. Chem Pap 2008, 62, 527–535. [Google Scholar]
- Dahiya, R; Kaur, K. Synthetic and Pharmacological Investigation of Segetalin C as a Novel Antifungal and Cytotoxic Agent. Arzneim Forsch 2008, 58, 29–34. [Google Scholar]
- Dahiya, R; Kumar, A; Gupta, R. Synthesis, Cytotoxic and Antimicrobial Screening of a Proline-Rich Cyclopolypeptide. Chem Pharm Bull 2009, 57, 214–217. [Google Scholar]
- Dahiya, R; Maheshwari, M; Kumar, A. Toward the Synthesis and Biological Evaluation of Hirsutide. Monatsh Chem 2009, 140, 121–127. [Google Scholar]
- Dahiya, R; Gautam, H. Synthetic and Pharmacological Studies on a Natural Cyclopeptide from Gypsophila arabica. J Med Plants Res 2010, 4, 1960–1966. [Google Scholar]
- Dahiya, R; Gautam, H. Total Synthesis and Antimicrobial Activity of a Natural Cycloheptapeptide of Marine-Origin. Mar Drugs 2010, 8, 2384–2394. [Google Scholar]
- Dahiya, R; Gautam, H. Toward the First Total Synthesis of Gypsin D: A Natural Cyclopolypeptide from Gypsophila arabica. Am J Sci Res 2010, 11, 150–158. [Google Scholar]
- Bodanzsky, M; Bodanzsky, A. The Practice of Peptide Synthesis; Springer: New York, NY, USA, 1984; pp. 68–143. [Google Scholar]
- Garg, LC; Atal, CK. Anthelmintic Activity of Myrsine africana Indian. J Pharm Sci 1963, 59, 240–245. [Google Scholar]
- Bauer, AW; Kirby, WM; Sherris, JC; Turck, M. Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am J Clin Pathol 1966, 45, 493–496. [Google Scholar]
| Compound | Physical state | M.p. (°C) | α[D] a (°) | Yield (%) | Rf b | Mol. Formula (Mr) | Elemental analysis Calcd./found (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| C | H | N | |||||||
| 1 | Viscous mass | - | +71.4 1 | 87 | 0.69 | C17H30N2O5 | 59.63 | 8.83 | 8.18 |
| (342) | 59.62 | 8.85 | 8.21 | ||||||
| 1a | White solid | 79–80 | +41.6 | 72 | 0.53 | C16H28N2O5 | 58.52 | 8.59 | 8.53 |
| (328) | 58.49 | 8.62 | 8.55 | ||||||
| 2 | Viscous mass | - | −121.8 | 93 | 0.87 | C15H28N2O5 | 56.94 | 8.92 | 8.85 |
| (316) | 56.95 | 8.94 | 8.83 | ||||||
| 2a | Semisolid mass | - | −89.2 | 78 | 0.66 | C10H20N2O3 | 55.53 | 9.32 | 12.95 |
| (216) | 55.52 | 9.35 | 12.97 | ||||||
| 3 | Semisolid mass | - | −55.1 2 | 91 | 0.81 | C26H46N4O7 | 59.29 | 8.80 | 10.64 |
| (526) | 59.26 | 8.79 | 10.66 | ||||||
| 4 | White solid | 198 (d) | −12.9 3 (−13.0) | 74 | 0.62 * | C20H34N4O4 | 60.89 | 8.69 | 14.20 |
| (394) | 60.91 | 8.70 | 14.18 | ||||||
| Compound | IR (CHCl3/KBr, v cm−1), 1H/13C NMR (CDCl3, δ ppm), ESIMS/MS (rel. int., m/z) |
|---|---|
| 1 | 3128, 3123 (N–H str, amide), 2998–2989 (C–H str, CH2, Pro), 2966, 2928 (C–H str, asym, CH3 and CH2), 1748 (C=O str, ester), 1675, 1643 (C=O str, tert and sec amide), 1539 (N–H def, sec amide), 1391, 1377 (C–H def, tert-Butyl), 1384, 1369 (C–H def, iso-propyl), 1267 (C–O str, ester) 6.65 (1H, br. s, NH), 4.40 (1H, q, H-α, Leu), 4.05 (1H, t, J = 7.2 Hz, H-α, Pro), 3.62 (3H, s, OCH3), 3.22 (2H, t, J = 7.2 Hz, H-δ, Pro), 2.58 (2H, q, H-β, Pro), 1.96–1.88 (2H, m, H-γ, Pro), 1.47 (9H, s, tert-Butyl), 1.45–1.36 (3H, m, H-β and H-γ, Leu), 0.94 (6H, d, J = 6.35 Hz, H-δ, Leu) |
| 1a | 3297–2478 (m/br, OH str, COOH), 3125, 3122 (N–H str, amide), 2999–2992 (C–H str, CH2, Pro), 2968, 2929 (C–H str, asym, CH3 and CH2), 1711 (s, C=O str, COOH), 1678, 1642 (C=O str, tert and sec amide), 1536 (N–H def, sec amide), 1393, 1375 (C–H def, tert-Butyl), 1385, 1368 (C–H def, iso-propyl) 12.52 (1H, s, OH, COOH), 6.73 (1H, br. s, NH), 4.47 (1H, q, H-α, Leu), 3.86 (1H, t, J = 7.2 Hz, H-α, Pro), 3.21 (2H, t, J = 7.15 Hz, H-δ, Pro), 2.55 (2H, q, H-β, Pro), 1.95–1.87 (3H, m, H-γ, Pro and Leu), 1.56 (2H, t, H-β, Leu), 1.49 (9H, s, tert-Butyl), 0.95 (6H, d, J = 6.4 Hz, H-δ, Leu) |
| 2 | 3502, 3396 (N–H str, sec amine), 3132, 3127 (N–H str, amide), 2965, 2928 (C–H str, asym, CH3 and CH2), 2874 (C–H str, sym, CH3), 1747 (C=O str, ester), 1646 (C=O str, sec amide), 1535 (N–H def, sec amide), 1269 (C–O str, ester), 1159 (C–N str, sec amine) 5.89 (1H, br. s, NH), 4.28 (1H, t, J = 6.4 Hz, H-α, Ile), 3.75–3.69 (1H, m, H-α, Ala), 3.51 (3H, s, OCH3), 2.03–1.95 (1H, m, H-β, Ile), 1.70–1.62 (2H, m, H-γ, Ile), 1.46 (2H, br. s, NH2), 1.15 (3H, d, J = 7.25 Hz, H-β, Ala), 0.94 (3H, t, J = 7.2 Hz, H-δ, Ile), 0.86 (3H, d, J = 6.4 Hz, H-γ′, Ile) |
| 3 | 3129–3122 (N–H str, amide), 2999–2993 (C–H str, CH2, Pro), 2968, 2929, 2925 (C–H str, asym, CH3 and CH2), 2878–2873 (C–H str, sym, CH3), 1745 (C=O str, ester), 1676, 1646–1641 (C=O str, tert and sec amide), 1538, 1535 (N–H def, sec amide), 1392, 1375 (C–H def, tert-Butyl), 1386, 1368 (C–H def, iso-propyl), 1272 (C–O str, ester) 8.48 (1H, br. s, NH), 7.45 (1H, br. s, NH), 5.14 (1H, br. s, NH), 4.52 (1H, q, H-α, Leu), 4.25 (1H, q, H-α, Ala), 4.14 (1H, t, J = 7.2 Hz, H-α, Pro), 3.73 (1H, t, J = 6.35 Hz, H-α, Ile), 3.49 (3H, s, OCH3), 3.43 (2H, t, J = 7.25 Hz, H-δ, Pro), 2.49 (2H, q, H-β, Pro), 2.05–1.97 (1H, m, H-β, Ile), 1.95–1.89 (2H, m, H-γ, Pro), 1.83–1.62 (4H, m, H-β, Leu and H-γ, Ile), 1.48 (9H, s, tert-Butyl), 1.48–1.43 (1H, m, H-γ, Leu), 1.29 (3H, d, J = 7.2 Hz, H-β, Ala), 0.99 (6H, d, J = 6.4 Hz, H-δ, Leu), 0.93 (3H, t, J = 7.15 Hz, H-δ, Ile), 0.89 (3H, d, J = 6.45 Hz, H-γ′, Ile) 174.8 (C=O, Leu), 172.9, 172.4 (2C, C=O, Pro and Ile), 169.8 (C=O, Ala), 156.2 (C=O, Boc), 79.8 (C-α, tert-Butyl), 59.7 (C-α, Pro), 58.5 (C-α, Ile), 54.7 (OCH3), 51.3 (C-α, Ala), 49.7 (C-α, Leu), 46.9 (C-δ, Pro), 37.6, 37.1 (2C, C-β, Ile and Leu), 28.9 (C-β, Pro), 28.2 (3C, C-β, tert-Butyl), 24.2, 23.9 (2C, C-γ, Ile and Leu), 23.7 (C-γ, Pro), 22.3 (2C, C-δ, Leu), 17.9 (C-β, Ala), 15.1 (C-γ′, Ile), 9.5 (C-δ, Ile) |
| 4 | 3128, 3123, 3119 (N–H str, amide), 2998, 2995–2992 (C–H str, CH2, Pro), 2969, 2928–2924 (C–H str, asym, CH3 and CH2), 2879, 2875 (C–H str, sym, CH3), 1675, 1648–1643 (C=O str, tert and sec amide), 1539–1536 (N–H def, sec amide), 1385, 1369 (C–H def, iso-propyl) 9.37 (1H, br. s, NH), 9.31 (1H, br. s, NH), 7.76 (1H, br. s, NH), 4.26 (1H, t, J = 7.15 Hz, H-α, Pro), 3.99 (1H, q, H-α, Ala), 3.95 (1H, q, H-α, Leu), 3.61 (1H, t, J = 6.4 Hz, H-α, Ile), 3.52 (2H, t, J = 7.3 Hz, H-δ, Pro), 2.35 (2H, q, H-β, Pro), 1.96–1.83 (2H, m, H-γ, Pro), 1.89–1.83 (1H, m, H-γ, Leu), 1.81–1.76 (1H, m, H-β, Ile), 1.73–1.64 (2H, m, H-β, Leu), 1.59–1.53 (2H, m, H-γ, Ile), 1.45 (3H, d, J = 7.25 Hz, H-β, Ala), 0.98 (6H, d, J = 6.35 Hz, H-δ, Leu), 0.95 (3H, d, J = 6.45 Hz, H-γ′, Ile), 0.89 (3H, t, J = 7.2 Hz, H-δ, Ile) 171.1 (C=O, Leu), 170.9 (C=O, Ala), 170.5, 168.1 (2C, C=O, Pro and Ile), 63.3 (C-α, Ile), 57.5 (C-α, Pro), 54.1 (C-α, Leu), 49.9 (C-α, Ala), 46.5 (C-δ, Pro), 43.8, 39.9 (2C, C-β, Leu and Ile), 29.9 (C-β, Pro), 24.8, 24.2 (2C, C-γ, Ile and Leu), 22.0 (2C, C-δ, Leu), 21.3 (C-γ, Pro), 19.2 (C-β, Ala), 15.4 (C-γ′, Ile), 9.8 (C-δ, Ile) 395.4 [(M + H)+, 100], 367.4 [(395.4 − CO)+, 11], 324.4 [(H-Ile-Pro-Leu)+, 32], 298.4 [(H-Leu-Ala-Ile)+, 39], 296.4 [(324.4 − CO)+, 10], 282.3 [(H-Ala-Ile-Pro)+, 29], 270.3 [(298.4 − CO)+, 17], 254.3 [(282.3 − CO)+, 15], 211.3 [(H-Ile-Pro)+, 19], 185.2 [(H-Leu-Ala)+, 76], 157.2 [(185.2 − CO)+, 12], 114.1 [(H-Leu)+, 19], 86.1 [Ile/Leu (C5H12N)+, 16], 70.1 [Pro (C4H8N)+, 10], 57.1 [(C4H9)+, 15], 44.1 [Ala (C2H6N)+, 12], 43.1 [(C3H7)+, 11], 29.1 [(C2H5)+, 9], 15.0 [(CH3)+, 13] |
| Compound | Earthworm species | |||||
|---|---|---|---|---|---|---|
| M. konk. | P. core. | E. euge. | ||||
| Mean paralyzing time (min) ‡ | Mean death time (min) ‡ | Mean paralyzing time (min) | Mean death time (min) | Mean paralyzing time (min) | Mean death time (min) | |
| 3 † | 14.25 ± 0.42 | 22.57 ± 0.36 | 18.11 ± 0.26 | 29.47 ± 0.14 | 14.12 ± 0.23 | 24.54 ± 0.12 |
| 4 † | 9.28 ± 0.20 | 18.27 ± 0.17 | 12.44 ± 0.19 | 23.55 ± 0.27 | 12.40 ± 0.13 | 22.05 ± 0.37 |
| Control # | - | - | - | - | - | - |
| Mebendazole † | 13.85 ± 0.64 | 22.85 ± 0.53 | 17.82 ± 0.43 | 29.60 ± 0.22 | 13.54 ± 0.45 | 24.05 ± 0.62 |
| Compound | Diameter of zone of inhibition (mm) | |||||||
|---|---|---|---|---|---|---|---|---|
| Bacterial strains | Fungal strains | |||||||
| B. sub. | S. aur. | P. aeru. | K. pneu. | C. alb. | M. audo. | A. niger | T. menta. | |
| 3 | - | - | 13(6) † | 16(6) | 12(6) | 18(6) | - | 19(6) |
| 4 | - | - | 18(6) | 20(6) | 15(6) | 23(6) | - | 25(6) |
| Control * | - | - | - | - | - | - | - | - |
| Gatifloxacin | 18(12.5) | 28(6) | 22(6) | 25(6) | - | - | - | - |
| Griseofulvin | - | - | - | - | 20(6) | 17(6) | 18(12.5) | 20(6) |
© 2011 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dahiya, R.; Gautam, H. Toward the Synthesis and Biological Screening of a Cyclotetrapeptide from Marine Bacteria. Mar. Drugs 2011, 9, 71-81. https://doi.org/10.3390/md9010071
Dahiya R, Gautam H. Toward the Synthesis and Biological Screening of a Cyclotetrapeptide from Marine Bacteria. Marine Drugs. 2011; 9(1):71-81. https://doi.org/10.3390/md9010071
Chicago/Turabian StyleDahiya, Rajiv, and Hemendra Gautam. 2011. "Toward the Synthesis and Biological Screening of a Cyclotetrapeptide from Marine Bacteria" Marine Drugs 9, no. 1: 71-81. https://doi.org/10.3390/md9010071
APA StyleDahiya, R., & Gautam, H. (2011). Toward the Synthesis and Biological Screening of a Cyclotetrapeptide from Marine Bacteria. Marine Drugs, 9(1), 71-81. https://doi.org/10.3390/md9010071