Single-Cell Transcriptomic Analysis Reveals Cell Heterogeneity and Altered Signaling Pathways in Jellyfish Sting Patients
Abstract
1. Introduction
2. Results
2.1. Overview of Single-Cell Transcriptomic Profiling of PBMCs
2.2. Altered Immune Cell Composition in Jellyfish Envenomation
2.3. Feature Plot of Canonical Marker Genes
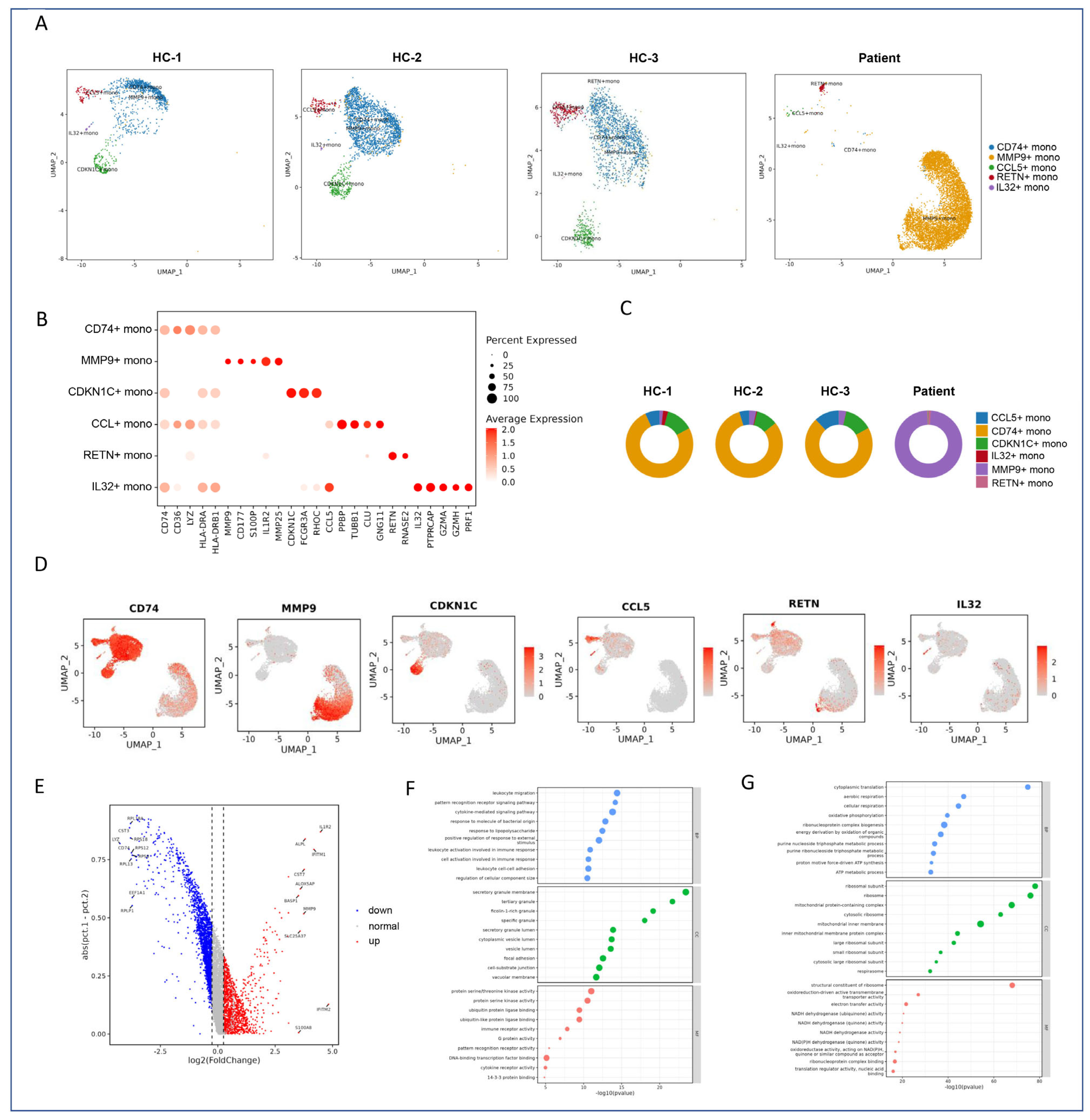
2.4. Subclustering and Functional Annotation of Monocytes
2.4.1. Subtype-Specific Gene Signatures of Monocytes
2.4.2. Differential Gene Expression Analysis
2.4.3. Classification of Monocyte Subclusters
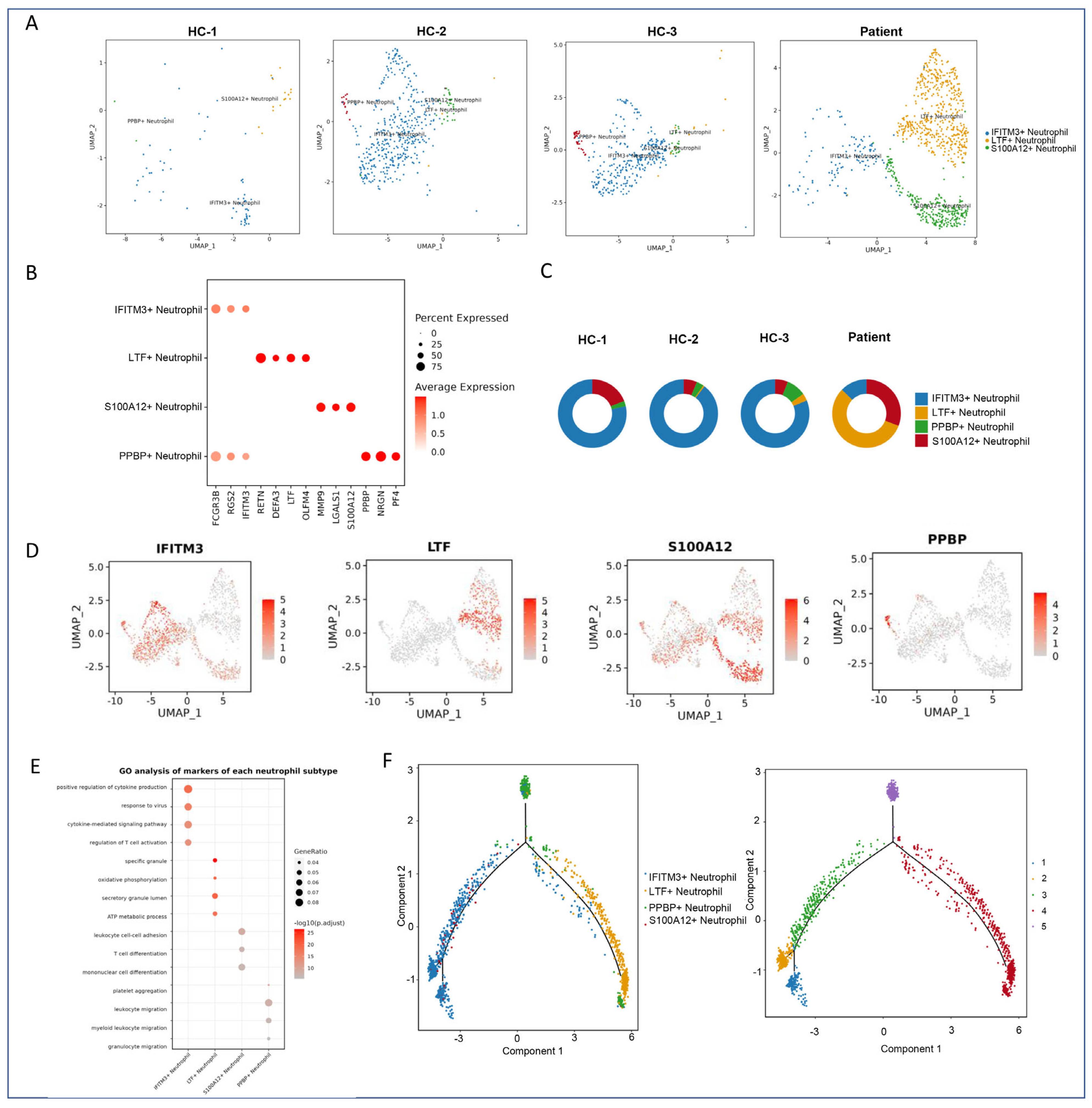
2.5. Subclustering and Functional Annotation of Neutrophils
2.5.1. Subtype-Specific Gene Signatures of Neutrophils
2.5.2. Functional Pathway Enrichment Analysis
2.5.3. Pseudotime Trajectory Analysis
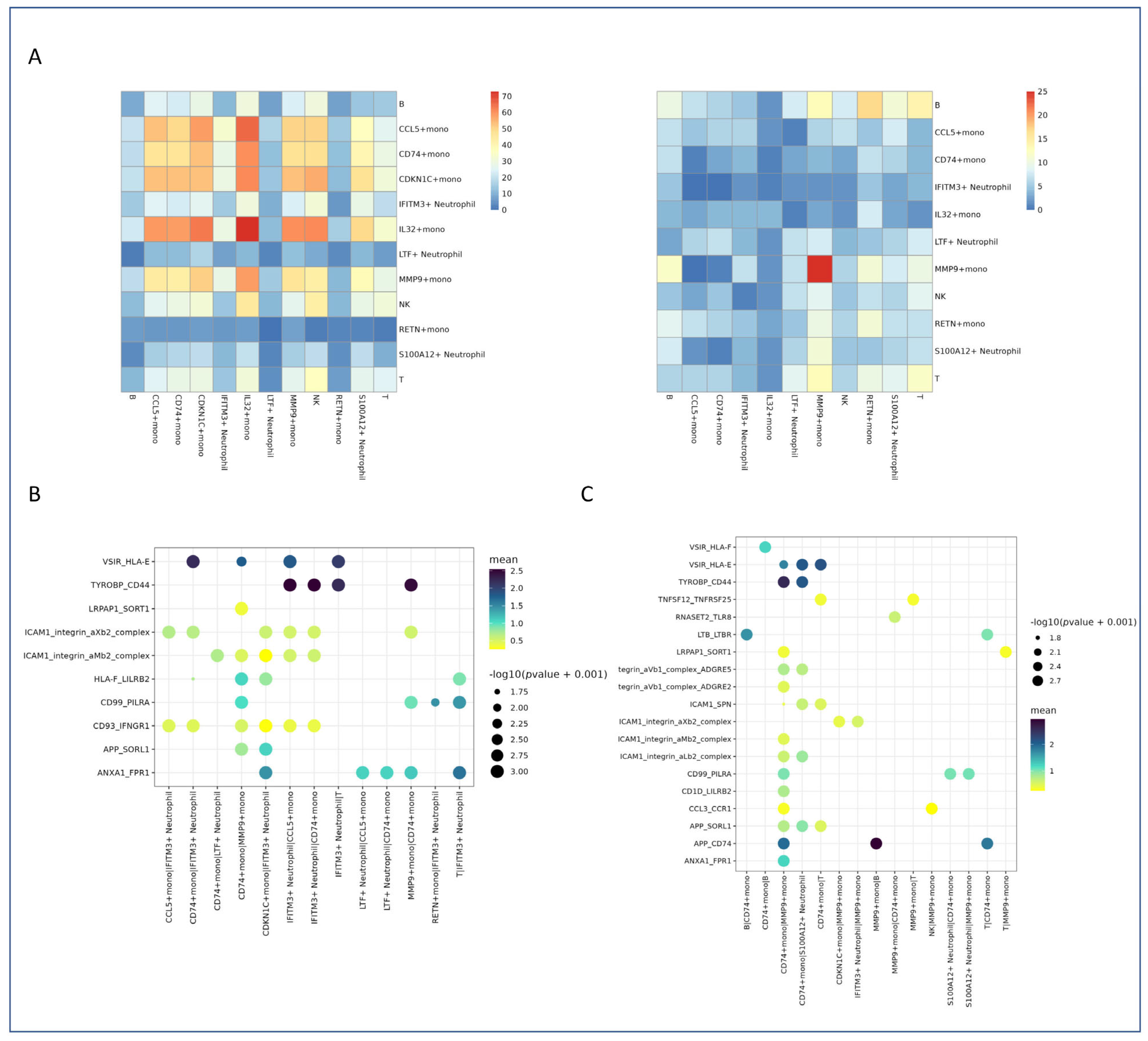
2.6. Altered Cell–Cell Communication Networks
2.6.1. Heatmap Visualization of Interaction Strength and Specificity
2.6.2. Dot Plot Analysis of Key Interaction Pairs in Healthy Controls
2.6.3. Dot Plot Analysis of Key Interaction Pairs in Patient Samples
3. Discussion
3.1. Immune Cell Composition Remodeling and Pro-Inflammatory Monocyte Expansion
3.2. Neutrophil Subsets and Impaired Maturation Trajectory
3.3. Altered Cell–Cell Communication and Inflammatory Synergy
3.4. Therapeutic Implications and Future Directions
3.5. Limitations and Outlook
4. Materials and Methods
4.1. Human Sample Collection
4.2. Isolation of PBMCs and Preparation of Single-Cell Suspensions
4.3. Processing scRNA-Seq Data
4.4. Identification of Marker Genes
4.5. Cell Type Annotation
4.6. Pathway Enrichment Analysis
4.7. Quality Control, Dimension Reduction, and Clustering
4.8. Differentially Expressed Gene (DEG) Analysis (Seurat)
4.9. UCell Gene Set Scoring
4.10. Cell–Cell Communication Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| scRNA-seq | Single-cell RNA sequencing |
| PBMC | Peripheral blood mononuclear cells |
| CF | Cardiac function |
| UMAP | Uniform Manifold Approximation and Projection |
| NK | Natural killer |
| DC | Dendritic cells |
| GSEA | Gene set enrichment analysis |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| HC | Healthy control |
| S100A12 | S100 calcium-binding protein A12 |
| MMP9 | Matrix metalloproteinase |
| DEGs | Differentially expressed genes |
References
- Naveed, M.; Chan, M.W.H.; Aslam, S.; Wang, F.; Sajjad, A.; Ullah, A.; Saleem, N.; Haider, M.S.; Arija, V. Nutritional composition assessment and antimicrobial activity of Catostylus perezi, jellyfish blooms along the coast of Pakistan: An awareness to avoid food neophobia in Pakistan. Nat. Prod. Res. 2024, 38, 3957–3963. [Google Scholar] [CrossRef]
- Killi, N.; Mariottini, G.L. Cnidarian Jellyfish: Ecological Aspects, Nematocyst Isolation, and Treatment Methods of Sting. Results Probl. Cell Differ. 2018, 65, 477–513. [Google Scholar]
- Malacarne, P.F.; Menezes, T.N.; Martins, C.W.; Naumann, G.B.; Gomes, H.L.; Pires, R.G.W.; Figueiredo, S.G.; Campos, F.V. Advances in the characterization of the Scorpaena plumieri cytolytic toxin (Sp-CTx). Toxicon 2018, 150, 220–227. [Google Scholar] [CrossRef]
- Baysoy, A.; Bai, Z.; Satija, R.; Fan, R. The technological landscape and applications of single-cell multi-omics. Nat. Rev. Mol. Cell Biol. 2023, 24, 695–713. [Google Scholar] [CrossRef]
- Ruwanpathirana, P.; Priyankara, D. Clinical manifestations of wasp stings: A case report and a review of literature. Trop. Med. Health 2022, 50, 82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, S.; Min, M.; Ni, Y.; Lu, Z.; Sun, X.; Wu, J.; Liu, B.; Ying, X.; Liu, Y. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut 2021, 70, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yu, Q.; Song, T.; Wang, Z.; Song, L.; Yang, Y.; Shao, J.; Li, J.; Ni, Y.; Chao, N.; et al. The heterogeneous immune landscape between lung adenocarcinoma and squamous carcinoma revealed by single-cell RNA sequencing. Signal Transduct. Target. Ther. 2022, 7, 289. [Google Scholar] [CrossRef] [PubMed]
- Peroni, E.; Randi, M.L.; Rosato, A.; Cagnin, S. Acute myeloid leukemia: From NGS, through scRNA-seq, to CAR-T. dissect cancer heterogeneity and tailor the treatment. J. Exp. Clin. Cancer Res. 2023, 42, 259. [Google Scholar] [CrossRef]
- Choi, Y.H.; Kim, J.K. Dissecting Cellular Heterogeneity Using Single-Cell RNA Sequencing. Mol. Cells 2019, 42, 189–199. [Google Scholar]
- Zhao, J.; Zhang, S.; Liu, Y.; He, X.; Qu, M.; Xu, G.; Wang, H.; Huang, M.; Pan, J.; Liu, Z.; et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 2020, 6, 22. [Google Scholar] [CrossRef]
- Massalha, H.; Bahar Halpern, K.; Abu-Gazala, S.; Jana, T.; Massasa, E.E.; Moor, A.E.; Buchauer, L.; Rozenberg, M.; Pikarsky, E.; Amit, I.; et al. A single cell atlas of the human liver tumor microenvironment. Mol. Syst. Biol. 2020, 16, e9682. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Luan, M.; Li, W.; Niu, M.; He, Q.; Zhao, Y.; Chen, J.; Mao, B.; Mou, K.; Li, P. Single-cell transcriptomics reveals peripheral immune responses in non-segmental vitiligo. Front. Immunol. 2023, 14, 1221260. [Google Scholar] [CrossRef]
- Abplanalp, W.T.; John, D.; Cremer, S.; Assmus, B.; Dorsheimer, L.; Hoffmann, J.; Becker-Pergola, G.; Rieger, M.A.; Zeiher, A.M.; Vasa-Nicotera, M.; et al. Single-cell RNA-sequencing reveals profound changes in circulating immune cells in patients with heart failure. Cardiovasc. Res. 2021, 117, 484–494. [Google Scholar] [CrossRef]
- Kim, D.; Kobayashi, T.; Voisin, B.; Jo, J.H.; Sakamoto, K.; Jin, S.P.; Kelly, M.; Pasieka, H.B.; Naff, J.L.; Meyerle, J.H.; et al. Targeted therapy guided by single-cell transcriptomic analysis in drug-induced hypersensitivity syndrome: A case report. Nat. Med. 2020, 26, 236–243. [Google Scholar] [CrossRef]
- McKellar, D.W.; Walter, L.D.; Song, L.T.; Mantri, M.; Wang, M.F.Z.; De Vlaminck, I.; Cosgrove, B.D. Large-scale integration of single-cell transcriptomic data captures transitional progenitor states in mouse skeletal muscle regeneration. Commun. Biol. 2021, 4, 1280. [Google Scholar] [CrossRef]
- Gotts, J.E.; Matthay, M.A. Sepsis: Pathophysiology and clinical management. BMJ 2016, 353, i1585. [Google Scholar] [CrossRef]
- Feio-Azevedo, R.; Boesch, M.; Radenkovic, S.; van Melkebeke, L.; Smets, L.; Wallays, M.; Boeckx, B.; Philips, G.; Prata de Oliveira, J.; Ghorbani, M.; et al. Distinct immunometabolic signatures in circulating immune cells define disease outcome in acute-on-chronic liver failure. Hepatology 2025, 81, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Kronsten, V.T.; Tranah, T.H.; Pariante, C.; Shawcross, D.L. Gut-derived systemic inflammation as a driver of depression in chronic liver disease. J. Hepatol. 2022, 76, 665–680. [Google Scholar] [CrossRef]
- Miyabe, C.; Miyabe, Y.; Nagai, J.; Miura, N.N.; Ohno, N.; Chun, J.; Tsuboi, R.; Ueda, H.; Miyasaka, M.; Miyasaka, N.; et al. Abrogation of lysophosphatidic acid receptor 1 ameliorates murine vasculitis. Arthritis Res. Ther. 2019, 21, 191. [Google Scholar] [CrossRef]
- Berling, I.; Isbister, G. Marine envenomations. Aust. Fam. Physician 2015, 44, 28–32. [Google Scholar] [PubMed]
- Galeano Niño, J.L.; Wu, H.; LaCourse, K.D.; Kempchinsky, A.G.; Baryiames, A.; Barber, B.; Futran, N.; Houlton, J.; Sather, C.; Sicinska, E.; et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 2022, 611, 810–817. [Google Scholar] [CrossRef]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef]
- Yan, N.; Xie, W.; Wang, D.; Fang, Q.; Guo, J.; Chen, Y.; Li, X.; Gong, L.; Wang, J.; Guo, W.; et al. Single-cell transcriptomic analysis reveals tumor cell heterogeneity and immune microenvironment features of pituitary neuroendocrine tumors. Genome Med. 2024, 16, 2. [Google Scholar] [CrossRef]
- Tegowski, M.; Flamand, M.N.; Meyer, K.D. scDART-seq reveals distinct m6A signatures and mRNA methylation heterogeneity in single cells. Mol. Cell 2022, 82, 868–878.e10. [Google Scholar] [CrossRef]
- Jin, H.; Li, M.; Jeong, E.; Castro-Martinez, F.; Zuker, C.S. A body-brain circuit that regulates body inflammatory responses. Nature 2024, 630, 695–703. [Google Scholar] [CrossRef]
- Mulder, K.; Patel, A.A.; Kong, W.T.; Piot, C.; Halitzki, E.; Dunsmore, G.; Khalilnezhad, S.; Irac, S.E.; Dubuisson, A.; Chevrier, M.; et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity 2021, 54, 1883–1900.e5. [Google Scholar] [CrossRef] [PubMed]
- Thakore, P.I.; Schnell, A.; Huang, L.; Zhao, M.; Hou, Y.; Christian, E.; Zaghouani, S.; Wang, C.; Singh, V.; Singaraju, A.; et al. BACH2 regulates diversification of regulatory and proinflammatory chromatin states in TH17 cells. Nat. Immunol. 2024, 25, 1395–1410. [Google Scholar] [CrossRef]
- Jin, C.; Jiang, P.; Zhang, Z.; Han, Y.; Wen, X.; Zheng, L.; Kuang, W.; Lian, J.; Yu, G.; Qian, X.; et al. Single-cell RNA sequencing reveals the pro-inflammatory roles of liver-resident Th1-like cells in primary biliary cholangitis. Nat. Commun. 2024, 15, 8690. [Google Scholar] [CrossRef] [PubMed]
- Hoogstrate, Y.; Draaisma, K.; Ghisai, S.A.; van Hijfte, L.; Barin, N.; de Heer, I.; Coppieters, W.; van den Bosch, T.P.P.; Bolleboom, A.; Gao, Z.; et al. Transcriptome analysis reveals tumor microenvironment changes in glioblastoma. Cancer Cell 2023, 41, 678–692.e7. [Google Scholar] [CrossRef]
- Guo, S.; Liu, X.; Zhang, J.; Huang, Z.; Ye, P.; Shi, J.; Stalin, A.; Wu, C.; Lu, S.; Zhang, F.; et al. Integrated analysis of single-cell RNA-seq and bulk RNA-seq unravels T cell-related prognostic risk model and tumor immune microenvironment modulation in triple-negative breast cancer. Comput. Biol. Med. 2023, 161, 107066. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Bian, X.; Meng, X.; Li, L.; Fu, L.; Zhang, Y.; Wang, L.; Zhang, Y.; Gao, D.; Guo, X.; et al. Unveiling inflammatory and prehypertrophic cell populations as key contributors to knee cartilage degeneration in osteoarthritis using multi-omics data integration. Ann. Rheum. Dis. 2024, 83, 926–944. [Google Scholar] [CrossRef]
- Wang, M.; Liu, X.; Chang, G.; Chen, Y.; An, G.; Yan, L.; Gao, S.; Xu, Y.; Cui, Y.; Dong, J.; et al. Single-Cell RNA Sequencing Analysis Reveals Sequential Cell Fate Transition during Human Spermatogenesis. Cell Stem Cell 2018, 23, 599–614.e4. [Google Scholar] [CrossRef]
- Honardoost, M.A.; Adinatha, A.; Schmidt, F.; Ranjan, B.; Ghaeidamini, M.; Arul Rayan, N.; Gek Liang Lim, M.; Joanito, I.; Xiao Xuan Lin, Q.; Rajagopalan, D.; et al. Systematic immune cell dysregulation and molecular subtypes revealed by single-cell RNA-seq of subjects with type 1 diabetes. Genome Med. 2024, 16, 45. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Larsson, L.; Swarbrick, A.; Lundeberg, J. Spatial landscapes of cancers: Insights and opportunities. Nat. Rev. Clin. Oncol. 2024, 21, 660–674. [Google Scholar] [CrossRef]
- Kim, H.; Kim, K.E.; Madan, E.; Martin, P.; Gogna, R.; Rhee, H.W.; Won, K.J. Unveiling contact-mediated cellular crosstalk. Trends Genet. 2024, 40, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, Z.; Qiao, Q.; Zou, Y.; Wang, L.; Wang, T.; Lou, B.; Li, G.; Xu, M.; Wang, Y.; et al. Immunometabolic changes and potential biomarkers in CFS peripheral immune cells revealed by single-cell RNA sequencing. J. Transl. Med. 2024, 22, 925. [Google Scholar] [CrossRef]
- Zhang, D.; Wen, Q.; Zhang, R.; Kou, K.; Lin, M.; Zhang, S.; Yang, J.; Shi, H.; Yang, Y.; Tan, X.; et al. From Cell to Gene: Deciphering the Mechanism of Heart Failure With Single-Cell Sequencing. Adv. Sci. 2024, 11, e2308900. [Google Scholar] [CrossRef]
- Yang, W.; Wang, P.; Xu, S.; Wang, T.; Luo, M.; Cai, Y.; Xu, C.; Xue, G.; Que, J.; Ding, Q.; et al. Deciphering cell-cell communication at single-cell resolution for spatial transcriptomics with subgraph-based graph attention network. Nat. Commun. 2024, 15, 7101. [Google Scholar] [CrossRef] [PubMed]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018, 18, 35–45. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Reyes, M.; Filbin, M.R.; Bhattacharyya, R.P.; Billman, K.; Eisenhaure, T.; Hung, D.T.; Levy, B.D.; Baron, R.M.; Blainey, P.C.; Goldberg, M.B.; et al. An immune-cell signature of bacterial sepsis. Nat. Med. 2020, 26, 333–340. [Google Scholar] [CrossRef]
- Peet, C.; Ivetic, A.; Bromage, D.I.; Shah, A.M. Cardiac monocytes and macrophages after myocardial infarction. Cardiovasc. Res. 2020, 116, 1101–1112. [Google Scholar] [CrossRef]
- Simats, A.; Zhang, S.; Messerer, D.; Chong, F.; Beşkardeş, S.; Chivukula, A.S.; Cao, J.; Besson-Girard, S.; Montellano, F.A.; Morbach, C.; et al. Innate immune memory after brain injury drives inflammatory cardiac dysfunction. Cell 2024, 187, 4637–4655.e26. [Google Scholar] [CrossRef]
- Zhao, G.; Lu, H.; Chang, Z.; Zhao, Y.; Zhu, T.; Chang, L.; Guo, Y.; Garcia-Barrio, M.T.; Chen, Y.E.; Zhang, J. Single-cell RNA sequencing reveals the cellular heterogeneity of aneurysmal infrarenal abdominal aorta. Cardiovasc. Res. 2021, 117, 1402–1416. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, G.; Gropper, J.; Piollet, M.; Vafadarnejad, E.; Rizakou, A.; Bandi, S.R.; Arampatzi, P.; Krammer, T.; DiFabion, N.; Dietrich, O.; et al. Dynamics of monocyte-derived macrophage diversity in experimental myocardial infarction. Cardiovasc. Res. 2023, 119, 772–785. [Google Scholar] [CrossRef]
- Wang, Z.; Xie, L.; Ding, G.; Song, S.; Chen, L.; Li, G.; Xia, M.; Han, D.; Zheng, Y.; Liu, J.; et al. Single-cell RNA sequencing of peripheral blood mononuclear cells from acute Kawasaki disease patients. Nat. Commun. 2021, 12, 5444. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.W.; Tong, Z.; Cao, P.; Wang, F.B.; Liu, Y.; Zheng, Z.Q.; Wang, S.Y.; Li, S.; Wang, Y.Y. Transcription-independent regulation of STING activation and innate immune responses by IRF8 in monocytes. Nat. Commun. 2022, 13, 4822. [Google Scholar] [CrossRef]
- Rentsendorj, A.; Sheyn, J.; Fuchs, D.T.; Daley, D.; Salumbides, B.C.; Schubloom, H.E.; Hart, N.J.; Li, S.; Hayden, E.Y.; Teplow, D.B.; et al. A novel role for osteopontin in macrophage-mediated amyloid-β clearance in Alzheimer’s models. Brain Behav. Immun. 2018, 67, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Capuccini, B.; Lin, J.; Talavera-López, C.; Khan, S.M.; Sodenkamp, J.; Spaccapelo, R.; Langhorne, J. Transcriptomic profiling of microglia reveals signatures of cell activation and immune response, during experimental cerebral malaria. Sci. Rep. 2016, 6, 39258. [Google Scholar] [CrossRef]
- Xu, C.; Xu, H.; Dai, X.; Gui, S.; Chen, J. Effects and mechanism of combination of Platycodon grandiflorum polysaccharides and Platycodon saponins in the treatment of chronic obstructive pulmonary disease rats through the gut-lung axis. J. Ethnopharmacol. 2025, 341, 119305. [Google Scholar] [CrossRef]
- Zhai, R.; Xu, H.; Hu, F.; Wu, J.; Kong, X.; Sun, X. Exendin-4, a GLP-1 receptor agonist regulates retinal capillary tone and restores microvascular patency after ischaemia-reperfusion injury. Br. J. Pharmacol. 2020, 177, 3389–3402. [Google Scholar] [CrossRef]
- Campregher, C.; Luciani, M.G.; Gasche, C. Activated neutrophils induce an hMSH2-dependent G2/M checkpoint arrest and replication errors at a (CA)13-repeat in colon epithelial cells. Gut 2008, 57, 780–787. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Daily, K.P.; Gray, M.C.; Ragland, S.A.; Werner, L.M.; Brittany Johnson, M.; Eby, J.C.; Hewlett, E.L.; Taylor, R.P.; Criss, A.K. Phagocytosis via complement receptor 3 enables microbes to evade killing by neutrophils. J. Leukoc. Biol. 2023, 114, 1–20. [Google Scholar] [CrossRef]
- Edmisson, J.S.; Tian, S.; Armstrong, C.L.; Vashishta, A.; Klaes, C.K.; Miralda, I.; Jimenez-Flores, E.; Le, J.; Wang, Q.; Lamont, R.J.; et al. Filifactor alocis modulates human neutrophil antimicrobial functional responses. Cell. Microbiol. 2018, 20, e12829. [Google Scholar] [CrossRef]
- Nazari, A.; Khorramdelazad, H.; Hassanshahi, G.; Day, A.S.; Sardoo, A.M.; Fard, E.T.; Abedinzadeh, M.; Nadimi, A.E. S100A12 in renal and cardiovascular diseases. Life Sci. 2017, 191, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Foell, D.; Kane, D.; Bresnihan, B.; Vogl, T.; Nacken, W.; Sorg, C.; Fitzgerald, O.; Roth, J. Expression of the pro-inflammatory protein S100A12 (EN-RAGE) in rheumatoid and psoriatic arthritis. Rheumatology 2003, 42, 1383–1389. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Chen, H.; Wang, M. Identification of the biological processes, immune cell landscape, and hub genes shared by acute anaphylaxis and ST-segment elevation myocardial infarction. Front. Pharmacol. 2023, 14, 1211332. [Google Scholar] [CrossRef]
- Chen, H.; Lu, S.; Guan, J.; Zhu, X.; Sun, F.; Huang, J.; Zhu, J.; Wang, J.; Zhen, Z.; Que, Y.; et al. Identification of prognostic immune-related genes in rhabdoid tumor of kidney based on TARGET database analysis. Aging 2021, 13, 5461–5474. [Google Scholar] [CrossRef]
- Kulohoma, B.W.; Marriage, F.; Vasieva, O.; Mankhambo, L.; Nguyen, K.; Molyneux, M.E.; Molyneux, E.M.; Day, P.J.R.; Carrol, E.D. Peripheral blood RNA gene expression in children with pneumococcal meningitis: A prospective case-control study. BMJ Paediatr. Open 2017, 1, e000092. [Google Scholar] [CrossRef]
- Thaikruea, L. Differences in clinical manifestations between cases stung by single-tentacle and multiple-tentacle box jellyfish over two decades. Heliyon 2023, 9, e16374. [Google Scholar] [CrossRef]
- de Bruin, A.M.; Libregts, S.F.; Valkhof, M.; Boon, L.; Touw, I.P.; Nolte, M.A. IFNγ induces monopoiesis and inhibits neutrophil development during inflammation. Blood 2012, 119, 1543–1554. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Shi, Q.; Wu, P.; Zhang, X.; Kambara, H.; Su, J.; Yu, H.; Park, S.Y.; Guo, R.; Ren, Q.; et al. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat. Immunol. 2020, 21, 1119–1133. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, Z.; Hao, Z.; Wang, C.; Lu, N.; Qiu, P.; Wang, S.; Yu, R. Single-Cell Transcriptomic Analysis Reveals Cell Heterogeneity and Altered Signaling Pathways in Jellyfish Sting Patients. Mar. Drugs 2025, 23, 358. https://doi.org/10.3390/md23090358
Qin Z, Hao Z, Wang C, Lu N, Qiu P, Wang S, Yu R. Single-Cell Transcriptomic Analysis Reveals Cell Heterogeneity and Altered Signaling Pathways in Jellyfish Sting Patients. Marine Drugs. 2025; 23(9):358. https://doi.org/10.3390/md23090358
Chicago/Turabian StyleQin, Zhen, Zhengfeng Hao, Chun Wang, Ning Lu, Peiju Qiu, Su Wang, and Rilei Yu. 2025. "Single-Cell Transcriptomic Analysis Reveals Cell Heterogeneity and Altered Signaling Pathways in Jellyfish Sting Patients" Marine Drugs 23, no. 9: 358. https://doi.org/10.3390/md23090358
APA StyleQin, Z., Hao, Z., Wang, C., Lu, N., Qiu, P., Wang, S., & Yu, R. (2025). Single-Cell Transcriptomic Analysis Reveals Cell Heterogeneity and Altered Signaling Pathways in Jellyfish Sting Patients. Marine Drugs, 23(9), 358. https://doi.org/10.3390/md23090358






