Novel Anti-MRSA Peptide from Mangrove-Derived Virgibacillus chiguensis FN33 Supported by Genomics and Molecular Dynamics
Abstract
1. Introduction
2. Results
2.1. Investigating the Antimicrobial Potential of Bacteria in Mangrove Sediments
2.2. Production Kinetics of Antibacterial Components of FN33 Isolate
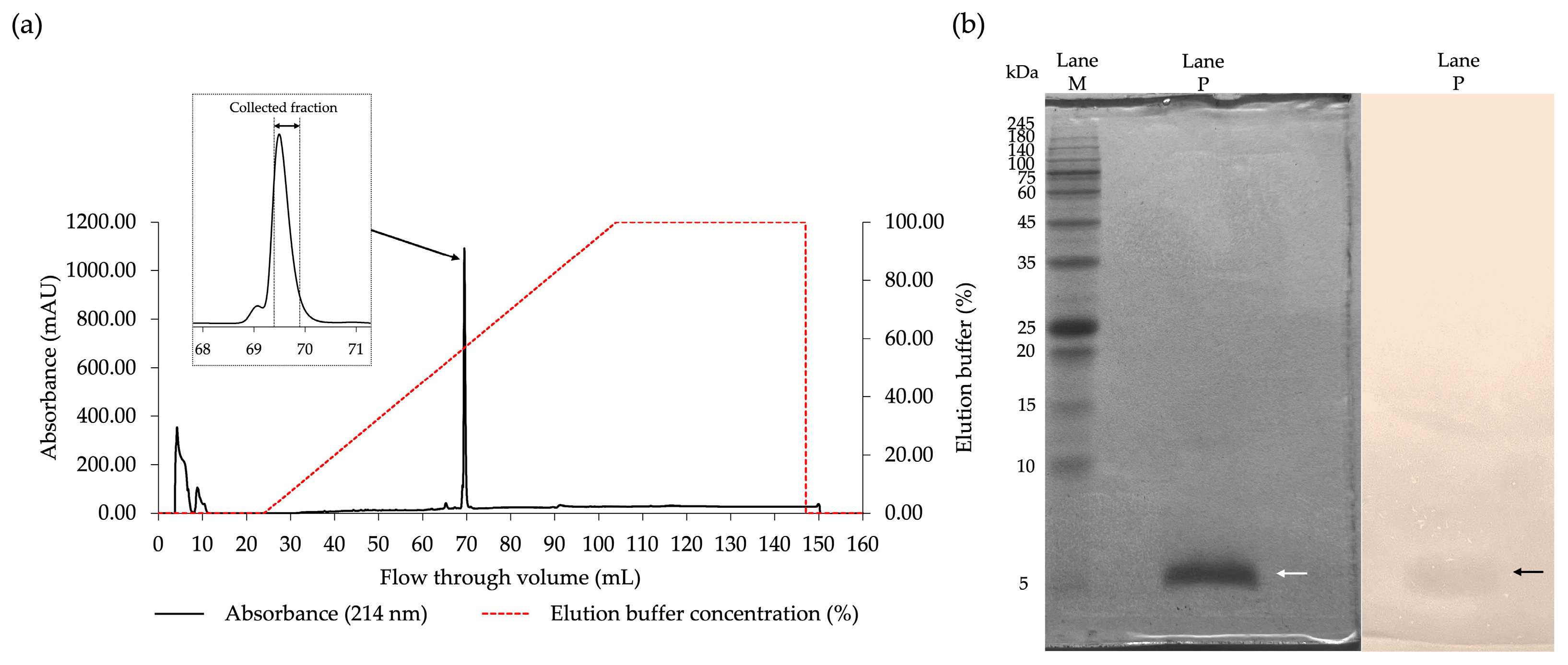
2.3. Purification of the Antibacterial Components of FN33 Isolate
2.4. De Novo Amino Acid Sequencing
2.5. Determination of Antibacterial Activity of FN33 AMP by Microdilution Assay
2.6. Killing Kinetic Studies
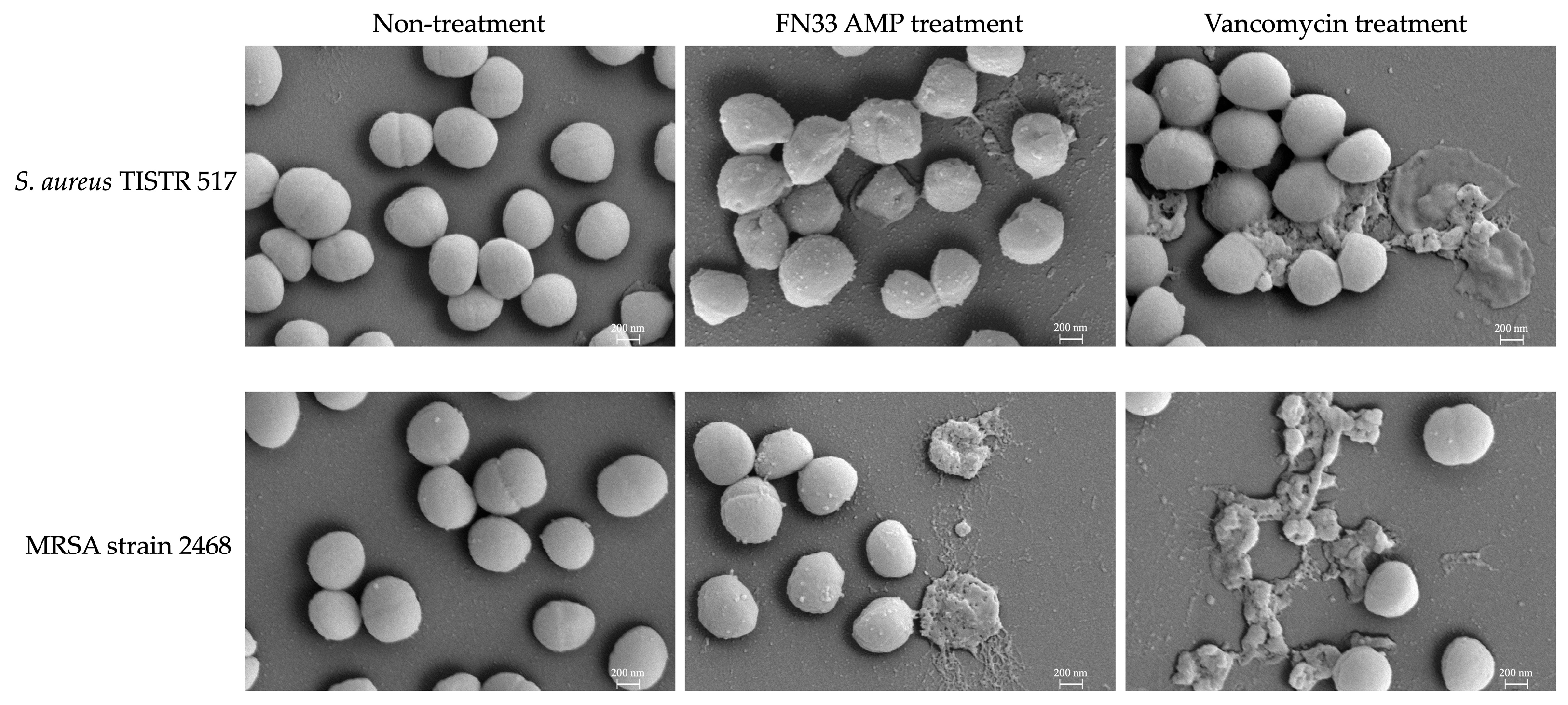
2.7. Investigation of Bacterial Pathogens Treated with FN33 AMP by Scanning Electron Microscopy (SEM)
2.8. Assay for Bacterial Membrane Permeabilization
2.9. Stability Studies of FN33 AMP
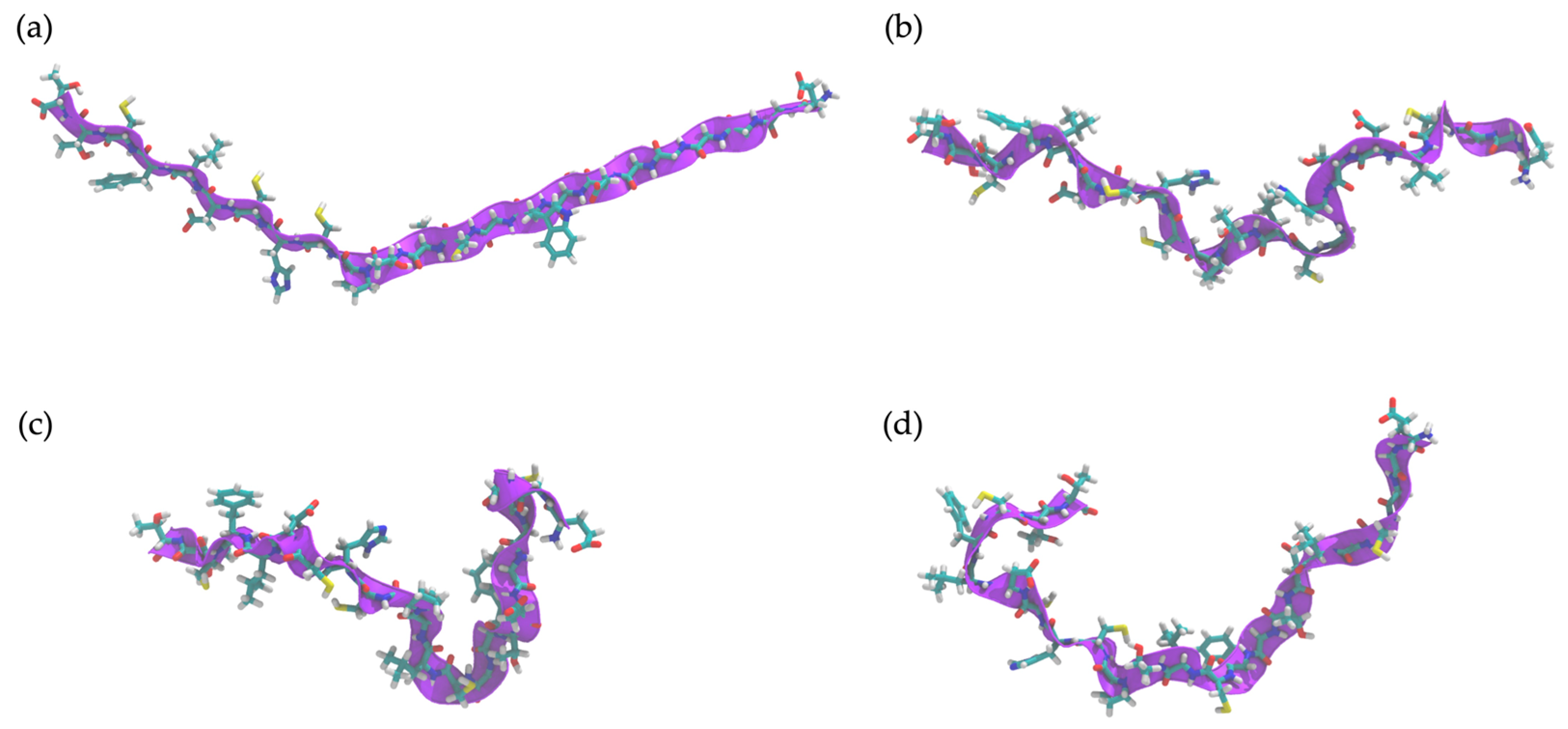
2.10. Determination of Peptide Secondary Structure by Molecular Dynamics (MD) Simulation
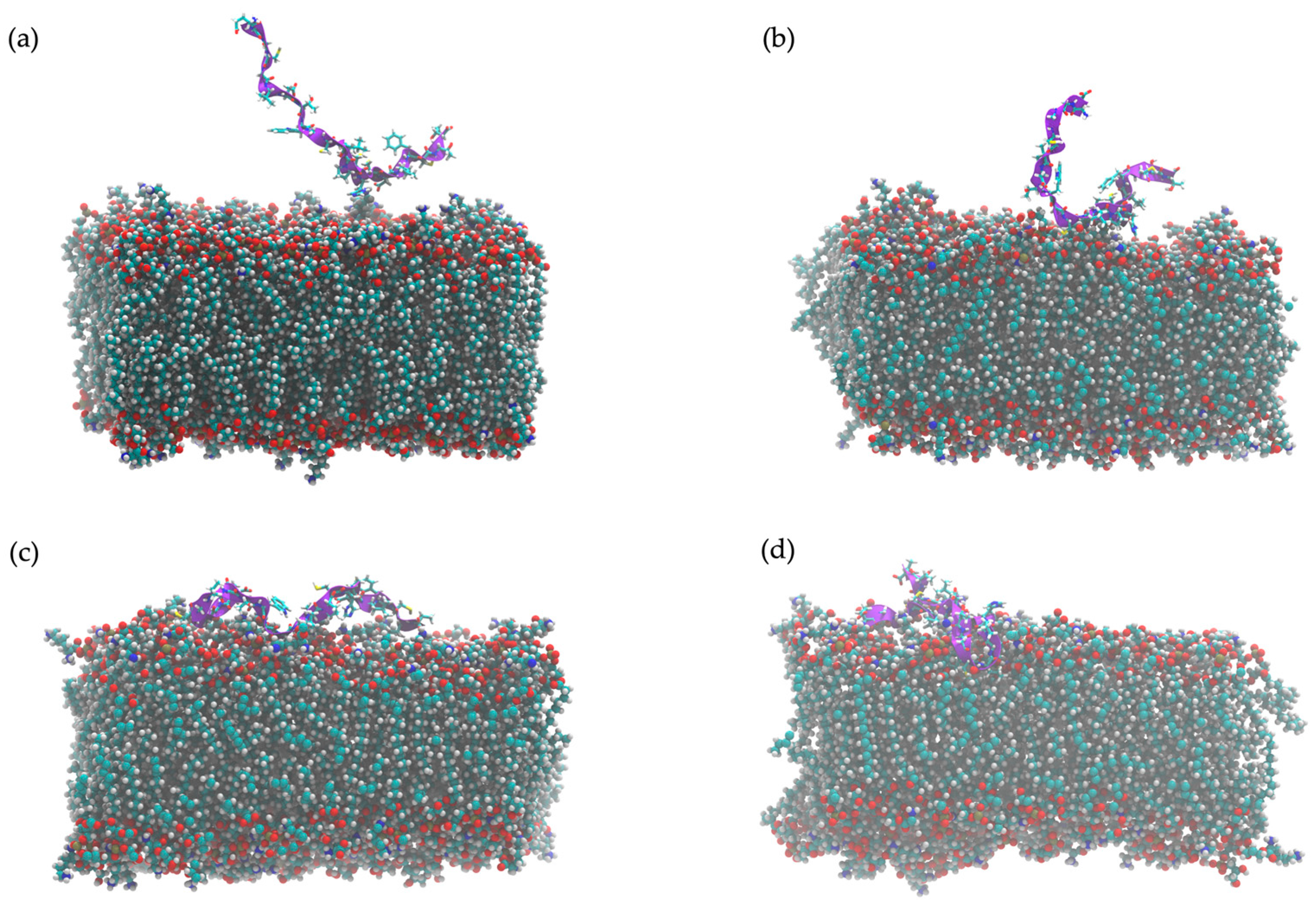
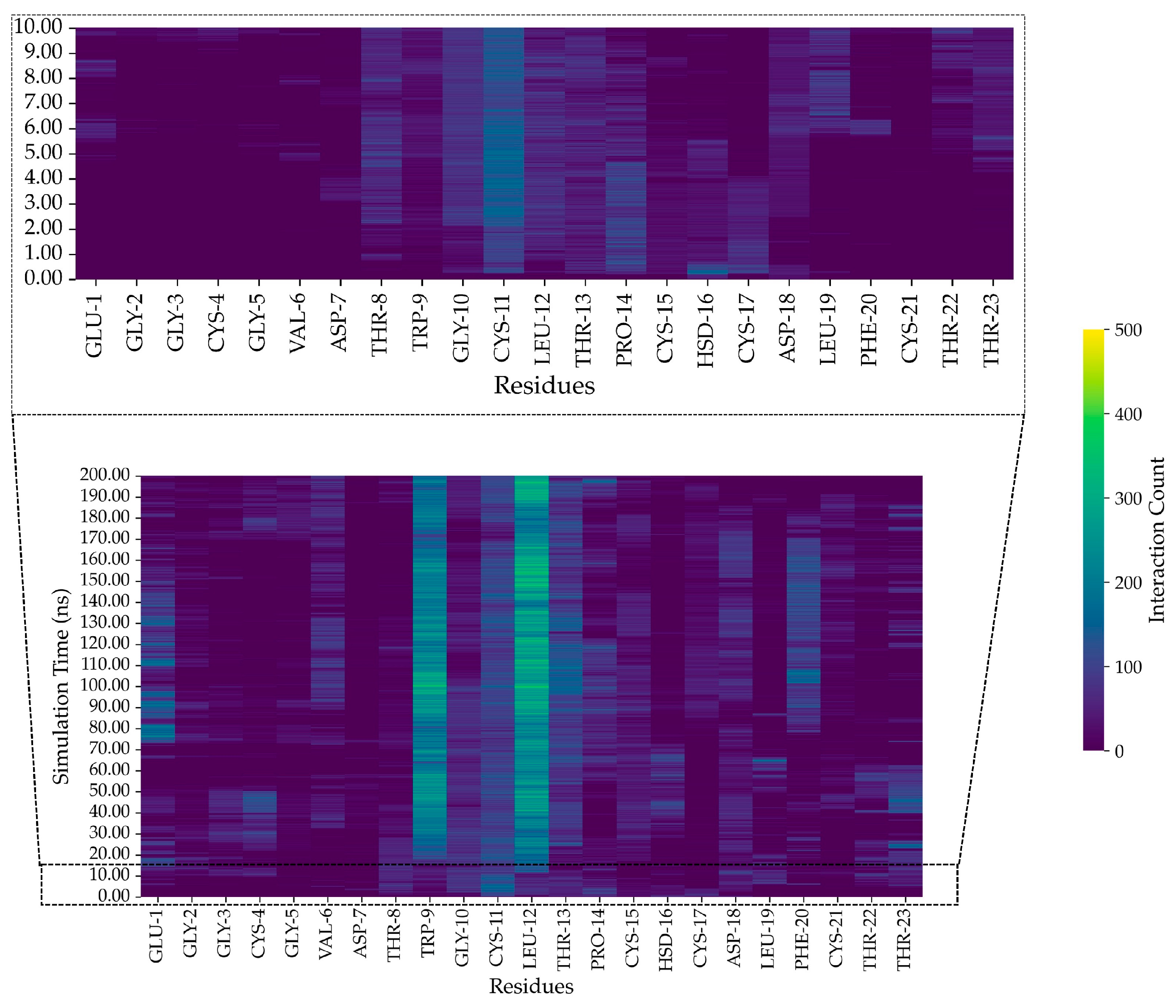
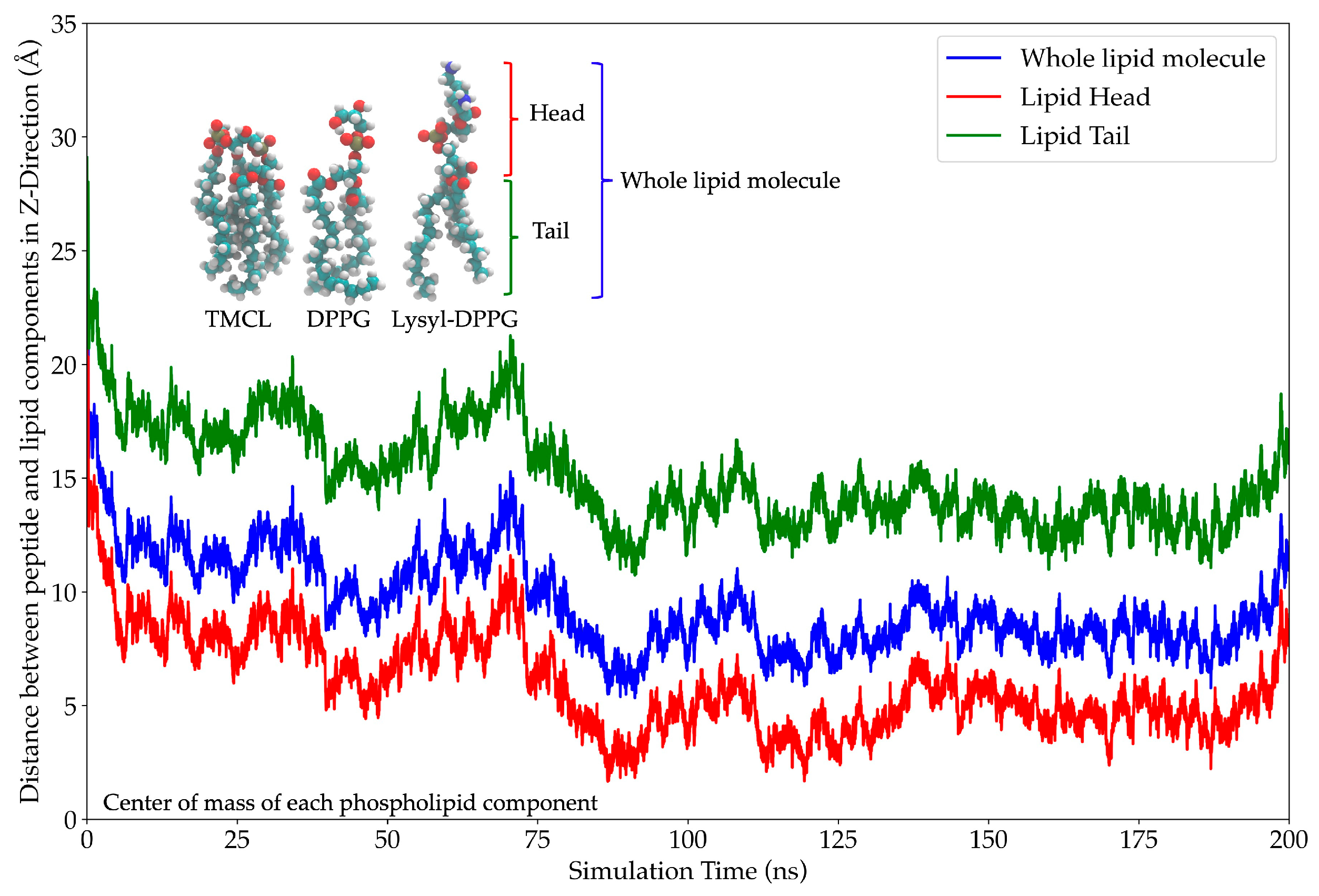
2.11. In Silico Study of AMP Dynamics in a Bacterial Membrane Model
2.12. Effect of AMP on Albumin Denaturation and 50% Inhibitory Concentration (IC50) of DPPH
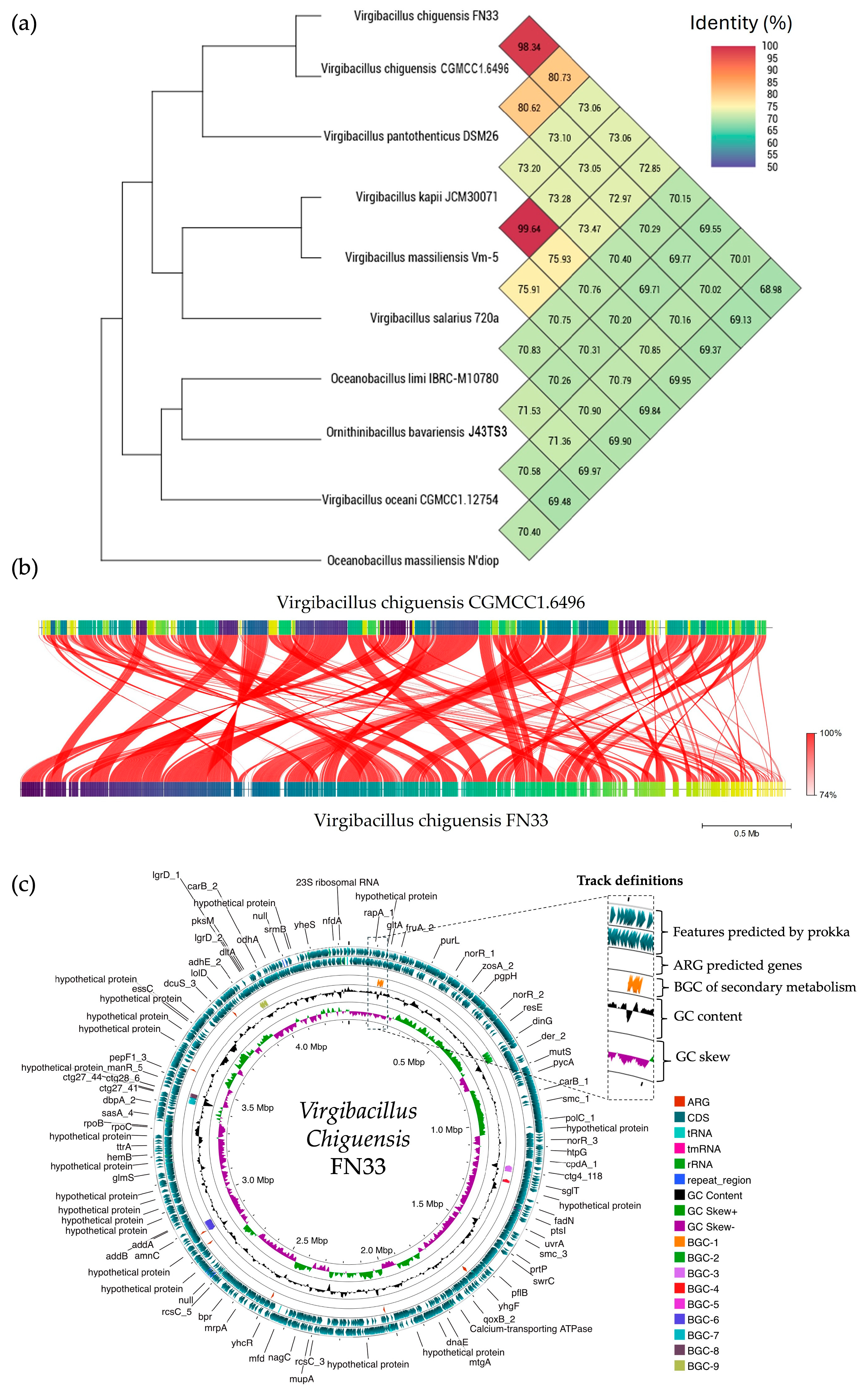
2.13. Sequencing of FN33 Genome
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Bacterial Isolation
4.2. Screening of Antibacterial Activity Using the Soft Agar Overlay Method Against MRSA Strain
4.3. Verification of Antibacterial Activity by Agar Well Diffusion Method
4.4. Production Kinetic Studies of Antimicrobial Compounds of FN33 Isolate
4.5. Purification of the AMP and Amino Acid Sequence Determination
4.6. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of the AMP
4.7. Effect of the AMP on Membrane Permeability
4.8. Stability Analysis of FN33 AMP Under Various Environmental Conditions
4.9. Computational Modeling of the Secondary Structure of FN33 AMP
4.10. In Silico Analysis of FN33 AMP Dynamics in a Bacterial Membrane Model
4.11. In Vitro Anti-Inflammatory Activity of FN33 AMP Assessed Using the Albumin Denaturation Assay
4.12. DPPH Free Radical Scavenging Activity of FN33 AMP
4.13. Bacterial Identification by Whole-Genome Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Antimicrobial Resistance (AMR). Br. J. Biomed. Sci. 2023, 80, 11387. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lou, W.; Liu, J.; Ding, B.; Fan, W.; Hong, J. A novel antimicrobial polymer efficiently treats multidrug-resistant MRSA-induced bloodstream infection. Biosci. Rep. 2019, 39, BSR20192354. [Google Scholar] [CrossRef] [PubMed]
- Arias-Ortiz, P.M.; Calderón, L.D.; Castillo, J.S.; Moreno, J.; Leal, A.L.; Cortés, J.A.; Álvarez, C.A.; Grebo, G. Risk factors for methicillin-resistant Staphylococcus aureus bacteremia: A multicenter matched case-control study. Biomedica 2016, 36, 612–619. [Google Scholar] [CrossRef]
- Al Bshabshe, A.; Joseph, M.R.P.; Awad El-Gied, A.A.; Fadul, A.N.; Chandramoorthy, H.C.; Hamid, M.E. Clinical relevance and antimicrobial profiling of methicillin-resistant Staphylococcus aureus (MRSA) on routine antibiotics and ethanol extract of mango kernel (Mangifera indica L.). Biomed. Res. Int. 2020, 2020, 4150678. [Google Scholar] [CrossRef]
- Karthik, Y.; Ishwara Kalyani, M.; Krishnappa, S.; Devappa, R.; Anjali Goud, C.; Ramakrishna, K.; Wani, M.A.; Alkafafy, M.; Hussen Abduljabbar, M.; Alswat, A.S.; et al. Antiproliferative activity of antimicrobial peptides and bioactive compounds from the mangrove Glutamicibacter mysorens. Front. Microbiol. 2023, 14, 1096826. [Google Scholar] [CrossRef]
- Karthik, Y.; Kalyani, M.I.; Sheetal, K.; Rakshitha, D.; Bineesha, B.K. Cytotoxic and antimicrobial activities of microbial proteins from mangrove soil actinomycetes of Mangalore, Dakshina Kannada. Biomedicine 2020, 40, 59–67. [Google Scholar]
- Chen, Y.H.; Shyu, Y.T.; Lin, S.S. Characterization of candidate genes involved in halotolerance using high-throughput omics in the halotolerant bacterium Virgibacillus chiguensis. PLoS ONE 2018, 13, e0201346. [Google Scholar] [CrossRef]
- Xu, B.; Hu, B.; Wang, J.; Lan, Y.; Zhu, Y.; Dai, X.; Huang, L.; Huang, Y.; Du, W. Virgibacillus indicus sp. nov. and Virgibacillus profundi sp. nov, two moderately halophilic bacteria isolated from marine sediment by using microfluidic streak plates. Int. J. Syst. Evol. Microbiol. 2018, 68, 2015–2023. [Google Scholar] [CrossRef]
- Al-Amoudi, S.; Essack, M.; Simões, M.F.; Bougouffa, S.; Soloviev, I.; Archer, J.A.; Lafi, F.F.; Bajic, V.B. Bioprospecting red sea coastal ecosystems for culturable microorganisms and their antimicrobial potential. Mar. Drugs 2016, 14, 165. [Google Scholar] [CrossRef]
- Nandhagopal, M.; Chandrasekaran, R.; Narayanasamy, M. Antimicrobial potential of secondary metabolites and DNA gyrase B blocking molecules produced by a halophilic bacterium Virgibacillus salarius (MML1918). J. Appl. Microbiol. 2023, 134, lxad286. [Google Scholar] [CrossRef]
- Galaviz-Silva, L.; Iracheta-Villarreal, J.M.; Molina-Garza, Z.J. Bacillus and Virgibacillus strains isolated from three Mexican coasts antagonize Staphylococcus aureus and Vibrio parahaemolyticus. FEMS Microbiol. Lett. 2018, 365, fny202. [Google Scholar] [CrossRef]
- Kristiana, R.; Ayuningrum, D.; Asagabaldan, M.A.; Nuryadi, H.; Sabdono, A.; Radjasa, O.K.; Trianto, A. Isolation and partial characterization of bacteria activity associated with Gorgonian Euplexaura sp. against methicillin-resistant Staphylococcus aureus (MRSA). In Proceedings of the 2nd International Conference on Tropical and Coastal Region Eco Development 2016, Diponegoro University, Kota Semarang, Indonesia, 25–27 October 2016; IOP Conference Series: Earth and Environmental Science. Volume 55, p. 012056. [Google Scholar]
- Rajan, L.; Chakraborty, K.; Chakraborty, R.D. Pharmacological properties of some mangrove sediment-associated bacillus isolates. Arch. Microbiol. 2021, 203, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Roversi, D.; Troiano, C.; Salnikov, E.; Giordano, L.; Riccitelli, F.; De Zotti, M.; Casciaro, B.; Loffredo, M.R.; Park, Y.; Formaggio, F.; et al. Effects of antimicrobial peptides on membrane dynamics: A comparison of fluorescence and NMR experiments. Biophys. Chem. 2023, 300, 107060. [Google Scholar] [CrossRef] [PubMed]
- Pandi, A.; Adam, D.; Zare, A.; Trinh, V.T.; Schaefer, S.L.; Burt, M.; Klabunde, B.; Bobkova, E.; Kushwaha, M.; Foroughijabbari, Y.; et al. Cell-free biosynthesis combined with deep learning accelerates de novo-development of antimicrobial peptides. Nat. Commun. 2023, 14, 7197. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.; Liu, H.; Hui Lee, W.; Zhang, Y. An anionic antimicrobial peptide from toad Bombina maxima. Biochem. Biophys. Res. Commun. 2002, 295, 796–799. [Google Scholar] [CrossRef]
- Li, J.; Hu, S.; Jian, W.; Xie, C.; Yang, X. Plant antimicrobial peptides: Structures, functions, and applications. Bot. Stud. 2021, 62, 5. [Google Scholar] [CrossRef]
- Paulmann, M.; Arnold, T.; Linke, D.; Özdirekcan, S.; Kopp, A.; Gutsmann, T.; Kalbacher, H.; Wanke, I.; Schuenemann, V.J.; Habeck, M.; et al. Structure-activity analysis of the dermcidin-derived peptide DCD-1L, an anionic antimicrobial peptide present in human sweat. J. Biol. Chem. 2012, 287, 8434–8443. [Google Scholar] [CrossRef]
- Dennison, S.R.; Howe, J.; Morton, L.H.; Brandenburg, K.; Harris, F.; Phoenix, D.A. Interactions of an anionic antimicrobial peptide with Staphylococcus aureus membranes. Biochem. Biophys. Res. Commun. 2006, 347, 1006–1010. [Google Scholar] [CrossRef]
- Palmer, N.; Maasch, J.R.M.A.; Torres, M.D.T.; de la Fuente-Nunez, C. Molecular dynamics for antimicrobial peptide discovery. Infect. Immun. 2021, 89, e00703-20. [Google Scholar] [CrossRef]
- Cao, Q.; Ge, C.; Wang, X.; Harvey, P.J.; Zhang, Z.; Ma, Y.; Wang, X.; Jia, X.; Mobli, M.; Craik, D.J.; et al. Designing antimicrobial peptides using deep learning and molecular dynamic simulations. Brief. Bioinform. 2023, 24, bbad058. [Google Scholar] [CrossRef]
- Panayi, T.; Diavoli, S.; Nicolaidou, V.; Papaneophytou, C.; Petrou, C.; Sarigiannis, Y. Short-chained linear scorpion peptides: A Pool for novel antimicrobials. Antibiotics 2024, 13, 422. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.S.; Hassan, F.U.; Rehman, Z.U.; Ishtiaq, I.; Rehman, S.U.; Liu, Q. Molecular characterization of TGF-beta gene family in buffalo to identify gene duplication and functional mutations. Genes 2022, 13, 1302. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, S.M.; Sando, L.; Ireland, D.C.; Colgrave, M.L.; Bharathi, R.; Göransson, U.; Craik, D.J. A continent of plant defense peptide diversity: Cyclotides in Australian Hybanthus (Violaceae). Plant Cell. 2005, 17, 3176–3189. [Google Scholar] [CrossRef] [PubMed]
- Rehal, R.P.; Marbach, H.; Hubbard, A.T.M.; Sacranie, A.A.; Sebastiani, F.; Fragneto, G.; Harvey, R.D. The influence of mild acidity on lysyl-phosphatidylglycerol biosynthesis and lipid membrane physico-chemical properties in methicillin-resistant Staphylococcus aureus. Chem. Phys. Lipids 2017, 206, 60–70. [Google Scholar] [CrossRef]
- Mirnejad, R.; Fasihi-Ramandi, M.; Behmard, E.; Najafi, A.; Moghaddam, M.M. Interaction of antibacterial CM11 peptide with the gram-positive and gram-negative bacterial membrane models: A molecular dynamics simulations study. Chem. Pap. 2023, 77, 3727–3735. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Hernández-Salmerón, J.E.; Moreno-Hagelsieb, G. FastANI, Mash and Dashing equally differentiate between Klebsiella species. PeerJ 2022, 10, e13784. [Google Scholar] [CrossRef]
- Ebu, S.M.; Ray, L.; Panda, A.N.; Gouda, S.K. De novo assembly and comparative genome analysis for polyhydroxyalkanoates-producing Bacillus sp. BNPI-92 strain. J. Genet. Eng. Biotechnol. 2023, 21, 132. [Google Scholar] [CrossRef]
- Guo, Y.; Song, G.; Sun, M.; Wang, J.; Wang, Y. Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2020, 10, 107. [Google Scholar] [CrossRef]
- Hartman, B.J.; Tomasz, A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 1984, 158, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Singh, S.S.; Sidhu, C.; Grover, V.; Pinnaka, A.K.; Korpole, S. Virgicin, a novel lanthipeptide from Virgibacillus sp. strain AK90 exhibits inhibitory activity against Gram-positive bacteria. World J. Microbiol. Biotechnol. 2019, 35, 133. [Google Scholar] [CrossRef]
- Mota-Meira, M.; Lacroix, C.; LaPointe, G.; Lavoie, M.C. Purification and structure of mutacin B-Ny266: A new lantibiotic produced by Streptococcus mutans. FEBS Lett. 1997, 410, 275–279. [Google Scholar] [CrossRef]
- Schnell, N.; Entian, K.D.; Schneider, U.; Götz, F.; Zähner, H.; Kellner, R.; Jung, G. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 1988, 333, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Schnell, N.; Entian, K.D.; Götz, F.; Hörner, T.; Kellner, R.; Jung, G. Structural gene isolation and prepeptide sequence of gallidermin, a new lanthionine containing antibiotic. FEMS Microbiol. Lett. 1989, 49, 263–267. [Google Scholar] [CrossRef][Green Version]
- Sandiford, S.K. Advances in the arsenal of tools available enabling the discovery of novel lantibiotics with therapeutic potential. Expert. Opin. Drug Discov. 2014, 9, 283–297. [Google Scholar] [CrossRef]
- Assoni, L.; Milani, B.; Carvalho, M.R.; Nepomuceno, L.N.; Waz, N.T.; Guerra, M.E.S.; Converso, T.R.; Darrieux, M. Resistance mechanisms to antimicrobial peptides in Gram-positive bacteria. Front. Microbiol. 2020, 11, 593215. [Google Scholar] [CrossRef]
- Charkhian, H.; Soleimannezhadbari, E.; Bodaqlouei, A.; Lotfollahi, L.; Lotfi, H.; Yousefi, N.; Shojadel, E.; Gholinejad, Z. Assessment of bacteriocin production by clinical Pseudomonas aeruginosa isolates and their potential as therapeutic agents. Microb. Cell Fact. 2024, 23, 175. [Google Scholar] [CrossRef]
- Rajasekaran, G.; Kim, E.Y.; Shin, S.Y. LL-37-derived membrane-active FK-13 analogs possessing cell selectivity, anti-biofilm activity and synergy with chloramphenicol and anti-inflammatory activity. Biochim. Biophys. Acta. Biomembr. 2017, 1859, 722–733. [Google Scholar] [CrossRef]
- Dasagrandhi, C.; Pandith, A.; Imran, K. Sclerotiorin: A novel azaphilone with demonstrated membrane targeting and DNA binding activity against methicillin-resistant Staphylococcus aureus. Microbiol. Biotechnol. Lett. 2020, 48, 429–438. [Google Scholar] [CrossRef]
- Yoshida, Y.; Matsuo, M.; Oogai, Y.; Kato, F.; Nakamura, N.; Sugai, M.; Komatsuzawa, H. Bacitracin sensing and resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 2011, 320, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, P.; Mudgil, P. The cell wall, cell membrane and virulence factors of Staphylococcus aureus and their role in antibiotic resistance. Microorganisms 2023, 11, 259. [Google Scholar] [CrossRef] [PubMed]
- Slavetinsky, C.; Kuhn, S.; Peschel, A. Bacterial aminoacyl phospholipids—Biosynthesis and role in basic cellular processes and pathogenicity. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids. 2017, 1862, 1310–1318. [Google Scholar] [CrossRef]
- Nikolic, P.; Mudgil, P.; Harman, D.G.; Whitehall, J. Untargeted lipidomic differences between clinical strains of methicillin-sensitive and methicillin-resistant Staphylococcus aureus. Infect Dis. 2022, 54, 497–507. [Google Scholar] [CrossRef]
- Keil, B. Specificity of Proteolysis, 1st ed.; Springer: Berlin/Heidelberg, Germany, 1992; pp. 43–227. [Google Scholar]
- Liu, K.; Yang, L.; Peng, X.; Gong, H.; Wang, J.; Lu, J.R.; Xu, H. Effects of conventional surfactants on the activity of designed antimicrobial peptide. Langmuir 2020, 36, 3531–3539. [Google Scholar] [CrossRef]
- Żamojć, K.; Wyrzykowski, D.; Chmurzyński, L. On the effect of pH, temperature, and surfactant structure on bovine serum albumin-cationic/anionic/nonionic surfactants interactions in cacodylate buffer-fluorescence quenching studies supported by UV spectrophotometry and CD spectroscopy. Int. J. Mol. Sci. 2021, 23, 41. [Google Scholar] [CrossRef]
- Green, R.J.; Su, T.J.; Joy, H.; Lu, J.R. Interaction of lysozyme and sodium dodecyl sulfate at the air−liquid interface. Langmuir 2000, 16, 5797–5805. [Google Scholar] [CrossRef]
- Matamoros-Recio, A.; Franco-Gonzalez, J.F.; Forgione, R.E.; Torres-Mozas, A.; Silipo, A.; Martín-Santamaría, S. Understanding the antibacterial resistance: Computational explorations in bacterial membranes. ACS Omega 2021, 6, 6041–6054. [Google Scholar] [CrossRef]
- Gagat, P.; Ostrówka, M.; Duda-Madej, A.; Mackiewicz, P. Enhancing antimicrobial peptide activity through modifications of charge, hydrophobicity, and structure. Int. J. Mol. Sci. 2024, 25, 10821. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Yan, Z.B.; Meng, Y.M.; Hong, X.Y.; Shao, G.; Ma, J.J.; Cheng, X.R.; Liu, J.; Kang, J.; Fu, C.Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Dennison, S.R.; Morton, L.H.; Harris, F.; Phoenix, D.A. Low pH enhances the action of maximin H5 against Staphylococcus aureus and helps mediate lysylated phosphatidylglycerol-induced resistance. Biochemistry 2016, 55, 3735–3751. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Wang, Y.; Han, Y.; Zhou, H.; Huang, Z.; Zhang, X.; Zhou, C.; Cao, J.; Zhou, T. Octominin: An antimicrobial peptide with antibacterial and anti-inflammatory activity against carbapenem-resistant Escherichia coli both in vitro and in vivo. J. Glob. Antimicrob. Resist. 2023, 35, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, L.; He, D.; Liu, Q.; Han, Q.; Zhang, J.; Zhang, A.M.; Song, Y. Exploration of the antibacterial and anti-inflammatory activity of a novel antimicrobial peptide brevinin-1BW. Molecules 2024, 29, 1534. [Google Scholar] [CrossRef]
- Kashfi, R.; Kelsey, C.; Gang, D.J.; Call, D.R.; Gang, D.R. Metabolomic diversity and identification of antibacterial activities of bacteria isolated from marine sediments in Hawai’i and Puerto Rico. Front. Mol. Biosci. 2020, 7, 23. [Google Scholar] [CrossRef]
- Hockett, K.L.; Baltrus, D.A. Use of the soft-agar overlay technique to screen for bacterially produced inhibitory compounds. J. Vis. Exp. 2017, 119, 55064. [Google Scholar]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Regmi, S.; Choi, Y.S.; Choi, Y.H.; Kim, Y.K.; Cho, S.S.; Yoo, J.C.; Suh, J.W. Antimicrobial peptide from Bacillus subtilis CSB138: Characterization, killing kinetics, and synergistic potency. Int. Microbiol. 2017, 20, 43–53. [Google Scholar]
- Sermkaew, N.; Atipairin, A.; Krobthong, S.; Aonbangkhen, C.; Yingchutrakul, Y.; Uchiyama, J.; Songnaka, N. Unveiling a new antimicrobial peptide with efficacy against P. aeruginosa and K. pneumoniae from mangrove-derived Paenibacillus thiaminolyticus NNS5-6 and genomic analysis. Antibiotics 2024, 13, 846. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the Expasy Server. In The Proteomics Protocols Handbook, 1st ed.; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Shi, G.; Kang, X.; Dong, F.; Liu, Y.; Zhu, N.; Hu, Y.; Xu, H.; Lao, X.; Zheng, H. DRAMP 3.0: An enhanced comprehensive data repository of antimicrobial peptides. Nucleic Acids Res. 2022, 50, D488–D496. [Google Scholar] [CrossRef]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bairoch, A. UniProtKB/Swiss-Prot. Methods Mol. Biol. 2007, 406, 89–112. [Google Scholar] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; pp. 15–52. [Google Scholar]
- Songnaka, N.; Lertcanawanichakul, M.; Hutapea, A.M.; Krobthong, S.; Yingchutrakul, Y.; Atipairin, A. Purification and characterization of novel anti-MRSA peptides produced by Brevibacillus sp. SPR-20. Molecules 2022, 27, 8452. [Google Scholar] [CrossRef]
- Xu, Y.; Shen, J.; Luo, X.; Zhu, W.; Chen, K.; Ma, J.; Jiang, H. Conformational transition of amyloid β-peptide. Proc. Natl. Acad. Sci. USA 2005, 102, 5403–5407. [Google Scholar] [CrossRef]
- Free Download: BIOVIA Discovery Studio Visualize. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 12 December 2024).
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Ribeiro, J.; JS, B.R.; Phillips, J. VMD psfgen Plugin, Version 2.0. 2020. Available online: https://www.ks.uiuc.edu/Research/vmd/plugins/psfgen/ (accessed on 20 December 2024).
- Stephani, J.C.; Gerhards, L.; Khairalla, B.; Solov’yov, I.A.; Brand, I. How do antimicrobial peptides interact with the outer membrane of Gram-negative bacteria? Role of lipopolysaccharides in peptide binding, anchoring, and penetration. ACS Infect. Dis. 2024, 10, 763–778. [Google Scholar] [CrossRef]
- Isralewitz, B. Timeline: A VMD plugin for trajectory analysis, Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign, NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute Computational Biophysics Workshop. 2011. Available online: https://www.ks.uiuc.edu/Training/Tutorials/science/timeline/tutorial_timeline.pdf (accessed on 28 August 2023).
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef]
- Maleš, M.; Zoranić, L. Simulation study of the effect of antimicrobial peptide associations on the mechanism of action with bacterial and eukaryotic membranes. Membranes 2022, 12, 891. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.Y.; Lee, H. All-atom simulations and free-energy calculations of coiled-coil peptides with lipid bilayers: Binding strength, structural transition, and effect on lipid dynamics. Sci. Rep. 2016, 6, 22299. [Google Scholar] [CrossRef] [PubMed]
- Fogolari, F.; Corazza, A.; Toppo, S.; Tosatto, S.C.; Viglino, P.; Ursini, F.; Esposito, G. Studying interactions by molecular dynamics simulations at high concentration. J. Biomed. Biotechnol. 2012, 2012, 303190. [Google Scholar] [CrossRef] [PubMed]
- Gowers, R.J.; Linke, M.; Barnoud, J.; Reddy, T.J.E.; Melo, M.N.; Seyler, S.L.; Dot-son, D.L.; Domanski, J.; Buchoux, S.; Kenney, I.M.; et al. MDAnalysis: A Python package for the rapid analysis of molecular dynamics simulations. In Proceedings of the 15th Python in Science Conference, Austin, TX, USA, 11–17 July 2016; Benthall, S., Rostrup, S., Eds.; pp. 98–105. [Google Scholar]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef]
- Chandra, S.; Chatterjee, P.; Dey, P.; Bhattacharya, S. Evaluation of in vitro anti-inflammatory activity of coffee against the denaturation of protein. Asian Pac. J. Trop. Biomed. 2012, 2, S178–S180. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C.L.W.T. Use of a free radical method to evaluate antioxidant activity. LWT—Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Tripathi, N.; Sapra, A. Gram Staining. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2020; [Updated 14 August 2023]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK562156/ (accessed on 28 August 2023).
- Hussey, M.A.; Zayaitz, A. Endospore stain protocol. Am. Soc. Microbiol. 2007, 8, 1–11. [Google Scholar]
- Galaxy Community. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- Babraham Bioinformatics: FastQC A Quality Control tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 23 July 2023).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Tseemann/Shovill: Faster SPAdes Assembly of Illumina Reads. Available online: https://github.com/tseemann/shovill (accessed on 23 July 2023).
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2209. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Terlouw, B.R.; Blin, K.; Navarro-Muñoz, J.C.; Avalon, N.E.; Chevrette, M.G.; Egbert, S.; Lee, S.; Meijer, D.; Recchia, M.J.J.; Reitz, Z.L.; et al. MIBiG 3.0: A community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 2023, 51, D603–D610. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]





| Tested Antimicrobial Agents | Zone of Inhibition (mm ± SD; n = 3) | ||||||
|---|---|---|---|---|---|---|---|
| S. aureus TISTR 517 | MRSA Strain 142 | MRSA Strain 1096 | MRSA Strain 2468 | E. coli TISTR 887 | K. pneumoniae TISTR 1383 | P. aeruginosa TISTR 357 | |
| CFS of FN33 culture | 12.41 ± 1.20 | 17.47 ± 1.03 | 17.05 ± 1.83 | 17.98 ± 2.07 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Vancomycin (30 µg) | 23.37 ± 1.27 | 24.13 ± 1.27 | 24.38 ± 0.51 | 24.64 ± 1.02 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Cefoxitin (30 µg) | 35.87 ± 0.46 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 35.12 ± 1.04 | 19.67 ± 0.88 | 0.00 ± 0.00 |
| Purification Procedures | Total Volume (mL) | Total Dried Weight (mg) | Arbitrary Activity (AU/mL) | Total Activity (AU) | Specific Activity (AU/mg) | Purification Factor (Folds) | Yield (%) |
|---|---|---|---|---|---|---|---|
| CFS of FN33 culture | 976.86 | 1873.92 | 20 | 19,537.20 | 10.43 | 1.00 | 100.00 |
| Protein precipitate of 50% ammonium sulfate saturation | 45.24 | 86.25 | 80 | 3619.20 | 41.96 | 4.02 | 18.52 |
| Active fraction of cation-exchange chromatography | 17.38 | 11.06 | 80 | 1390.40 | 125.71 | 12.06 | 7.12 |
| Active fraction of reversed-phase chromatography | 3.22 | 0.84 | 320 | 1030.40 | 1226.67 | 117.66 | 5.27 |
| Tested Samples | S. aureus TISTR 517 | MRSA Strain 142 | MRSA Strain 1096 | MRSA Strain 2468 | |
|---|---|---|---|---|---|
| FN33 AMP | MIC (μg/mL) | 16 | 8 | 8 | 8 |
| MBC (μg/mL) | 64 | 16 | 16 | 16 | |
| Vancomycin | MIC (μg/mL) | 2 | 2 | 2 | 2 |
| MBC (μg/mL) | 2 | 2 | 2 | 2 | |
| Cefoxitin | MIC (μg/mL) | 2 | >64 | >64 | >64 |
| MBC (μg/mL) | 2 | >64 | >64 | >64 | |
| Treatment Conditions | Antibacterial Activity Retention of FN33 AMP (%) Against MRSA Strain 2468 (Mean ± SD; n = 3) | ||
|---|---|---|---|
| Control | 1 h | 6 h | 12 h |
| 100.00 ± 0.84 | 100.00 ± 0.76 | 100.00 ± 0.42 | |
| Effect of temperature variation | 1 h | 6 h | 12 h |
| 30 °C | 100.00 ± 0.66 | 100.00 ± 0.17 | 99.79 ± 0.47 |
| 40 °C | 100.08 ± 0.48 | 100.02 ± 0.35 | 100.23 ± 0.28 |
| 60 °C | 98.58 ± 1.74 | 93.53 ± 1.27 *,† | 86.98 ± 0.79 *,† |
| 80 °C | 95.17 ± 0.43 * | 90.91 ± 0.26 *,† | 82.46 ± 0.55 *,† |
| 100 °C | 90.83 ± 0.67 * | 83.38 ± 0.83 *,† | 79.97 ± 0.06 *,† |
| Effect of autoclave condition | 15 min | 30 min | |
| 121 °C and 15 psi | 72.82 ± 0.53 * | 65.87 ± 0.82 *,† | |
| Effect of protease digestion | 1 h | 6 h | 12 h |
| FN33 AMP with proteinase K (1 mg/mL) | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * |
| FN33 AMP with trypsin (1 mg/mL) | 99.60 ± 0.6 | 98.82 ± 0.67 | 99.04 ± 1.02 |
| FN33 AMP with α-chymotrypsin (1 mg/mL) | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * |
| Effect of surfactant | 1 h | 6 h | 12 h |
| FN33 AMP with CTAB (1% w/v) | 93.19 ± 0.52 * | 93.17 ± 0.48 * | 92.51 ± 0.69 *,† |
| FN33 AMP with SDS (1% w/v) | 117.08 ± 0.78 * | 117.39 ± 0.96 * | 115.95 ± 0.57 * |
| FN33 AMP with Triton X-100 (1% w/v) | 81.23 ± 1.04 * | 81.69 ± 0.38 * | 81.30 ± 0.72 * |
| CTAB (1% w/v) alone | 98.62 ± 0.69 | 98.89 ± 1.10 | 98.42 ± 0.72 |
| SDS (1% w/v) alone | 110.10 ± 0.84 * | 109.30 ± 0.56 * | 108.93 ± 0.57 * |
| Triton X-100 (1% w/v) alone | 0.00 ± 0.00 * | 0.00 ± 0.00 * | 0.00 ± 0.00 * |
| Effect of pH variation | 1 h | 6 h | 12 h |
| pH 1 | 74.56 ± 0.69 * | 76.51 ± 0.39 * | 72.85 ± 0.22 *,† |
| pH 4 | 97.42 ± 0.65 * | 96.19 ± 0.21 * | 95.30 ± 0.69 *,† |
| pH 8 | 99.53 ± 0.41 | 98.03 ± 0.65 | 98.76 ± 0.81 |
| pH 10 | 74.63 ± 0.79 * | 73.12 ± 1.04 *,† | 69.46 ± 0.77 *,† |
| pH 14 | 68.58 ± 0.45 * | 66.80 ± 0.27 *,† | 65.80 ± 0.84 *,† |
| Group | Concentration (µg/mL) | % Inhibition of Albumin Denaturation |
|---|---|---|
| FN33 AMP | 50 | 4.85 ± 0.93 *,† |
| 100 | 17.75 ± 0.97 *,† | |
| 250 | 34.39 ± 1.82 * | |
| 500 | 57.16 ± 1.06 † | |
| Diclofenac sodium (the concentration was equivalent to diclofenac) | 50 | 3.09 ± 0.44 *,† |
| 100 | 12.24 ± 1.50 *,† | |
| 250 | 25.31 ± 0.75 *,† | |
| 500 | 34.85 ± 0.81 *,† |
| Group | IC50 (µg/mL) |
|---|---|
| FN33 AMP | 11.66 ± 2.87 |
| Ascorbic acid | 6.78 ± 1.58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sermkaew, N.; Atipairin, A.; Boonruamkaew, P.; Krobthong, S.; Aonbangkhen, C.; Uchiyama, J.; Yingchutrakul, Y.; Songnaka, N. Novel Anti-MRSA Peptide from Mangrove-Derived Virgibacillus chiguensis FN33 Supported by Genomics and Molecular Dynamics. Mar. Drugs 2025, 23, 209. https://doi.org/10.3390/md23050209
Sermkaew N, Atipairin A, Boonruamkaew P, Krobthong S, Aonbangkhen C, Uchiyama J, Yingchutrakul Y, Songnaka N. Novel Anti-MRSA Peptide from Mangrove-Derived Virgibacillus chiguensis FN33 Supported by Genomics and Molecular Dynamics. Marine Drugs. 2025; 23(5):209. https://doi.org/10.3390/md23050209
Chicago/Turabian StyleSermkaew, Namfa, Apichart Atipairin, Phetcharat Boonruamkaew, Sucheewin Krobthong, Chanat Aonbangkhen, Jumpei Uchiyama, Yodying Yingchutrakul, and Nuttapon Songnaka. 2025. "Novel Anti-MRSA Peptide from Mangrove-Derived Virgibacillus chiguensis FN33 Supported by Genomics and Molecular Dynamics" Marine Drugs 23, no. 5: 209. https://doi.org/10.3390/md23050209
APA StyleSermkaew, N., Atipairin, A., Boonruamkaew, P., Krobthong, S., Aonbangkhen, C., Uchiyama, J., Yingchutrakul, Y., & Songnaka, N. (2025). Novel Anti-MRSA Peptide from Mangrove-Derived Virgibacillus chiguensis FN33 Supported by Genomics and Molecular Dynamics. Marine Drugs, 23(5), 209. https://doi.org/10.3390/md23050209










