Whole-Genome Sequence Analysis, Probiotic Potential, and Safety Assessment of the Marine Bacterium Paraliobacillus zengyii CGMCC1.16464
Abstract
1. Introduction
2. Results
2.1. Genome Characteristics and Homology Analysis of P. zengyii
2.2. Gene Annotation for P. zengyii
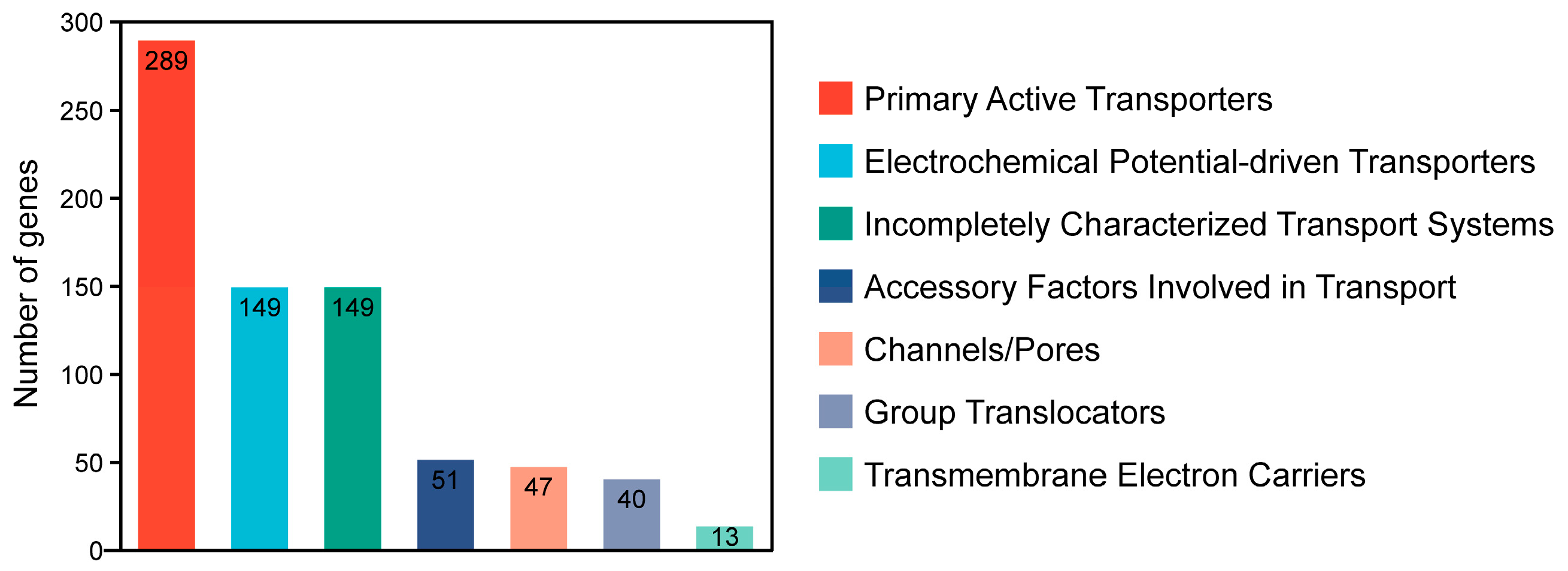
2.2.1. TCDB Annotation
2.2.2. CAZyme Database Annotation
2.2.3. antiSMASH Database Annotation
2.2.4. VFDB Annotation
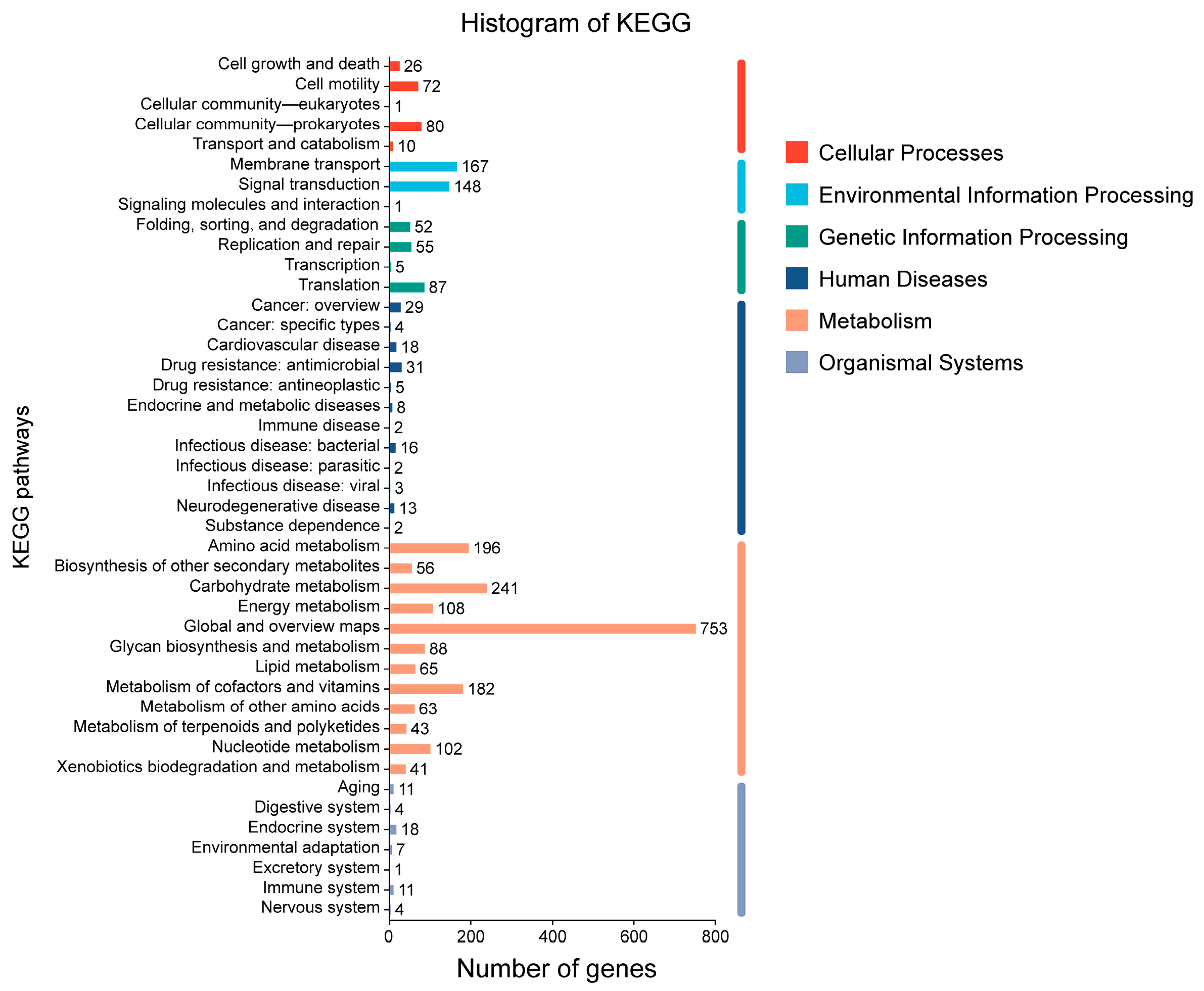
2.2.5. KEGG Database Annotation
2.2.6. GO Database Annotation
2.3. Antibiotic Sensitivity Analysis and Plasmid Detection Analysis of P. zengyii
2.4. Hemolytic Activity and Gelatinase Activity of P. zengyii
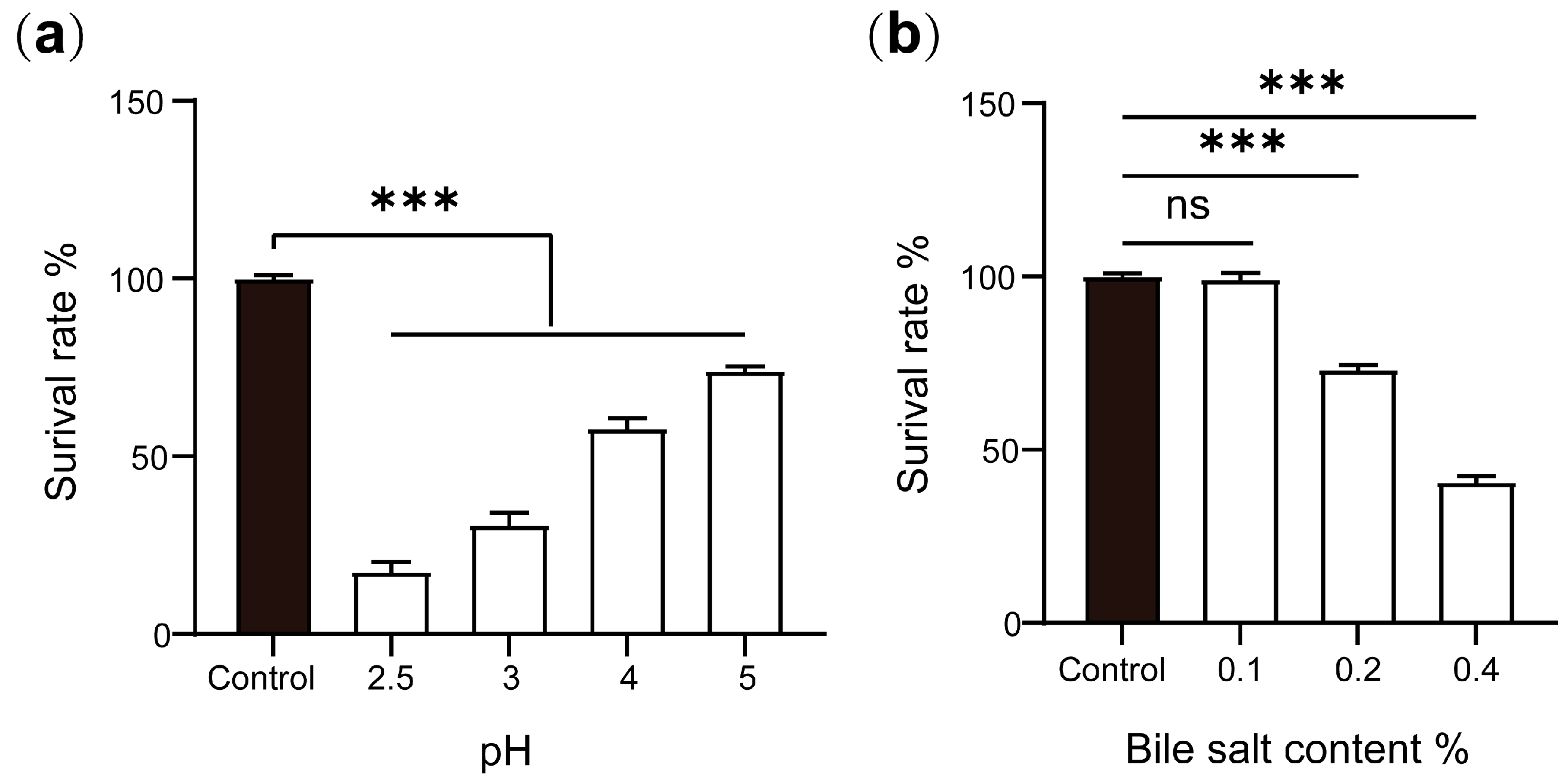
2.5. Acid and Bile Salt Tolerances of P. zengyii
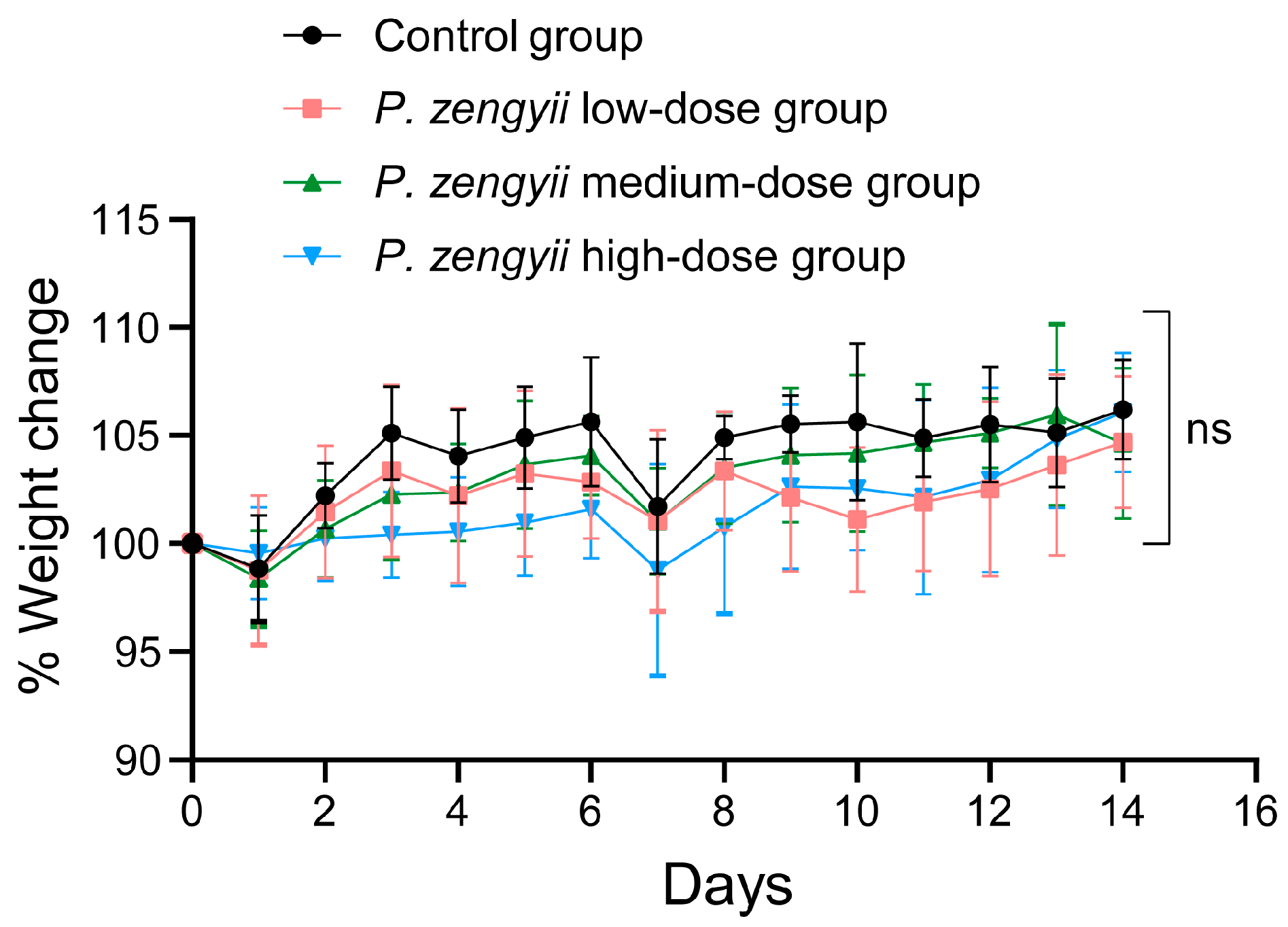
2.6. In Vivo Safety Evaluation of P. zengyii
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Genomic Features and Phylogenetic Analysis
4.3. Genome Functional Annotation
4.4. Antibiotic Sensitivity Testing
4.5. Plasmid Extraction and Detection
4.6. Hemolysis Test
4.7. Gelatinase Activity Test
4.8. Acid Resistance Test
4.9. Bile Salt Tolerance Test
4.10. Animal Experiments
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ishikawa, M.; Ishizaki, S.; Yamamoto, Y.; Yamasato, K. Paraliobacillus ryukyuensis gen. nov., sp. nov., a new Gram-positive, slightly halophilic, extremely halotolerant, facultative anaerobe isolated from a decomposing marine alga. J. Gen. Appl. Microbiol. 2002, 48, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Bock, P.M.; Martins, A.F.; Schaan, B.D. Understanding how pre- and probiotics affect the gut microbiome and metabolic health. Am. J. Physiol. Endocrinol. Metab. 2024, 327, E89–E102. [Google Scholar] [CrossRef]
- Gao, Y.; Yao, Q.; Meng, L.; Wang, J.; Zheng, N. Double-side role of short chain fatty acids on host health via the gut-organ axes. Anim. Nutr. 2024, 18, 322–339. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.J.; Li, B.J.; Lu, S.M.; Yue, K.; Luo, X.L.; Xu, J.G. Study on the Anti⁃Influenza Virus of Marine Probiotic Paraliobacillus zengyii CGMCC1.16464. Bingduxuebao 2025, 41, 374–382. [Google Scholar] [CrossRef]
- Hua, Z.; Liu, S.; Yang, G.; Hou, X.; Fang, Y. Next-generation probiotics: Innovations in safety assessments. Curr. Opin. Food Sci. 2025, 61, 101238. [Google Scholar] [CrossRef]
- Chen, T.; Zhao, Y.; Fan, Y.; Dong, Y.; Gai, Z. Genome sequence and evaluation of safety and probiotic potential of Lacticaseibacillus paracasei LC86 and Lacticaseibacillus casei LC89. Front. Microbiol. 2024, 15, 1501502. [Google Scholar] [CrossRef]
- Liu, A.; Liu, X.; Lu, Y.; Gao, Z.; Tang, R.; Huang, Y.; Zheng, L.; Fan, Z.; He, M. Two chronically misdiagnosed patients infected with Nocardia cyriacigeorgica accurately diagnosed by whole genome resequencing. Front. Cell. Infect. Microbiol. 2022, 12, 1032669. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, S.; Li, H.; Zhu, J.; Liu, Z.; Hu, X.; Yi, J. Assessments of Probiotic Potentials of Lactiplantibacillus plantarum Strains Isolated From Chinese Traditional Fermented Food: Phenotypic and Genomic Analysis. Front. Microbiol. 2022, 13, 895132. [Google Scholar] [CrossRef]
- Kim, J.; Jo, J.; Cho, S.; Kim, H. Genomic insights and functional evaluation of Lacticaseibacillus paracasei EG005: A promising probiotic with enhanced antioxidant activity. Front. Microbiol. 2024, 15, 1477152. [Google Scholar] [CrossRef]
- Joseph, L.C.; Shi, J.; Nguyen, Q.N.; Pensiero, V.; Goulbourne, C.; Bauer, R.C.; Zhang, H.; Morrow, J.P. Combined metabolomic and transcriptomic profiling approaches reveal the cardiac response to high-fat diet. iScience 2022, 25, 104184. [Google Scholar] [CrossRef]
- Ortiz de Choudens, S.; Sparapani, R.; Narayanan, J.; Lohr, N.; Gao, F.; Fish, B.L.; Zielonka, M.; Gasperetti, T.; Veley, D.; Beyer, A.; et al. Lisinopril Mitigates Radiation-Induced Mitochondrial Defects in Rat Heart and Blood Cells. Front. Oncol. 2022, 12, 828177. [Google Scholar] [CrossRef]
- Liou, G.G.; Hsieh, C.C.; Lee, Y.J.; Li, P.H.; Tsai, M.S.; Li, C.T.; Wang, S.H. N-Acetyl Cysteine Overdose Inducing Hepatic Steatosis and Systemic Inflammation in Both Propacetamol-Induced Hepatotoxic and Normal Mice. Antioxidants 2021, 10, 442. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, Z.; Dai, X.; Zhang, D.; Zeng, Y.; Ni, X.; Pan, K. Evaluation of probiotic properties of a Brevibacillus laterosporus strain. Faseb J. 2024, 38, e23530. [Google Scholar] [CrossRef]
- Lu, H.; Zhao, W.; Liu, W.H.; Sun, T.; Lou, H.; Wei, T.; Hung, W.L.; Chen, Q. Safety Evaluation of Bifidobacterium lactis BL-99 and Lacticaseibacillus paracasei K56 and ET-22 in vitro and in vivo. Front. Microbiol. 2021, 12, 686541. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, J.; Lu, S.; Lai, X.H.; Jin, D.; Pu, J.; Niu, L.; Zhu, W.; Liang, J.; Huang, Y.; et al. Paraliobacillus zengyii sp. nov., a slightly halophilic and extremely halotolerant bacterium isolated from Tibetan antelope faeces. Int. J. Syst. Evol. Microbiol. 2019, 69, 1426–1432. [Google Scholar] [CrossRef]
- Cao, W.R.; Guo, L.Y.; Du, Z.J.; Das, A.; Saren, G.; Jiang, M.Y.; Dunlap, C.A.; Rooney, A.P.; Yu, X.K.; Li, T.G. Corrigendum: Paraliobacillus sediminis sp. nov., isolated from East China sea sediment. Int. J. Syst. Evol. Microbiol. 2017, 67, 3135. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Han, S.; Zhang, K.; Xu, G.; Zhang, H.; Chen, F.; Wang, L.; Liu, Q.; Guo, Z.; Zhang, J.; et al. Genome Analysis and Safety Assessment of Achromobacter marplatensis Strain YKS2 Strain Isolated from the Rumen of Yaks in China. Probiotics Antimicrob. Proteins 2024, 16, 1638–1656. [Google Scholar] [CrossRef]
- Nakajima, Y.; Ishibashi, J.; Yukuhiro, F.; Asaoka, A.; Taylor, D.; Yamakawa, M. Antibacterial activity and mechanism of action of tick defensin against Gram-positive bacteria. Biochim. Biophys. Acta 2003, 1624, 125–130. [Google Scholar] [CrossRef]
- Beecher, D.J.; Schoeni, J.L.; Wong, A.C. Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect. Immun. 1995, 63, 4423–4428. [Google Scholar] [CrossRef]
- Gao, X.; Li, C.; He, R.; Zhang, Y.; Wang, B.; Zhang, Z.-H.; Ho, C.-T. Research advances on biogenic amines in traditional fermented foods: Emphasis on formation mechanism, detection and control methods. Food Chem. 2023, 405, 134911. [Google Scholar] [CrossRef]
- Abarquero, D.; Bodelón, R.; Flórez, A.B.; Fresno, J.M.; Renes, E.; Mayo, B.; Tornadijo, M.E. Technological and safety assessment of selected lactic acid bacteria for cheese starter cultures design: Enzymatic and antimicrobial activity, antibiotic resistance and biogenic amine production. LWT 2023, 180, 114709. [Google Scholar] [CrossRef]
- Deng, L.; Liu, L.; Fu, T.; Li, C.; Jin, N.; Zhang, H.; Li, C.; Liu, Y.; Zhao, C. Genome Sequence and Evaluation of Safety and Probiotic Potential of Lactiplantibacillus plantarum LPJZ-658. Microorganisms 2023, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Yang, S.M.; Kim, D.; Kim, H.Y. Complete Genome Sequencing and Comparative Genomics of Three Potential Probiotic Strains, Lacticaseibacillus casei FBL6, Lacticaseibacillus chiayiensis FBL7, and Lacticaseibacillus zeae FBL8. Front. Microbiol. 2021, 12, 794315. [Google Scholar] [CrossRef]
- Kandasamy, S.; Yoo, J.; Yun, J.; Lee, K.-H.; Kang, H.-B.; Kim, J.-E.; Oh, M.-H.; Ham, J.-S. Probiogenomic In-Silico Analysis and Safety Assessment of Lactiplantibacillus plantarum DJF10 Strain Isolated from Korean Raw Milk. Int. J. Mol. Sci. 2022, 23, 14494. [Google Scholar] [CrossRef]
- Hussein, M.; Kang, Z.; Neville, S.L.; Allobawi, R.; Thrombare, V.; Koh, A.J.J.; Wilksch, J.; Crawford, S.; Mohammed, M.K.; McDevitt, C.A.; et al. Metabolic profiling unveils enhanced antibacterial synergy of polymyxin B and teixobactin against multi-drug resistant Acinetobacter baumannii. Sci. Rep. 2024, 14, 27145. [Google Scholar] [CrossRef]
- Sun, Z.; Palzkill, T. Deep Mutational Scanning Reveals the Active-Site Sequence Requirements for the Colistin Antibiotic Resistance Enzyme MCR-1. mBio 2021, 12, e0277621. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Cheah, S.E.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Velkov, T.; Du, Z.; Johnson, M.D.; Li, J. Transcriptomic Analysis of the Activity of a Novel Polymyxin against Staphylococcus aureus. mSphere 2016, 1, e00119-16. [Google Scholar] [CrossRef] [PubMed]
- Materon, I.C.; Palzkill, T. Structural biology of MCR-1-mediated resistance to polymyxin antibiotics. Curr. Opin. Struct. Biol. 2023, 82, 102647. [Google Scholar] [CrossRef]
- Kintses, B.; Méhi, O.; Ari, E.; Számel, M.; Györkei, Á.; Jangir, P.K.; Nagy, I.; Pál, F.; Fekete, G.; Tengölics, R.; et al. Phylogenetic barriers to horizontal transfer of antimicrobial peptide resistance genes in the human gut microbiota. Nat. Microbiol. 2019, 4, 447–458. [Google Scholar] [CrossRef]
- Oliveira, F.S.; da Silva Rodrigues, R.; de Carvalho, A.F.; Nero, L.A. Genomic Analyses of Pediococcus pentosaceus ST65ACC, a Bacteriocinogenic Strain Isolated from Artisanal Raw-Milk Cheese. Probiotics Antimicrob. Proteins 2023, 15, 630–645. [Google Scholar] [CrossRef]
- Zaghloul, E.H.; Halfawy, N.M.E. Marine Pediococcus pentosaceus E3 Probiotic Properties, Whole-Genome Sequence Analysis, and Safety Assessment. Probiotics Antimicrob. Proteins 2024, 16, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Nowak, A.; Zakłos-Szyda, M.; Rosicka-Kaczmarek, J.; Motyl, I. Anticancer Potential of Post-Fermentation Media and Cell Extracts of Probiotic Strains: An In Vitro Study. Cancers 2022, 14, 1853. [Google Scholar] [CrossRef] [PubMed]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Dove, N.C.; Taş, N.; Hart, S.C. Ecological and genomic responses of soil microbiomes to high-severity wildfire: Linking community assembly to functional potential. ISME J. 2022, 16, 1853–1863. [Google Scholar] [CrossRef]
- Milke, L.; Kabuu, M.; Zschoche, R.; Gätgens, J.; Krumbach, K.; Carlstedt, K.-L.; Wurzbacher, C.E.; Balluff, S.; Beemelmanns, C.; Jogler, C.; et al. A type III polyketide synthase cluster in the phylum Planctomycetota is involved in alkylresorcinol biosynthesis. Appl. Microbiol. Biotechnol. 2024, 108, 239. [Google Scholar] [CrossRef]
- Xu, B.; Wang, L.; Yang, C.; Yan, R.; Zhang, P.; Jin, M.; Du, H.; Wang, Y. Specifically targeted antimicrobial peptides synergize with bacterial-entrapping peptide against systemic MRSA infections. J. Adv. Res. 2025, 67, 301–315. [Google Scholar] [CrossRef]
- Al Musaimi, O. Lasso peptides realm: Insights and applications. Peptides 2024, 182, 171317. [Google Scholar] [CrossRef]
- Duan, Y.; Niu, W.; Pang, L.; Bian, X.; Zhang, Y.; Zhong, G. Unusual Post-Translational Modifications in the Biosynthesis of Lasso Peptides. Int. J. Mol. Sci. 2022, 23, 7231. [Google Scholar] [CrossRef]
- Imai, M.; Colas, K.; Suga, H. Protein Grafting Techniques: From Peptide Epitopes to Lasso-Grafted Neobiologics. Chempluschem 2024, 89, e202400152. [Google Scholar] [CrossRef]
- Gómez Del Pulgar, E.M.; Benítez-Páez, A.; Sanz, Y. Safety Assessment of Bacteroides Uniformis CECT 7771, a Symbiont of the Gut Microbiota in Infants. Nutrients 2020, 12, 551. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, X.; Hao, Y.; Ding, J.; Shen, J.; Xue, Z.; Qi, W.; Li, Z.; Song, Y.; Zhang, T.; et al. Protective effects of a novel probiotic strain, Lactococcus lactis ML2018, in colitis: In vivo and in vitro evidence. Food Funct. 2019, 10, 1132–1145. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.; Gao, H.; He, L.; Liu, Y.; Fan, H.; Xu, Q.; Yang, S. Ginsenoside Rb1 is an immune-stimulatory agent with antiviral activity against enterovirus 71. J. Ethnopharmacol. 2021, 266, 113401. [Google Scholar] [CrossRef] [PubMed]





| Database | TCDB | CAZyme | antiSMASH | VFDB | KEGG | GO |
|---|---|---|---|---|---|---|
| Number of genes | 738 | 113 | 5 | 413 | 2618 | 458 |
| Cluster ID | Type | Start-End | Similar Cluster | Similarity (%) |
|---|---|---|---|---|
| Cluster 1 | terpene | 294,564–314,354 | - | - |
| Cluster 2 | ectoine | 315,647–326,034 | ectoine | 75 |
| Cluster 3 | terpene | 1,809,113–1,829,932 | - | - |
| Cluster 4 | T3PKS | 1,961,117–2,002,290 | - | - |
| Cluster 5 | lasso peptide | 3,062,131–3,084,506 | paeninodin | 50 |
| Gene ID | VFDB ID | VF | VF Category | Related Gene | Identity (%) |
|---|---|---|---|---|---|
| Gene 0139 | VFG000079 (gb|NP_463763) | ClpC (VF0072) | Stress survival | ClpC | 75.9 |
| Gene 0167 | VFG046465 (gb|WP_003028672) | EF-Tu (VF0460) | Adherence | TufA | 75.6 |
| Gene 0501 | VFG012095 (gb|WP_003435012) | GroEL (VF0594) | Adherence | GroEL | 70.6 |
| Gene 2841 | VFG000077 (gb|NP_465991) | ClpP (VF0074) | Stress survival | ClpP | 73.6 |
| Gene 3151 | VFG001301 (gb|WP_000459062) | Capsule (VF0003) | Immune modulation | Cap8E | 71.8 |
| Antibiotic Class | Antibiotic | MIC (μg/mL) | Sensitivity 1 |
|---|---|---|---|
| tetracyclines | tetracycline | 0.0940 | S |
| lincomycin | clindamycin | 0.050 | S |
| polypeptide antibiotics | polymyxin | 6 | R |
| macrolides | erythromycin | 0.016 | S |
| glycylcyclines | tigecycline | 0.023 | S |
| aminoglycosides | amikacin | 16 | S |
| sulfonamides | cotrimoxazole | 0.019 | S |
| macrolides | azithromycin | 0.38 | S |
| chloramphenicol | chloramphenicol | 3 | S |
| cephalosporins | ceftriaxone | 0.016 | S |
| quinolones | ciprofloxacin | 0.064 | S |
| rifamycin | rifampicin | 0.016 | S |
| streptogramins | quinupristin–dalfopristin | 0.75 | S |
| Group | Heart (%) | Liver (%) | Spleen (%) | Lung (%) | Kidney (%) |
|---|---|---|---|---|---|
| Control group | 0.52 ± 0.06 | 3.66 ± 0.35 | 0.25 ± 0.05 | 0.65 ± 0.08 | 0.9 ± 0.18 |
| P. zengyii low-dose group | 0.54 ± 0.05 | 3.64 ± 0.14 | 0.29 ± 0.04 | 0.62 ± 0.09 | 0.92 ± 0.06 |
| P. zengyii medium-dose group | 0.55 ± 0.04 | 3.53 ± 0.17 | 0.29 ± 0.1 | 0.65 ± 0.05 | 0.86 ± 0.09 |
| P. zengyii high-dose group | 0.57 ± 0.04 | 3.57 ± 0.1 | 0.24 ± 0.03 | 0.63 ± 0.15 | 0.87 ± 0.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Q.; Jiao, M.; Huangfu, H.; Chen, L.; Li, B.; Cao, Z.; Luo, X.; Xu, J. Whole-Genome Sequence Analysis, Probiotic Potential, and Safety Assessment of the Marine Bacterium Paraliobacillus zengyii CGMCC1.16464. Mar. Drugs 2025, 23, 202. https://doi.org/10.3390/md23050202
Fan Q, Jiao M, Huangfu H, Chen L, Li B, Cao Z, Luo X, Xu J. Whole-Genome Sequence Analysis, Probiotic Potential, and Safety Assessment of the Marine Bacterium Paraliobacillus zengyii CGMCC1.16464. Marine Drugs. 2025; 23(5):202. https://doi.org/10.3390/md23050202
Chicago/Turabian StyleFan, Qianjin, Mengqi Jiao, Haoyue Huangfu, Lan Chen, Beijie Li, Zhijie Cao, Xuelian Luo, and Jianguo Xu. 2025. "Whole-Genome Sequence Analysis, Probiotic Potential, and Safety Assessment of the Marine Bacterium Paraliobacillus zengyii CGMCC1.16464" Marine Drugs 23, no. 5: 202. https://doi.org/10.3390/md23050202
APA StyleFan, Q., Jiao, M., Huangfu, H., Chen, L., Li, B., Cao, Z., Luo, X., & Xu, J. (2025). Whole-Genome Sequence Analysis, Probiotic Potential, and Safety Assessment of the Marine Bacterium Paraliobacillus zengyii CGMCC1.16464. Marine Drugs, 23(5), 202. https://doi.org/10.3390/md23050202






