1. Introduction
Pyropia haitanensis is a commercially significant red alga (Rhodophyta, Bangiaceae) extensively cultivated along the southeastern coast of China, accounting for approximately three-quarters of the country’s nori production. Its life cycle alternates between two distinct stages: the macroscopic blade gametophyte (thallus) and the microscopic filamentous sporophyte (conchocelis) [
1,
2]. The transition from conchocelis to conchosporangia is a pivotal phase for producing conchospores, which directly influences the quality and yield of gametophytic thalli. In large-scale cultivation of
P. haitanensis, environmental factors, including light intensity, temperature, and phosphorus levels, are meticulously controlled to enhance conchosporangial development [
3,
4,
5]. Despite these advances, the molecular mechanisms governing this transition remain largely uncharacterized.
Recent studies highlight the involvement of phytohormones like jasmonic acid, ethylene, and indole-3-acetic acid in the reproductive processes of algae [
6,
7,
8]. For instance, the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) has been shown to induce the transformation of conchocelis into conchosporangia in
P. haitanensis [
2]. Furthermore, polyamines, small nitrogenous compounds such as putrescine (PUT), spermidine (SPD), and spermine (SPM), are increasingly recognized for their roles in algal reproduction. Distinct from higher plants and other red algae,
P. haitanensis has developed a unique stress-adaptation strategy that integrates S-adenosylmethionine (SAM)-dependent SPD biosynthesis with the glutathione (GSH) antioxidant system, enabling rapid acclimation to intertidal wet–dry fluctuations [
9]. Although polyamines have been shown to promote carpospore output and growth in
Gracilaria cornea (formerly
Hydropuntia cornea) [
10] and to enhance spore release, survival, and germination in
Gracilariopsis lemaneiformis [
11], their potential role in regulating conchosporangial formation in
P. haitanensis remains uninvestigated, highlighting a compelling direction for future research.
Reactive oxygen species (ROS), including H
2O
2 and O
2·
−, are pivotal signaling molecules involved in regulating plant growth, development, and stress responses [
12]. In
P. haitanensis, H
2O
2 has been identified as a mediator of ACC-induced conchosporangial formation [
2]. ROS are predominantly generated by NADPH oxidases (NOXs), which initiate the production of O
2·
−, subsequently dismutated into H
2O
2 by SOD [
13]. Notably, polyamines play a dual role in ROS dynamics: they can generate and regulate ROS through catabolic pathways, maintaining a balance between oxidative stress and cellular signaling [
14,
15]. While the signaling role of H
2O
2 is well-documented, the involvement of O
2·
− in algal development remains less understood. Emerging evidence suggests its significance in key processes, such as cell cycle progression and gametophyte formation, highlighting its potential as a regulatory molecule [
16,
17,
18,
19].
This study explored the role of SPM in regulating conchosporangial development in P. haitanensis. Our findings revealed O2·− as a critical signaling molecule activated by SPM, which facilitated the formation of conchosporangia. Transcriptomic analyses further demonstrated that SPM promoted this developmental transition by enhancing photosynthesis and carbon assimilation and maintaining oxidative–antioxidative balance. These results shed new light on the molecular mechanisms underlying algal reproduction and offered valuable insights for optimizing P. haitanensis cultivation strategies.
2. Results
2.1. Polyamines Enhance Conchosporangial Formation in P. haitanensis
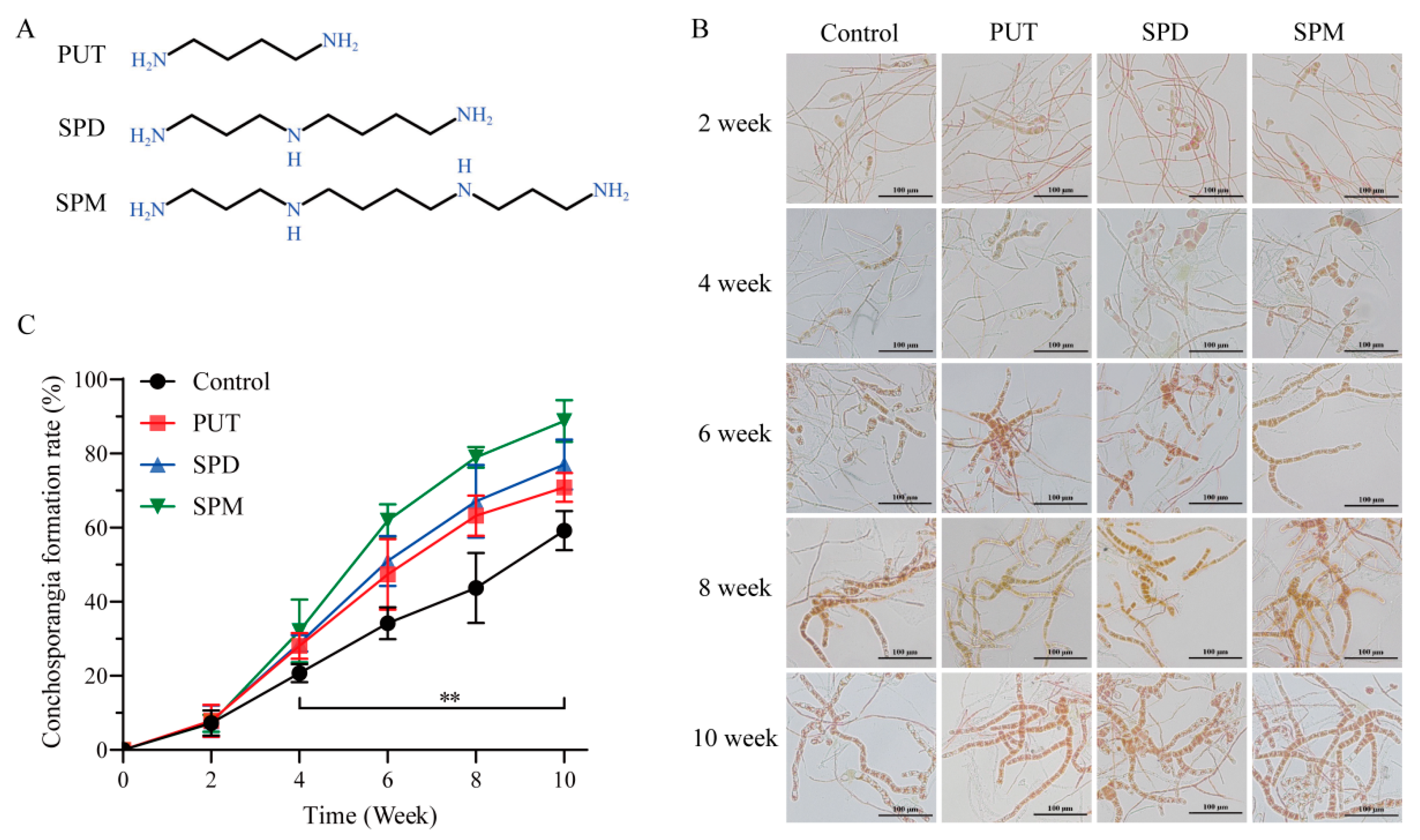
To clarify the involvement of polyamines in conchosporangial development, we first measured the endogenous levels of PUT, SPD, and SPM in
P. haitanensis (
Figure S1). The basal concentrations were quantified as 28.33 μg/g fresh weight (FW) for PUT, 2.09 μg/g FW for SPD, and 10.39 μg/g FW for SPM. To evaluate their physiological roles, exogenous polyamines were administered at concentrations ranging from 1 μM (10
−6 M) to 100 μM (10
−4 M). As shown in
Figure 1 and
Figure S2, following a 2-week treatment period, the emergence of conchosporangia was first observed, with their proportion progressively increasing over time. By the 10th week, the formation rate reached 70–90%, exhibiting a steady upward trend throughout the experimental duration.
Among the polyamines tested, SPM demonstrated the strongest promotive effect on conchosporangial formation. The SPM-treated group achieved a formation rate of 88.78% by the 10th week, the highest observed across all treatments. In comparison, PUT and SPD resulted in relatively lower formation rates. These findings underscored the superior efficacy of SPM in facilitating conchosporangial development. Consequently, SPM was selected for subsequent experiments aimed at elucidating the mechanisms underlying its role in promoting conchosporangial formation.
2.2. SPM Enhances Conchosporangial Formation in Both Shell-Living and Free-Living Conchocelis
The effects of SPM on conchosporangial formation in shell-living and free-living conchocelis of
P. haitanensis are presented in
Figure 2. In shell-living conchocelis, SPM treatment at concentrations of 0.2, 1, and 2.5 μM significantly stimulated conchosporangial formation compared to the control group. Conchosporangial clusters first appeared on the shell surface by the 3rd week of treatment, and by the 6th week, the surface was almost fully covered. Among the tested concentrations, 1 μM SPM was the most effective, achieving a formation rate of 73.4%, significantly exceeding that of the control group (
p < 0.05). Although 0.2 μM and 2.5 μM SPM also enhanced conchosporangial formation, their effects were slightly less pronounced than that of 1 μM (
p < 0.05).
In free-living conchocelis, treatment with 1 μM SPM similarly resulted in significant promotion of conchosporangial formation, reaching a formation rate of 69.49% after 8 weeks. This rate was markedly higher than that observed in the control group (p < 0.05), where conchosporangial formation was minimal. While 0.2 μM and 2.5 μM SPM also increased formation rates relative to the control, their effects were not as robust as those of 1 μM.
These findings conclusively demonstrated that SPM effectively promoted conchosporangial formation in both shell-living and free-living conchocelis, with 1 μM SPM being the optimal concentration. Consequently, 1 μM SPM was selected for subsequent experiments to investigate the mechanisms underlying its promotive effects on conchosporangial formation.
2.3. Role of O2·− in SPM-Induced Conchosporangial Formation
The role of SPM in generating ROS during conchosporangial formation in
P. haitanensis was investigated (
Figure 3). NOX activity assays exhibited a biphasic response following SPM treatment, with distinct peaks at 2 h (96.12 U/mg protein) and 6 h (184.19 U/mg protein), before gradually returning to baseline by 8 h. Notably, co-treatment with the NOX inhibitor DPI completely suppressed the SPM-induced enhancement of NOX activity (
Figure 3A). Consistently, RT-PCR analysis revealed that SPM upregulated the transcript levels of
NOXA and
NOX5, consistent with the observed enzyme activity dynamics (
Figure S3). To assess the functional impact of NOX activation, O
2·
− and H
2O
2 levels were measured in conchocelis exposed to 1 μM SPM over a 24 h period. A significant increase in O
2·
− levels was observed during the first 8 h, reaching a peak concentration of 12.36 μmol/g·wt. A secondary peak (11.43 μmol/g·wt) occurred at 16 h, after which O
2·
− levels gradually returned to baseline (
Figure 3B). In contrast, H
2O
2 levels remained relatively stable throughout the treatment period (
Figure S4), indicating that the observed increase in ROS was primarily attributable to O
2·
− production rather than H
2O
2.
Fluorescence microscopy using lucigenin further confirmed the production of O
2·
−, as evidenced by strong green fluorescence in conchocelis cells. This fluorescence reached its peak intensity after 8 h of SPM treatment and was notably diminished upon the addition of the DPI, demonstrating the involvement of NOX in O
2·
− generation during this process (
Figure 3C).
To establish the role of O2·− in SPM-induced conchosporangial formation, conchocelis were treated with 1 μM SPM in the presence of DPI. DPI treatment significantly suppressed conchosporangial formation, reducing the formation rate by 50.2% compared to SPM treatment alone (p < 0.05). This marked inhibition underscored the pivotal role of O2·− as a signaling molecule in SPM-mediated conchosporangial formation. The reduction in conchosporangial formation correlated strongly with decreased O2·− levels, further supporting the hypothesis that O2·− served as a critical mediator of this developmental process.
2.4. Transcriptomic Profiling of Conchosporangia in P. haitanensis in Response to SPM
To elucidate the molecular mechanisms underlying SPM-induced conchosporangial formation, transcriptome sequencing was performed on
P. haitanensis conchocelis treated with 1 μM SPM for 24 h. The sequencing generated approximately 2.5 × 10
7 clean reads with an average length of 150 bp, of which 88.8% were successfully mapped to 11,129 genes in the
P. haitanensis reference genome. Validation of RNA-seq data through qRT-PCR analysis of 10 randomly selected unigenes demonstrated a high degree of consistency between the transcriptome and qRT-PCR results (
Figure S5).
Comparative analysis identified 2310 Differentially expressed genes (DEGs) following SPM treatment, including 1255 upregulated and 1055 downregulated genes (
Supplementary Table S1). KEGG pathway enrichment analysis, ranked by significance, revealed that these DEGs were predominantly associated with pathways involved in photosynthetic carbon fixation, photosynthesis, acetate and dicarboxylate metabolism, pyruvate metabolism, carotenoid biosynthesis, alanine, aspartate, and glutamate metabolism, as well as peroxisome functions (
Figure S6).
2.5. SPM Enhances Photosynthesis in P. haitanensis Conchosporangia
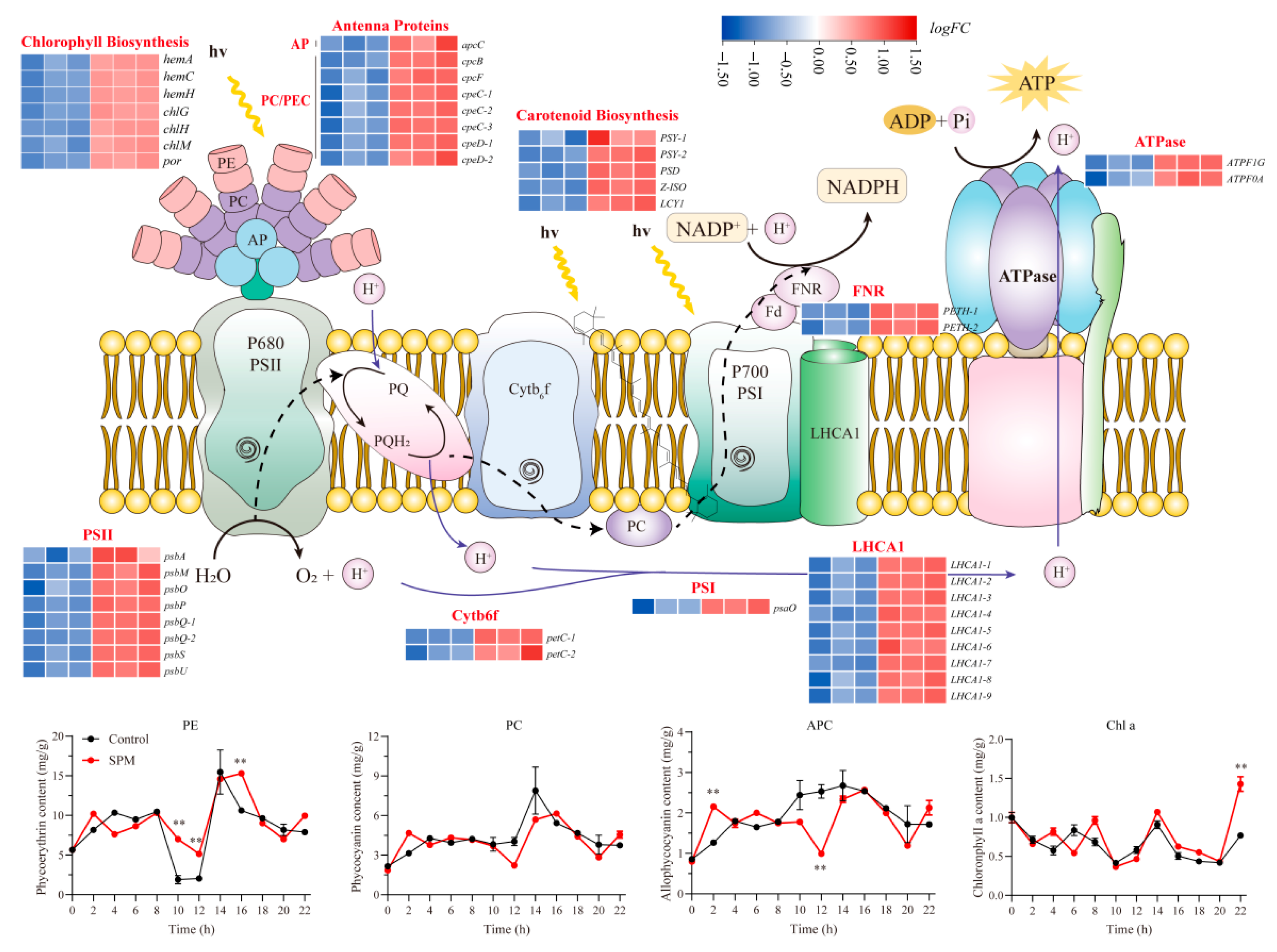
SPM treatment significantly impacted the expression of genes related to photosynthesis and photosynthetic carbon fixation pathways (
Figure 4). A total of 44 DEGs involved in photosynthesis were identified, all of which were markedly upregulated compared to the control group. Notably, genes encoding components of photosystem I (
psaO) and photosystem II (
psbA,
psbM,
psbO,
psbP,
psbQ,
psbS, and
psbU) exhibited substantial upregulation. Similarly, genes associated with the cytochrome b6f complex (
petC), ferredoxin-NADP
+ reductase (
petH), and ATP synthase complex (
atpF) were significantly upregulated (
p < 0.05), indicating enhanced light reaction activity.
Additionally, SPM treatment elevated the expression of genes encoding light-harvesting complex proteins (LHCA) and phycobiliproteins, including allophycocyanin (apcC) and phycocyanin (cpcB, cpcF, cpeC, and cpeD). Importantly, this upregulation extended to the major components of the photosynthetic pigment biosynthesis pathway, with key genes involved in chlorophyll a production (hemA, hemC, hemH, chlG, chlH, chlM, and por) and carotenoid synthesis (PSY, PSD, Z-ISO, and LCY) being coordinately activated.
Pigment content analysis confirmed these changes, with phycoerythrin (PE) being 2.70–3.68 times that of the control at 10–12 h, while phycocyanin (PC) and allophycocyanin (APC) reached 1.48 times and 1.7 times the control levels at 2 h, respectively. Chlorophyll a content peaked at 1.86 times the control at 24 h. These findings suggested that SPM enhanced photosynthetic efficiency and pigment biosynthesis, contributing to the observed promotion of conchosporangial formation.
2.6. SPM Enhances Carbon Assimilation by Regulating the Calvin Cycle and C2 Photorespiration Pathways
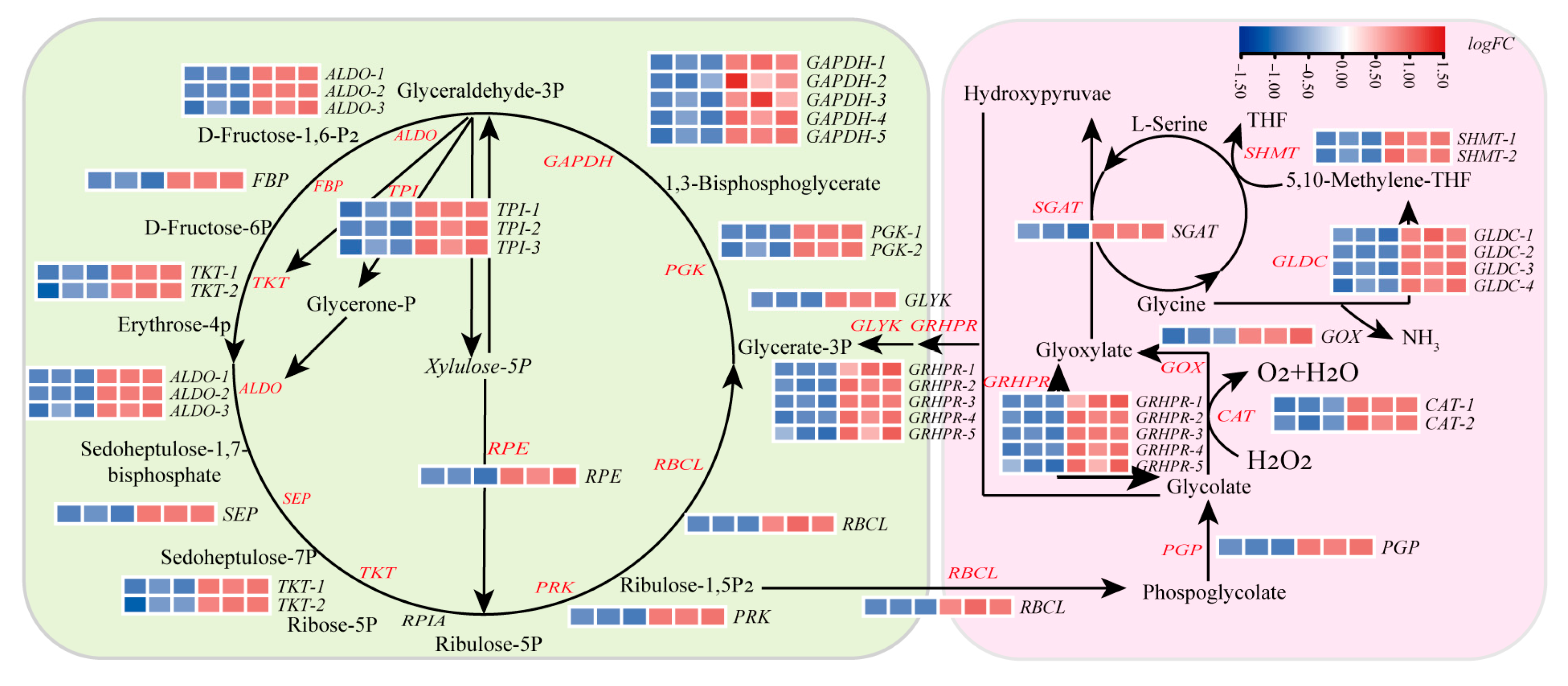
Transcriptomic analysis of
P. haitanensis conchocelis treated with 1 μM SPM for 24 h revealed significant upregulation of genes associated with both the Calvin cycle and C
2 photorespiration pathways, pivotal processes for carbon assimilation during conchosporangial formation (
Figure 5). In the Calvin cycle, 25 DEGs involved in carbon fixation were identified. Notably, key genes in the carboxylation and reduction phases, including ribulose-bisphosphate carboxylase (
RBCL), phosphoglycerate kinase (
PGK), and glyceraldehyde-3-phosphate dehydrogenase (
GAPDH), were significantly upregulated (
p < 0.05), suggesting enhanced carbon fixation efficiency.
Concurrently, SPM treatment upregulated genes associated with the C
2 photorespiration pathway (which mediates glyoxylate metabolism), including glycolate oxidase (
GOX), serine-glyoxylate aminotransferase (
SGAT), and glyoxylate/hydroxypyruvate reductase (
GRHPR). In contrast, transcripts of key tricarboxylic acid (TCA) cycle genes (e.g., aconitase (
ACO), ATP-citrate lyase (
ACLY), and malate dehydrogenase 1 (
MDH1)) were markedly downregulated (
Figure S7). This coordinated transcriptional shift suggested that enhanced C
2 pathway activity might functionally complement suppressed mitochondrial respiration, potentially optimizing carbon flux under SPM influence.
2.7. SPM Promotes Peroxisomal Biogenesis and Modulates Antioxidant Systems in P. haitanensis
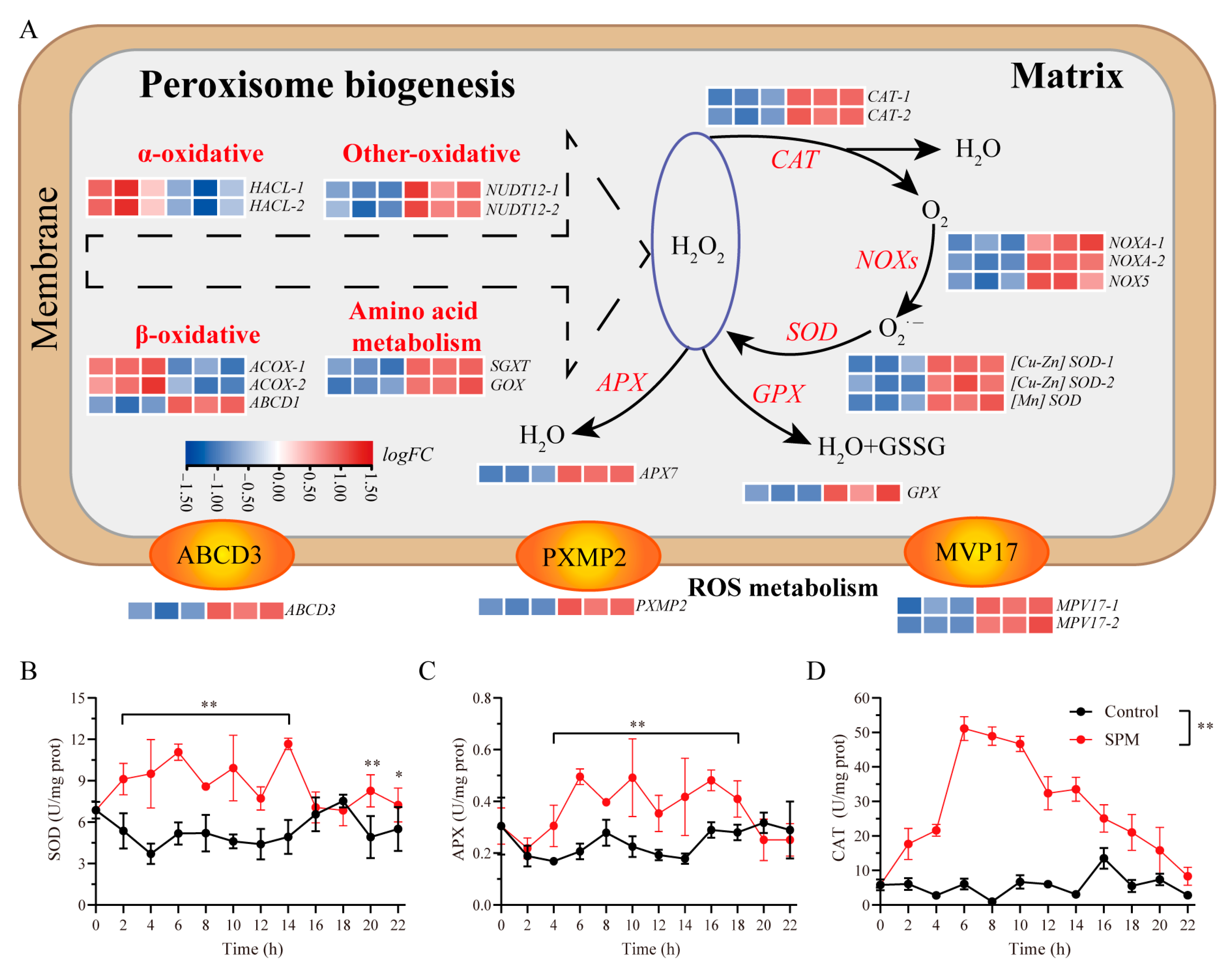
To investigate the role of SPM in managing oxidative stress during conchosporangial formation, the expression of genes related to peroxisomal biogenesis and antioxidant defense mechanisms was analyzed (
Figure 6). Genes critical for peroxisomal membrane protein transport, including ATP-binding cassette subfamily D member 3 (
abcD3) and peroxisomal membrane protein 2 (
pxmp2), were significantly upregulated (
p < 0.05). Additionally, enzymes involved in photorespiration, such as
GOX and
SGAT, showed increased expression, further supporting the role of peroxisomes in ROS production under SPM treatment.
Transcriptomic data revealed a marked upregulation of genes associated with both enzymatic and non-enzymatic antioxidant defenses in response to SPM. Key antioxidant enzymes, including SOD, CAT, APX, and glutathione peroxidase (GPX), displayed significantly elevated expression levels (p < 0.05).
The enzyme activity profiles mirrored these transcriptional changes. During conchosporangial formation under 1 μM SPM treatment, activities of SOD, APX, and CAT remained elevated over the 24 h period. SOD activity began to increase at 2 h, stabilizing at 7.72–11.67 U/mg protein before returning to baseline at 16–18 h, followed by a second peak at 20–22 h (
p < 0.05). APX activity rose at 4 h, maintaining a range of 0.31–0.49 U/mg protein (
p < 0.05) before gradually declining after 18 h. Similarly, CAT activity peaked at 6 h (51.14 ± 3.48 U/mg protein) and then progressively decreased to 8.30 ± 2.63 U/mg protein at 22 h (
p < 0.05;
Figure 6B–D).
These findings highlighted SPM’s dual role in promoting peroxisomal biogenesis and bolstering antioxidant systems, thereby managing ROS levels to support the formation of conchosporangia in P. haitanensis.
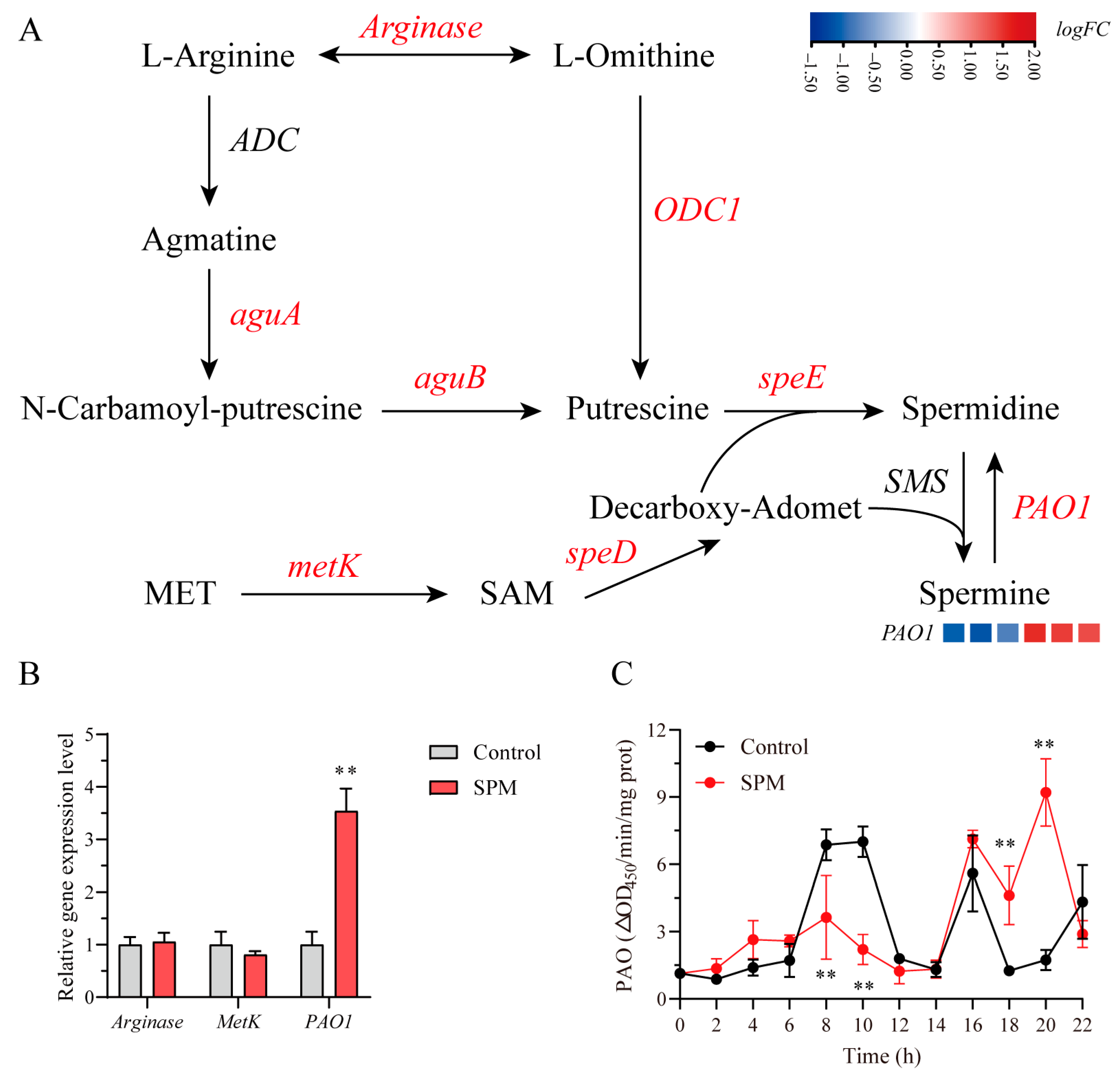
2.8. SPM-Mediated Regulation of Polyamine Metabolism in P. haitanensis Conchosporangia
To systematically dissect the regulatory effects of SPM on polyamine metabolism in
P. haitanensis conchosporangia, we employed an integrative approach combining transcriptome profiling, qPCR validation, and enzymatic assays. Transcriptomic analysis revealed that the majority of genes involved in polyamine biosynthesis and catabolism, such as
arginase and
metK, remained transcriptionally unchanged (|logFC| < 1,
p > 0.05), whereas polyamine oxidase 1 (
PAO1) was selectively upregulated (logFC = 1.482,
p < 0.001;
Figure 7A,
Supplementary Table S2), a result further confirmed by qPCR (
Figure 7B). Enzyme activity measurements revealed a biphasic PAO1 response to SPM exposure, characterized by an initial suppression at 8 h (
p < 0.01) followed by a pronounced activation at 22 h (9.21 ± 1.45 ΔOD
450/min/mg protein) (
Figure 7C). Collectively, these findings suggested that SPM preferentially promoted its own back-conversion to SPD via PAO1 induction, while exerting minimal influence on upstream biosynthetic pathways.
2.9. SPM-Induced Metabolic Reprogramming Sustains Long-Term Biomass Accumulation
Building upon the SPM-induced transcriptional activation of photosynthesis, carbon assimilation, and antioxidant systems, we assessed the long-term physiological outcomes of this metabolic reprogramming. Treatment with high SPM concentrations (1 μM and 2.5 μM) significantly increased the average daily mass growth rate of conchosporangia (
p < 0.05), achieving rates approximately 2.03 and 1.74 times higher than those of the control group. Conversely, pre-incubation with the NOX inhibitor DPI or the polyamine biosynthesis inhibitor mitoguazone (MGBG) led to a substantial reduction in the average daily mass growth rate, decreasing by 27.5% and 38.4%, respectively (
Figure S8).
3. Discussion
This study offered valuable insights into the molecular mechanisms driving conchosporangial formation in
P. haitanensis, with a particular focus on the role of SPM and its involvement in signaling pathways, especially those associated with ROS. Among the polyamines evaluated, SPM demonstrated the strongest promotive effect on conchosporangial formation, consistent with prior research on red algae. Previous studies have shown that polyamines like SPD and SPM enhance reproductive development in similar systems (
G. cornea and
G. lemaneiformis, etc.) [
10,
11,
20,
21].
The superior efficacy of SPM compared to PUT and SPD suggested that the polyamine’s structure and chain length were critical determinants of its biological activity. As a larger and more complex polyamine, SPM may engage more effectively with molecular targets [
22,
23], facilitating the transition from conchocelis to conchosporangia. This hypothesis is supported by findings in other algal species, where polyamines with longer chains and greater molecular complexity are often more potent in regulating developmental transitions [
24]. These observations underscore the intricate relationship between polyamine structure and function in modulating reproductive processes in algae.
The most remarkable discovery in this study was the identification of O
2·
− as a novel signaling molecule in SPM-mediated conchosporangial formation. Unlike earlier research in
P. haitanensis, which has highlighted H
2O
2 as a key signaling molecule in this process [
2], our findings revealed that O
2·
− assumed a pivotal role in initiating this developmental transition. While O
2·
− are well-documented signaling molecules in plant and algal growth and stress responses [
25,
26], their involvement in reproductive processes within marine red algae has remained elusive. Our data demonstrated that the application of the NOX inhibitor DPI markedly reduced both O
2·
− production and conchosporangial formation, confirming that NOX-driven O
2·
− generation was integral to SPM-induced conchosporangial development. This finding aligned with studies in higher plants, where NOX-derived O
2·
− has been implicated in regulating crucial developmental events, including seed germination, root elongation, and floral induction [
27,
28,
29]. Additionally, the temporal dynamics of O
2·
− production observed in our study, characterized by two distinct peaks at 8 and 16 h, suggested that O
2·
− functioned as a transient signal, orchestrating key events during the developmental progression.
Our study also provided compelling evidence that SPM enhanced photosynthetic efficiency and carbon assimilation in
P. haitanensis. Transcriptomic analysis revealed the significant upregulation of genes associated with the light reactions of photosynthesis, including those encoding components of photosystem I (
psaO) and photosystem II (
psbA,
psbM,
psbO,
psbP,
psbQ,
psbS, and
psbU), as well as genes involved in electron transport and ATP synthesis, such as
petC and
atpF. These results were consistent with previous studies demonstrating the role of polyamines in modulating photosynthesis, particularly under stress conditions [
30,
31,
32]. Polyamines like SPM are known to stabilize chloroplast membranes by binding to thylakoid structures, thereby enhancing the efficiency of light absorption and electron transport [
33,
34].
In the context of conchosporangial formation, enhanced photosynthetic activity likely fulfills the energy demands of this metabolically intensive developmental process [
35], particularly under suboptimal environmental conditions. The upregulation of genes (including
LHCA,
apc,
cpc and
hemA, etc.) and increased pigment levels demonstrate improved light-harvesting capacity, ensuring efficient light utilization. Additionally, the increased expression of carotenoid biosynthesis genes, including
PSY and
LCY, underscored a protective mechanism by which carotenoids mitigate oxidative stress during this critical developmental stage [
36,
37]. Since conchosporangial formation frequently occurs under environmental stress, these enhancements in photosynthetic efficiency and photoprotection are likely vital for sustaining the metabolic requirements of this process.
These findings suggested that SPM functioned not only as a signaling molecule but also as a modulator of photosynthetic efficiency, providing the energy and reducing power essential for conchosporangial development. Future research could investigate the interplay between SPM-mediated ROS signaling and the regulation of photosynthetic pathways, offering deeper insights into the molecular mechanisms governing algal reproduction.
Our findings revealed that SPM treatment significantly upregulated genes involved in both the Calvin cycle and the C
2 photorespiration pathways, underscoring its role in enhancing carbon assimilation. Specifically, key enzymes such as
RBCL,
PGK, and
GAPDH were markedly upregulated, indicating elevated carbon fixation activity. These results are aligned with prior studies demonstrating that polyamines enhance carbon fixation across various plant species, including algae [
38,
39]. By improving Calvin cycle efficiency, SPM ensures that
P. haitanensis can effectively incorporate carbon into organic compounds, providing both the energy and reducing power necessary for the developmental transition from conchocelis to conchosporangia.
Beyond the Calvin cycle, SPM treatment also upregulated genes involved in the C
2 photorespiration pathway, such as
GOX,
SGAT, and
GRHPR, highlighting increased photorespiratory activity. The C
2 cycle mitigated oxidative stress associated with photosynthesis by processing glycolate and glyoxylate from RuBisCO-mediated oxygenation reactions [
40]. Meanwhile, the concurrent downregulation of TCA cycle genes (
ACO,
ACLY, and
MDH1) implied a redirection of mitochondrial carbon flux toward the photorespiratory pathway. This metabolic reprogramming not only facilitated carbon fixation but also contributed to the maintenance of redox homeostasis during conchosporangial development, particularly under photorespiratory stress. Taken together, these findings suggested that SPM enhanced carbon assimilation by coordinately activating both the Calvin cycle and the C
2 pathway, thereby securing critical metabolic resources for conchosporangial formation, likely via energy redistribution and stress-responsive signaling mechanisms.
To mitigate the NOX-derived O
2·
− bursts, SPM concurrently stimulated peroxisome biogenesis and antioxidant defenses, establishing a tightly regulated ROS homeostasis cycle. The significant upregulation of peroxisomal membrane proteins such as
abcD3 and
pxmp2, along with enzymes involved in photorespiration, pointed to an active role for peroxisomes in ROS production, including H
2O
2. These findings were consistent with research in plants, where peroxisomes are central to ROS generation and oxidative stress regulation [
41,
42]. Interestingly, despite the increased peroxisomal activity, H
2O
2 levels remained stable following SPM treatment, suggesting the presence of an efficient antioxidant system. Key antioxidant enzymes, including SOD, APX, and CAT, as well as genes encoding
CAT,
APX, and
GPX, were significantly upregulated, playing a vital role in scavenging excess ROS.
This robust antioxidant response is crucial, as oxidative stress often accompanies heightened photosynthetic activity [
43]. In
P. haitanensis, the antioxidant system appears to operate in a highly coordinated manner, selectively regulating ROS levels. The differential control of ROS suggests that SPM orchestrates redox balance during conchosporangial formation. O
2·
− might serve as signaling molecules to drive developmental processes, while H
2O
2 levels were tightly regulated to prevent cellular damage. This selective modulation aligned with findings in plants, where polyamines modulate antioxidant enzyme activity to safeguard cells from oxidative damage while enabling ROS to function as signaling molecules [
44,
45]. The intricate regulation of O
2·
− and H
2O
2 highlighted the complex interplay between oxidative stress management and developmental signaling in
P. haitanensis, showcasing SPM’s critical role in this process. This context-dependent modulation of ROS aligned with the stress-development crosstalk model proposed by Liu et al., albeit here repurposed to regulate reproductive transitions [
46].
Polyamine homeostasis, pivotal to maintaining redox equilibrium, is tightly controlled. Although the majority of genes involved in polyamine biosynthesis remained transcriptionally unaltered under SPM treatment, the significant upregulation and biphasic activity pattern of
PAO1, a key catabolic enzyme that generates H
2O
2 from SPM, indicated that SPM preferentially modulated polyamine levels via degradation (
Figure 7). This catabolic emphasis likely fulfilled dual functions: preserving intracellular polyamine reserves under stress while enabling metabolic adaptability essential for developmental transitions, a mechanism reminiscent of phytohormone-regulated ROS signaling observed in higher plants [
47].
Interestingly, in addition to the polyamines (PUT, SPD, and SPM) identified in this study, previous research has shown that ACC and methyl jasmonate also promote conchosporangial formation in
P. haitanensis [
2]. These findings suggest that a variety of substances can induce this developmental event. Despite the widespread presence of phytohormones in
Pyropia, their functional roles may not be as precisely differentiated as those in higher plants, likely due to the relatively primitive evolutionary status of
Pyropia. Instead, these phytohormones may act primarily as elicitors. Notably, while multiple phytohormones can trigger conchosporangial formation through ROS signaling, they appear to utilize distinct ROS. The mechanisms underlying this specificity remain an intriguing avenue for future research.
4. Materials and Methods
4.1. Experimental Materials
Preparation of Free-Living Conchocelis: The free-living conchocelis of
P. haitanensis (ZD-2 strain) were obtained and conserved by the Key Laboratory of Marine Biotechnology, Zhejiang Province, China. The cultivation of conchocelis and the induction of conchosporangial formation were conducted following the method described by Niu et al. (2024) [
2].
Preparation of Shell-Living Conchocelis: Segments of free-living conchocelis, measuring 200–300 μm, were sprayed onto shell surfaces at a transplantation density of 100 mg/m2. These shells were initially kept in low-light conditions for 3 days. Following this, they were exposed to light and maintained at a constant temperature of 20 °C with a photosynthetic photon flux density (PPFD) of 15 μmol/(m2·s) under a photoperiod of 12 h of light and 12 h of darkness for two additional weeks. After this period, excess conchocelis filaments were carefully brushed off, the culture medium was replaced, and the light intensity was increased to 30 μmol/(m2·s). Once the conchocelis adequately covered the shell surfaces, the experiments to induce conchosporangial formation commenced.
4.2. Conchosporangia Formation Ratio Detection
Free-living conchocelis were cultivated in unsealed, transparent small glass bottles (15 mL capacity, 10 mL working volume) under various treatments, including 1 μM PUT, SPD, and SPM (Sigma-Aldrich, St. Louis, MO, USA) for 10 weeks; 0.2, 1, and 2.5 μM SPM for 8 weeks; and 1 μM SPM alone or combined with 0.1 μM DPI for 6 weeks. The cultures were maintained at 29 ± 0.5 °C with a light intensity of 20 μmol/(m
2·s) under a photoperiod of 8 h light and 16 h darkness. The medium, supplemented with K
2HPO
4 to a final concentration of 110 nmol/L, was refreshed weekly during the conchosporangial maturation phase. The conchocelis and conchosporangia were monitored weekly under a microscope (Nikon Eclipse 80i, Tokyo, Japan) to assess their developmental progression. The conchosporangial formation ratio was determined using the following formula:
Shell-living conchocelis were treated with final concentrations of 0.2, 1, and 2.5 μM SPM for a duration of 6 weeks (n = 6). Weekly, the shells were carefully removed and examined using a stereo microscope (Nikon, DS-Ri2, Tokyo, Japan). For each observation, 10 random fields of view were selected and imaged at 135× magnification. The captured images were processed and analyzed using ImageJ software (v.1.4.3.67) to quantify the proportion of conchosporangia branch formation. The proportion of conchosporangial branch formation was calculated using the following formula:
4.3. Relative Mass Growth Rate
Free-living conchocelis cultures were maintained for 6 weeks under various treatments, including final concentrations of 0.2, 1, and 2.5 μM SPM, as well as combinations of 1 μM SPM with either 0.1 μM DPI (MedChemExpress LLC, Monmouth Junction, NJ, USA) or 0.1 μM MGBG (MedChemExpress LLC, Monmouth Junction, NJ, USA). Each treatment group was established in triplicate, with an initial fresh weight of 0.03 g/L for the conchocelis. The culture medium was partially renewed every 7 days, with half the volume replaced during each change. Upon completing the 6-week experiment, the relative mass growth rate was determined using the following formula:
where W
0 is the initial FW (g), and W
t is the FW at 6 weeks (g).
4.4. Detection of H2O2, O2·− Contents, and Related Enzyme Activities
Free-living conchocelis were cultivated with a final concentration of 1 μM SPM for 24 h, and fresh samples weighing 0.03 g were collected at 2 h intervals. The collected samples were homogenized in a cell lysis buffer (Beyotime Biotechnology, Shanghai, China) for 10 min to prepare a 10% homogenate. After centrifugation at 2500× g for 10 min at 4 °C, the supernatants containing enzymes, H2O2, and O2·− were collected for further analysis. The levels of H2O2, SOD (EC 1.15.1.1), CAT (EC 1.11.1.6), and APX (EC 1.11.1.11) were quantified using assay kits provided by Nanjing Jiancheng Bioengineering Institute (Nanjing, China), while O2·−, NOX (EC 1.6.3.1), and PAO (EC 1.5.3.11) were assessed using assay kits from Suzhou Grace Biotechnology Co., Ltd. (Suzhou, China).
For cellular O2·− detection, conchocelis were treated with 1 μM SPM or a combination of 1 μM SPM and 0.1 μM DPI for 8 h. The samples were then rinsed three times with sterile seawater. Subsequently, 0.1 μM lucigenin (MedChemExpress LLC, Monmouth Junction, NJ, USA) was added, and the samples were incubated for an additional 30 min. Fluorescence microscopy was employed for imaging, with an excitation wavelength of λex = 405 nm and an emission wavelength of λem = 492 nm.
4.5. Determination of Endogenous PUT, SPD, and SPM Levels in P. haitanensis
Standard solutions of PUT, SPD, and SPM (2 mg/mL in ultrapure water) were derivatized with benzoyl chloride in 2 M NaOH at 37 °C for 20 min, followed by extraction with and vacuum concentration. The resulting derivatives were reconstituted in methanol, filtered through a 0.22 μm membrane, and serially diluted (0.5–8 μg/mL) to generate calibration curves. For sample preparation, 10 mg of lyophilized powder was extracted with 5% perchloric acid for 1 h in an ice bath and centrifuged at 15,000× g for 30 min, and the supernatant was derivatized using the same protocol. Chromatographic analysis was performed using UPLC equipped with a Waters ACQUITY HSS T3 column (Milford, MA, USA) (1.8 μm, 30 °C), employing isocratic elution with acetonitrile:water (44:56, v/v) at a flow rate of 0.45 mL/min and detection at 220–230 nm. Data acquisition and quantification were conducted using MassLynx software (v.4.2), and results were exported to Excel for statistical analysis. All measurements represent the mean ± SD from three biological replicates.
4.6. Phycobiliprotein and Chlorophyll a Quantification
Phycobiliproteins (PE, PC, APC) were extracted from 0.02 g algal samples using PBS buffer after liquid nitrogen grinding and freeze–thaw cycles. Absorbance was measured at 565 nm, 615 nm, and 650 nm, with concentrations calculated as follows:
Chlorophyll
a was extracted in 5 mL absolute methanol (24 h, 4 °C in darkness), with absorbance measured at 665 nm, 750 nm, and 652 nm. Pigment contents were determined using:
where V is extract volume (mL) and FW is fresh weight (g) [
48].
4.7. RNA Isolation, Transcriptome Sequencing, and Analysis
Approximately 100 mg of P. haitanensis tissue from each group was collected, snap-frozen in liquid nitrogen, and ground into a fine powder. Total RNA was extracted using the Plant RNA Isolation Kit (Meiji Bio, Guangzhou, China). The integrity of the RNA was evaluated via agarose gel electrophoresis, and its concentration and purity were determined using a NanoDrop™ One spectrophotometer (Wilmington, DE, USA). Only RNA samples meeting stringent quality standards were used for subsequent analyses.
Transcriptome sequencing was performed on the Illumina HiSeq platform at Wuhan Feisha Gene Information Co., Ltd. (Wuhan, China). After removing low-quality reads, the clean reads were aligned to the P. haitanensis reference genome using HISAT2 (v.2.2.1). Further mapping to the reference transcriptome was conducted with Bowtie2 software (v.2.5.1), and transcript quantification was carried out using RSEM. DEGs were identified through DESeq2 (v.1.22.2), with the criteria of |log2FC| > 1 and a false discovery rate (FDR) significance threshold (padj) < 0.05.
Functional annotation and enrichment analysis of the transcriptome were performed using the KEGG database (
www.KEGG.jp). Enriched pathways were identified via hypergeometric testing, with pathway enrichment analysis carried out using custom scripts in R (v3.3.2,
www.r-project.org). The Benjamini–Hochberg (BH) correction was applied to control the FDR.
To visualize transcriptome data, fuzzy C-means clustering was performed using the TCseq and Cairo packages in R. Heatmaps were generated using TBtools (
https://github.com/CJ-Chen/TBtools-II/releases; accessed on 28 July 2023), while KEGG bubble plots were created with Oebiotech’s online platform (
https://cloud.oebiotech.com/task/; accessed on 25 June 2024). Bar charts were generated using GraphPad Prism (v.8.0.2), and pathway diagrams were illustrated with Adobe Illustrator (v.2021).
4.8. Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA was extracted from samples of each group using the Plant RNA Extraction Kit (Magen, Guangzhou, China). Single-stranded cDNA was synthesized using the PrimeScript™ RT Master Mix (TaKaRa Biotechnology Co., Ltd., Dalian, China), following the manufacturer’s protocol. qRT-PCR was conducted on a LightCycler
® 96 Real-Time PCR System (Roche, Basel, Switzerland). The primer sequences used in this study are detailed in
Supplementary Table S3. The amplification protocol consisted of an initial denaturation at 94 °C for 30 s, followed by 40 cycles of 94 °C for 5 s, 60 °C for 15 s, and 72 °C for 10 s. Relative gene expression levels were calculated using the 2
−ΔΔCt method, with β-actin serving as the internal reference gene.
4.9. Statistical Analysis
All experiments were conducted in triplicate, with results expressed as the mean ± SD. Statistical analyses were performed using SPSS software (version 22.0; SPSS Inc., Chicago, IL, USA). Differences among treatment groups were evaluated using one-way ANOVA, followed by Tukey’s HSD test for multiple comparisons. A p-value of <0.05 was considered statistically significant.