Lipotrichaibol A and Trichoderpeptides A–D: Five New Peptaibiotics from a Sponge-Derived Trichoderma sp. GXIMD 01001
Abstract
1. Introduction
2. Results
2.1. Isolation and Structural Elucidation
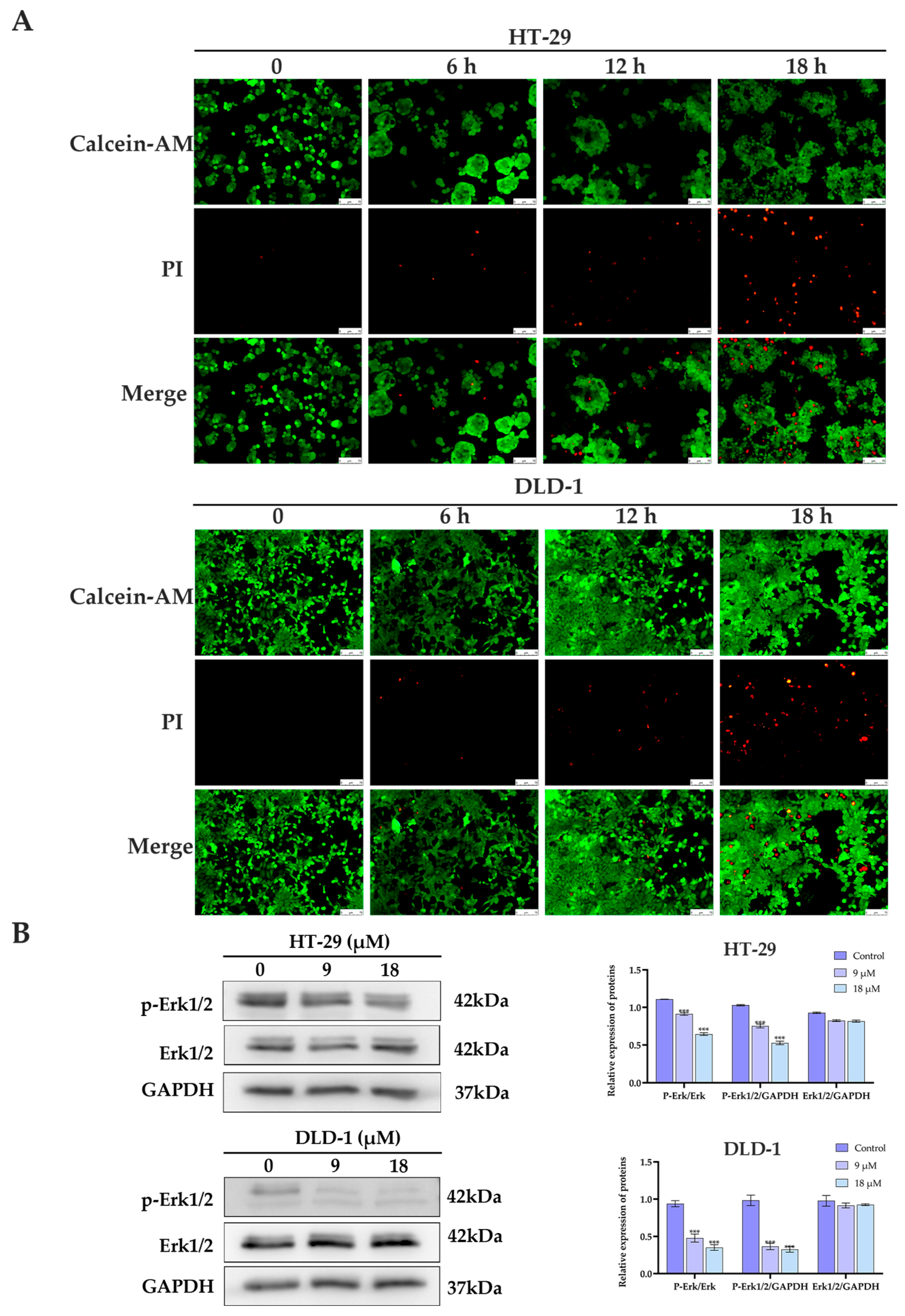
2.2. Biological Activity Assessment
2.3. ADMET Properties
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Extraction and Separation of Crude Extracts
3.3. Marfey’s Analysis
3.4. ECD Analysis
3.5. CCK8 Assay
3.6. Cell Clone Formation Experiment
3.7. Calcein-AM/PI Staining Assay
3.8. Cell Cycle Experiment
3.9. Western Blotting Analysis
3.10. Prediction of ADMET
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hafez Ghoran, S.; Taktaz, F.; Sousa, E.; Fernandes, C.; Kijjoa, A. Peptides from Marine-Derived Fungi: Chemistry and Biological Activities. Mar. Drugs 2023, 21, 510. [Google Scholar] [CrossRef] [PubMed]
- Eghtedari, M.; Porzani, S.J.; Nowruzi, B. Anticancer Potential of Natural Peptides from Terrestrial and Marine Environments: A Review. Phytochem. Lett. 2021, 42, 87–103. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide Therapeutics: Current Status and Future Directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef]
- Degenkolb, T.; Brückner, H. Peptaibiomics: Towards a Myriad of Bioactive Peptides Containing Cα-Dialkylamino Acids? Chem. Biodivers. 2008, 5, 1817–1843. [Google Scholar] [CrossRef]
- Castro, G.S.; Sousa, T.F.; da Silva, G.F.; Pedroso, R.C.N.; Menezes, K.S.; Soares, M.A.; Dias, G.M.; Santos, A.O.; Yamagishi, M.E.B.; Faria, J.V.; et al. Characterization of Peptaibols Produced by a Marine Strain of the Fungus Trichoderma Endophyticum via Mass Spectrometry, Genome Mining and Phylogeny-Based Prediction. Metabolites 2023, 13, 221. [Google Scholar] [CrossRef] [PubMed]
- Neumann, N.K.N.; Stoppacher, N.; Zeilinger, S.; Degenkolb, T.; Brückner, H.; Schuhmacher, R. The Peptaibiotics Database—A Comprehensive Online Resource. Chem. Biodivers. 2015, 12, 743–751. [Google Scholar] [CrossRef]
- Song, Y.-P.; Ji, N.-Y. Chemistry and Biology of Marine-Derived Trichoderma Metabolites. Nat. Prod. Bioprospect 2024, 14, 14. [Google Scholar] [CrossRef]
- Zhao, P.; Xue, Y.; Li, X.; Li, J.; Zhao, Z.; Quan, C.; Gao, W.; Zu, X.; Bai, X.; Feng, S. Fungi-Derived Lipopeptide Antibiotics Developed since 2000. Peptides 2019, 113, 52–65. [Google Scholar] [CrossRef]
- Setargie, A.; Wang, C.; Zhang, L.; Xu, Y. Chromatographic and Mass Spectroscopic Guided Discovery of Trichoderma Peptaibiotics and Their Bioactivity. Eng. Microbiol. 2024, 4, 100135. [Google Scholar] [CrossRef]
- Gavryushina, I.A.; Georgieva, M.L.; Kuvarina, A.E.; Sadykova, V.S. Peptaibols as Potential Antifungal and Anticancer Antibiotics: Current and Foreseeable Development (Review). Appl. Biochem. Microbiol. 2021, 57, 556–563. [Google Scholar] [CrossRef]
- Yang, G.; Lin, M.; Kaliaperumal, K.; Lu, Y.; Qi, X.; Jiang, X.; Xu, X.; Gao, C.; Liu, Y.; Luo, X. Recent Advances in Anti-Inflammatory Compounds from Marine Microorganisms. Mar. Drugs 2024, 22, 424. [Google Scholar] [CrossRef] [PubMed]
- Chugh, J.K.; Wallace, B.A. Peptaibols: Models for Ion Channels. Biochem. Soc. Trans. 2001, 29, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Tang, Z.; Gan, Y.; Li, Z.; Luo, X.; Gao, C.; Zhao, L.; Chai, L.; Liu, Y. 18-Residue Peptaibols Produced by the Sponge-Derived Trichoderma Sp. GXIMD 01001. J. Nat. Prod. 2023, 86, 994–1002. [Google Scholar] [CrossRef]
- Singh, V.P.; Yedukondalu, N.; Sharma, V.; Kushwaha, M.; Sharma, R.; Chaubey, A.; Kumar, A.; Singh, D.; Vishwakarma, R.A. Lipovelutibols A–D: Cytotoxic Lipopeptaibols from the Himalayan Cold Habitat Fungus Trichoderma velutinum. J. Nat. Prod. 2018, 81, 219–226. [Google Scholar] [CrossRef]
- Xiao, D.; Zhang, M.; Wu, P.; Li, T.; Li, W.; Zhang, L.; Yue, Q.; Chen, X.; Wei, X.; Xu, Y.; et al. Halovirs I–K, Antibacterial and Cytotoxic Lipopeptaibols from the Plant Pathogenic Fungus Paramyrothecium Roridum NRRL 2183. J. Antibiot. 2022, 75, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, R.; Brückner, H. Marfey’s Reagent for Chiral Amino Acid Analysis: A Review. Amino Acids 2004, 27, 231–247. [Google Scholar] [CrossRef]
- Du, L.; Risinger, A.L.; Mitchell, C.A.; You, J.; Stamps, B.W.; Pan, N.; King, J.B.; Bopassa, J.C.; Judge, S.I.V.; Yang, Z.; et al. Unique Amalgamation of Primary and Secondary Structural Elements Transform Peptaibols into Potent Bioactive Cell-Penetrating Peptides. Proc. Natl. Acad. Sci. USA 2017, 114, E8957–E8966. [Google Scholar] [CrossRef]
- Shi, M.; Wang, H.-N.; Xie, S.-T.; Luo, Y.; Sun, C.-Y.; Chen, X.-L.; Zhang, Y.-Z. Antimicrobial Peptaibols, Novel Suppressors of Tumor Cells, Targeted Calcium-Mediated Apoptosis and Autophagy in Human Hepatocellular Carcinoma Cells. Mol. Cancer 2010, 9, 26. [Google Scholar] [CrossRef]
- Pashirzad, M.; Khorasanian, R.; Fard, M.M.; Arjmand, M.-H.; Langari, H.; Khazaei, M.; Soleimanpour, S.; Rezayi, M.; Ferns, G.A.; Hassanian, S.M.; et al. The Therapeutic Potential of MAPK/ERK Inhibitors in the Treatment of Colorectal Cancer. Curr. Cancer Drug Targets 2021, 21, 932–943. [Google Scholar] [CrossRef]
- Li, Q.Z.; Chen, Y.Y.; Liu, Q.P.; Feng, Z.H.; Zhang, L.; Zhang, H. Cucurbitacin B Suppresses Hepatocellular Carcinoma Progression through Inducing DNA Damage-Dependent Cell Cycle Arrest. Phytomedicine 2024, 126, 155177. [Google Scholar] [CrossRef]
- Xu, C.; Cao, G.; Zhang, H.; Bai, M.; Yi, X.; Qu, X. Avellanin A Has an Antiproliferative Effect on TP-Induced RWPE-1 Cells via the PI3K-Akt Signalling Pathway. Mar. Drugs 2024, 22, 275. [Google Scholar] [CrossRef] [PubMed]
- Degenkolb, T.; Gräfenhan, T.; Berg, A.; Nirenberg, H.I.; Gams, W.; Brückner, H. Peptaibiomics: Screening for Polypeptide Antibiotics (Peptaibiotics) from Plant-Protective Trichoderma Species. Chem. Biodivers. 2006, 3, 593–610. [Google Scholar] [CrossRef] [PubMed]
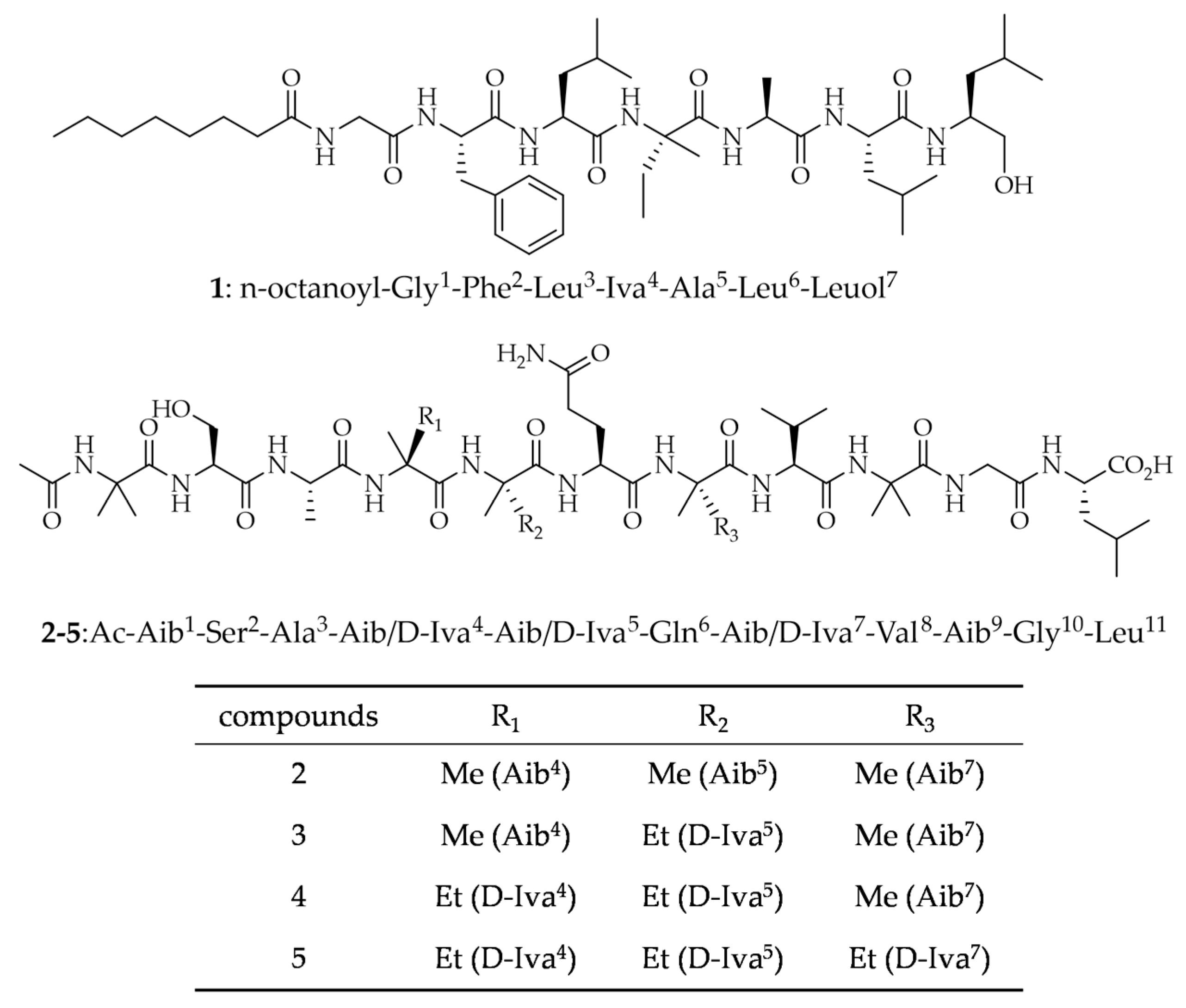
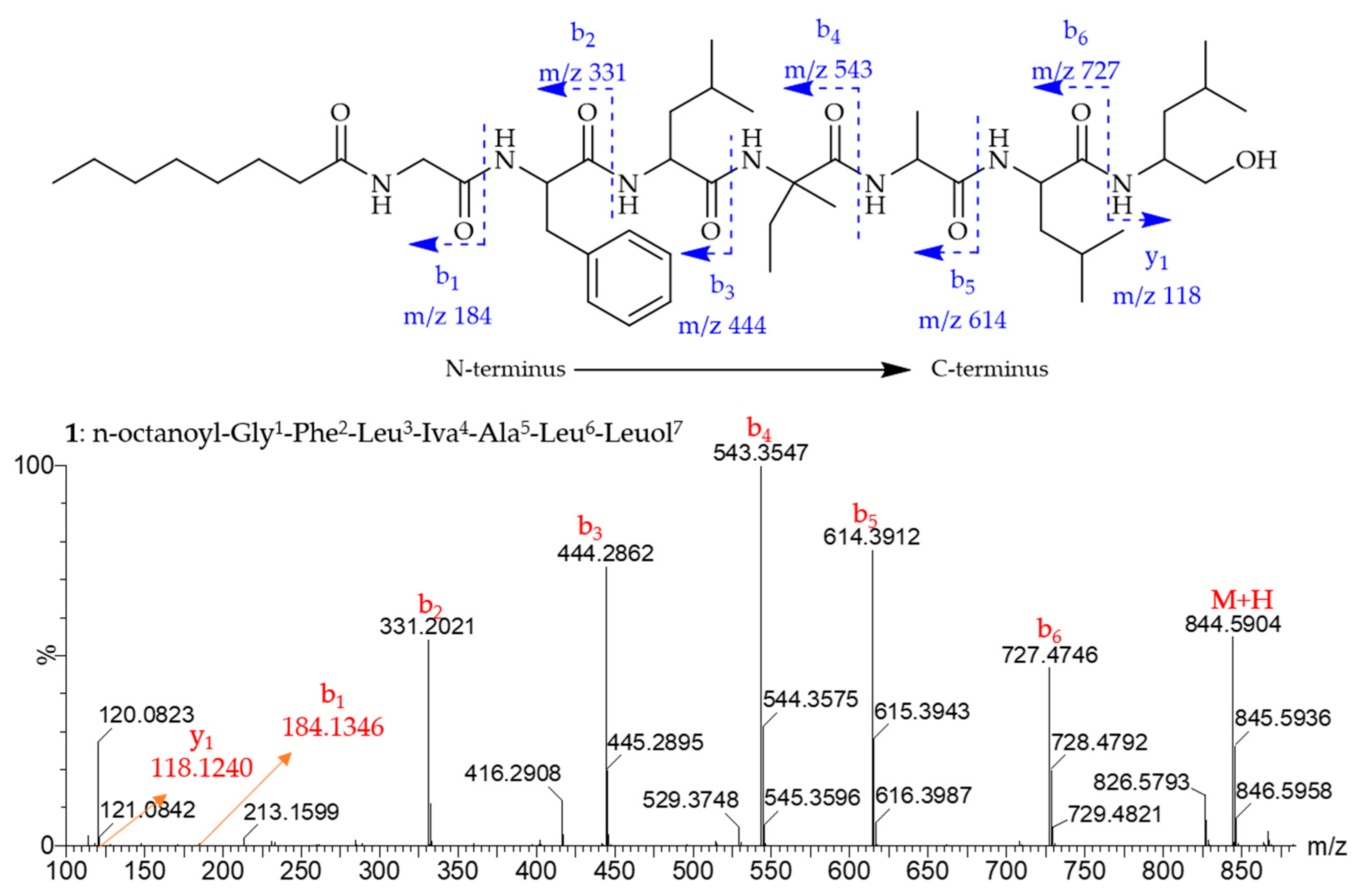

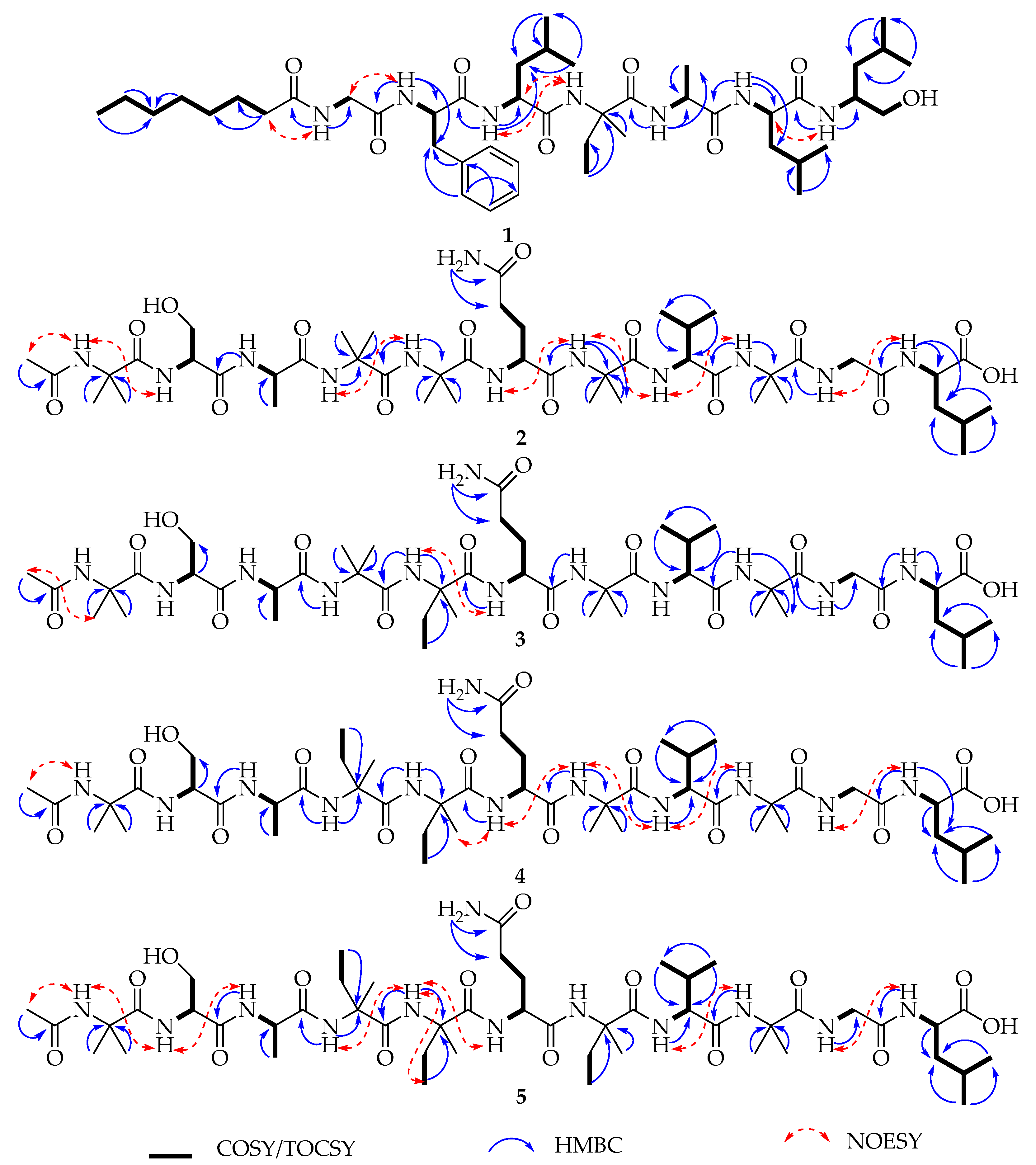

| Pos. | δC, Type | δH, (J in Hz) | Pos. | δC, Type | δH, (J in Hz) | Pos. | δC, Type | δH, (J in Hz) |
|---|---|---|---|---|---|---|---|---|
| n-Oct | 3Leu | 6Leu | ||||||
| 1 | 173.2, C | 1 | 172.9, C | 1 | 171.5, C | |||
| 2 | 35.0, CH2 | 2.10 a, m | 2 | 51.8, CH | 4.07 f, m | 2 | 53.1, CH | 4.06 f, m |
| 3 | 25.0, CH2 | 1.47 b, m | 3a | 40.1, CH2 | 1.33, m | 3a | 39.2, CH2 | 1.61 g m |
| 4 | 28.6, CH2 | 1.23 c, m | 3b | 1.25 c, m | 3b | 1.53, m | ||
| 5 | 28.5, CH2 | 1.23 c, m | 4 | 24.3, CH | 1.63 g, m | 4 | 24.0, CH | 1.56, m |
| 6 | 31.1, CH2 | 1.22 c, m | 5 | 22.7, CH3 | 0.91, d, (6.1) | 5 | 23.2, CH3 | 0.87, d, (6.4) |
| 7 | 22.1, CH2 | 1.23 c, m | 6 | 21.6, CH3 | 0.85, m | 6 | 21.5, CH3 | 0.76, d, (6.5) |
| 8 | 13.9, CH3 | 0.85, m | NH | 7.53, m | NH | 8.05, d, (6.3) | ||
| 1Gly | 4Iva | 7Leuol | ||||||
| 1 | 169.9, C | 1 | 175.1, C | 1 | 48.4, CH | 3.79, m | ||
| 2a | 42.4, CH2 | 3.73, dd, (16.3, 5.6) | 2 | 58.7, C | 2a | 39.9, CH2 | 1.61 g, m | |
| 2b | 3.52, dd, (16.3, 5.4) | 3a | 26.6, CH2 | 2.05 a, m | 2b | 1.53, m | ||
| NH | 8.15, t, (5.6) | 3b | 1.71, m | 3 | 24.1, CH | 1.61 g, m | ||
| 4 | 7.3, CH3 | 0.71, t, (7.5) | 4 | 23.4, CH3 | 0.80, m | |||
| 5 | 22.2, CH3 | 1.28 c, m | 5 | 21.2, CH3 | 0.78, s | |||
| NH | 7.90, s | 6a | 64.1, CH2 | 3.26, m | ||||
| 6b | 3.14, m | |||||||
| NH | 6.88, d, (9.2) | |||||||
| OH | 4.48 e, m | |||||||
| 2Phe | 5Ala | |||||||
| 1 | 172.3 d, C | 1 | 172.3 d, C | |||||
| 2 | 54.5, CH | 4.48 e, m | 2 | 50.0, CH | 3.96, m | |||
| 3a | 54.5, CH2 | 3.06, dd, (14.0, 4.2) | 3 | 17.1, CH3 | 1.27 c, m | |||
| 3b | 2.81, dd, (14.0, 9.7) | NH | 7.52, m | |||||
| 1′ | 137.7, C | |||||||
| 2′,6′ | 129.1, CH | 7.26, m | ||||||
| 3′,5′ | 128.1, CH | 7.24, m | ||||||
| 4′ | 126.4, CH | 7.19, m | ||||||
| NH | 8.22, d, (7.5) |
| Pos. | δC, Type | δH, (J in Hz) | Pos. | δC, Type | δH, (J in Hz) | Pos. | δC, Type | δH, (J in Hz) |
|---|---|---|---|---|---|---|---|---|
| Ac-1Aib | 5Aib | 9Aib | ||||||
| COCH3 | 171.1 a, C | 1 | 174.1, C | 1 | 173.5, C | |||
| COCH3 | 22.8 b, CH3 | 1.90, s | 2 | 55.8 c, C | 2 | 56.1 c, C | ||
| 1 | 175.7, C | 3 | 26.7 d, CH3 | 1.36 h, m | 3 | 23.0 d, CH3 | 1.37 h, m | |
| 2 | 55.8 c, C | 4 | 25.9 e, CH3 | 1.40 h, m | 4 | 25.9 e, CH3 | 1.43 h, m | |
| 3 | 24.0 d, CH3 | 1.30 h, m | NH | 7.55, s | NH | 7.56, s | ||
| 4 | 26.1 e, CH3 | 1.40 h, m | ||||||
| NH | 8.64, br s | |||||||
| 2Ser | 6Gln | 10Gly | ||||||
| 1 | 171.5, C | 1 | 173.3, C | 1 | 168.6, C | |||
| 2 | 57.6 f, C | 4.06, m | 2 | 56.1 c, C | 3.77, m | 2a | 42.5, CH2 | 3.57, m |
| 3a | 60.5, CH2 | 3.68, m | 3 | 26.0, CH2 | 2.00, m | 2b | ||
| 3b | 3.74, m | 4a | 31.4, CH2 | 2.18, m | NH | 7.82, m | ||
| OH | 5.36, m | 4b | 2.28, m | |||||
| NH | 8.18, br s | 5 | 173.5, C | |||||
| 6-NH2a | 7.20, s | |||||||
| 6-NH2b | 6.75, s | |||||||
| NH | 7.90, s | |||||||
| 3Ala | 7Aib | 11Leu | ||||||
| 1 | 174.3, C | 1 | 174.6, C | 1 | 170.9, C | |||
| 2 | 50.7 g, C | 4.05, m | 2 | 56.2 c, C | 2 | 50.8 g, C | 4.04, m | |
| 3 | 16.1, CH3 | 1.30, m | 3 | 23.1 d, CH3 | 1.37 h, m | 3a | 40, CH2 | 1.47, m |
| NH | 7.89, s | 4 | 26.4 e, CH3 | 1.44 h, m | 3b | 1.62, m | ||
| NH | 7.68, s | 4 | 24.1, CH | 1.67, m | ||||
| 5 | 21.5, CH3 | 0.82, d, (6.5) | ||||||
| 6 | 22.8 b, CH3 | 0.87, d, (6.6) | ||||||
| NH | 7.48, d, (7.7) | |||||||
| 4Aib | 8Val | |||||||
| 1 | 174.8, C | 1 | 176.8 a, C | |||||
| 2 | 55.8 c, C | 2 | 57.6 f, C | 4.12, m | ||||
| 3 | 23.3 d, CH3 | 1.35 h, m | 3 | 28.5, CH | 2.27, m | |||
| 4 | 26.1 e, CH3 | 1.43 h, m | 4 | 17.6, CH3 | 0.84, d, (6.8) | |||
| NH | 7.83, s | 5 | 19.4, CH3 | 0.80, d, (6.9) | ||||
| NH | 6.93, d, (9.1) |
| Compounds | Cytotoxicity, IC50 (μM) | ||
|---|---|---|---|
| HT-29 | DLD-1 | SW620 | |
| 1 | 10.3 ± 1.9 | 12.3 ± 1.5 | 20.0 ± 0.6 |
| 2–5 | >40 | >40 | >40 |
| cisplatin | 18.3 ± 0.5 | 14.1 ± 0.4 | 15.2 ± 1.2 |
| Properties | Parameters | Predicted Values |
|---|---|---|
| Absorption | HIA (Human Intestinal Absorption) | 1.0 |
| Distribution | VD (Volume Distribution) | 0.455 L/kg |
| Metabolism | CYP1A2 inhibitor | 0.0 |
| CYP1A2 substrate | 0.0 | |
| CYP2C19 inhibitor | 0.64 | |
| CYP2C19 substrate | 0.0 | |
| CYP2C9 inhibitor | 0.0 | |
| CYP2C9 substrate | 0.0 | |
| CYP2D6 inhibitor | 0.0 | |
| CYP2D6 substrate | 0.0 | |
| Elimination | T 1/2 (Half-Life Time) | 0.486 h |
| CL (Clearance Rate) | 5.44 mL/min/kg | |
| Toxicity | hERG (hERG Blockers) | 0.196 |
| H-HT (Human Hepatotoxicity) | 0.676 | |
| AMES (Ames Mutagenicity) | 0.05 | |
| DILI (Drug-Induced Liver Injury) | 0.06 | |
| ROAT (Rat Oral Acute Toxicity) | 0.113 | |
| Carcinogenicity | 0.011 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Tang, Z.; Luo, X.; Gan, Y.; Bai, M.; Lin, H.; Gao, C.; Chai, L.; Lin, X. Lipotrichaibol A and Trichoderpeptides A–D: Five New Peptaibiotics from a Sponge-Derived Trichoderma sp. GXIMD 01001. Mar. Drugs 2025, 23, 264. https://doi.org/10.3390/md23070264
Yang W, Tang Z, Luo X, Gan Y, Bai M, Lin H, Gao C, Chai L, Lin X. Lipotrichaibol A and Trichoderpeptides A–D: Five New Peptaibiotics from a Sponge-Derived Trichoderma sp. GXIMD 01001. Marine Drugs. 2025; 23(7):264. https://doi.org/10.3390/md23070264
Chicago/Turabian StyleYang, Weichan, Zhenzhou Tang, Xiaowei Luo, Yuman Gan, Meng Bai, Houwen Lin, Chenghai Gao, Ling Chai, and Xiao Lin. 2025. "Lipotrichaibol A and Trichoderpeptides A–D: Five New Peptaibiotics from a Sponge-Derived Trichoderma sp. GXIMD 01001" Marine Drugs 23, no. 7: 264. https://doi.org/10.3390/md23070264
APA StyleYang, W., Tang, Z., Luo, X., Gan, Y., Bai, M., Lin, H., Gao, C., Chai, L., & Lin, X. (2025). Lipotrichaibol A and Trichoderpeptides A–D: Five New Peptaibiotics from a Sponge-Derived Trichoderma sp. GXIMD 01001. Marine Drugs, 23(7), 264. https://doi.org/10.3390/md23070264






