Characterization of the VOC Promoter That Is Active Under Low-Salinity Conditions in the Diatom Phaeodactylum tricornutum
Abstract
:1. Introduction
2. Results and Discussion
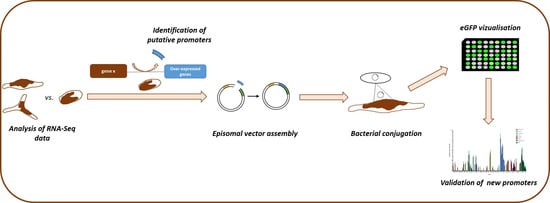
2.1. Identification and Cloning of Potential Promoter Sequences
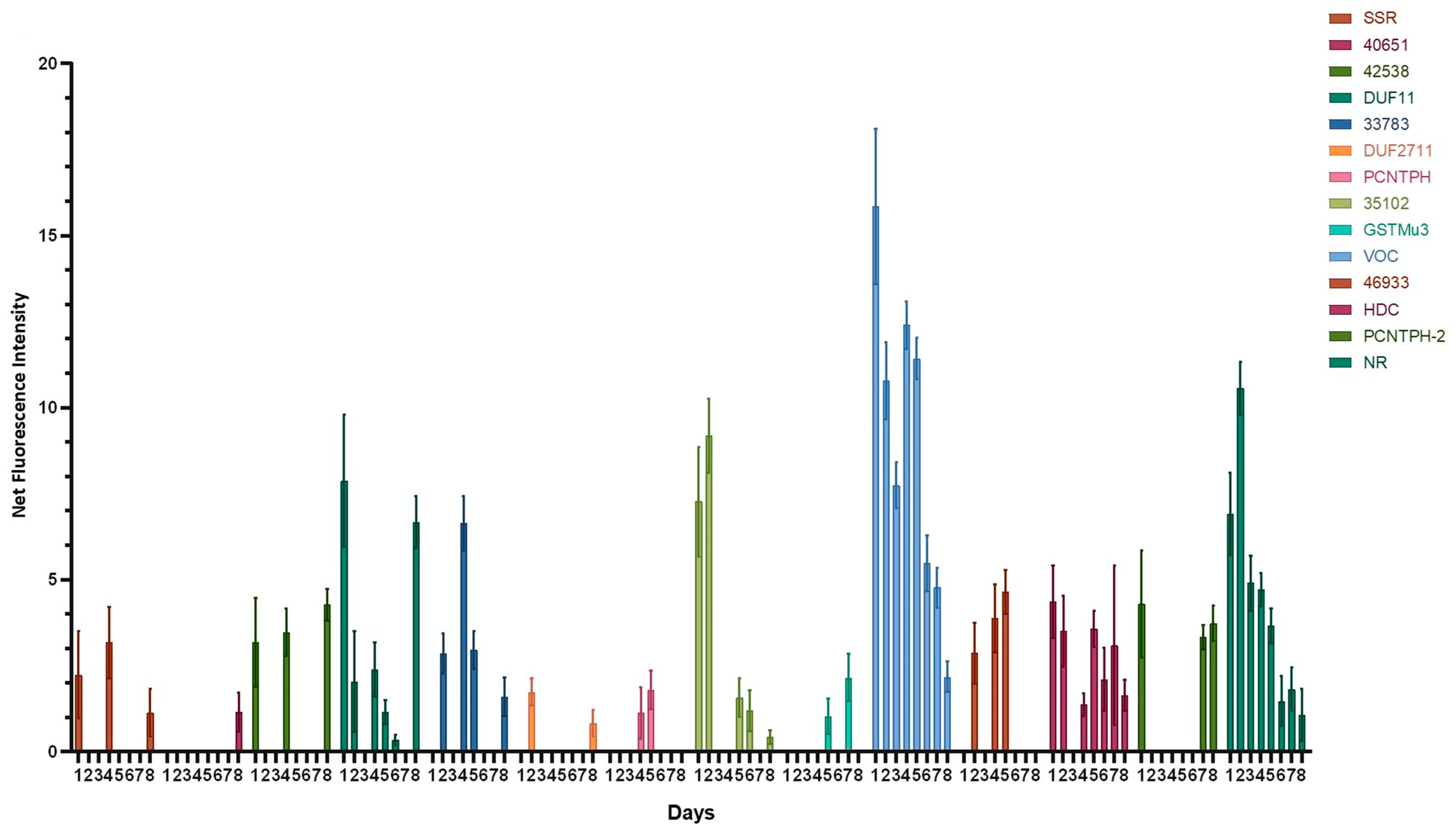
2.2. Assessment of the Ability of the Potential Promoter Sequences to Drive eGFP Expression
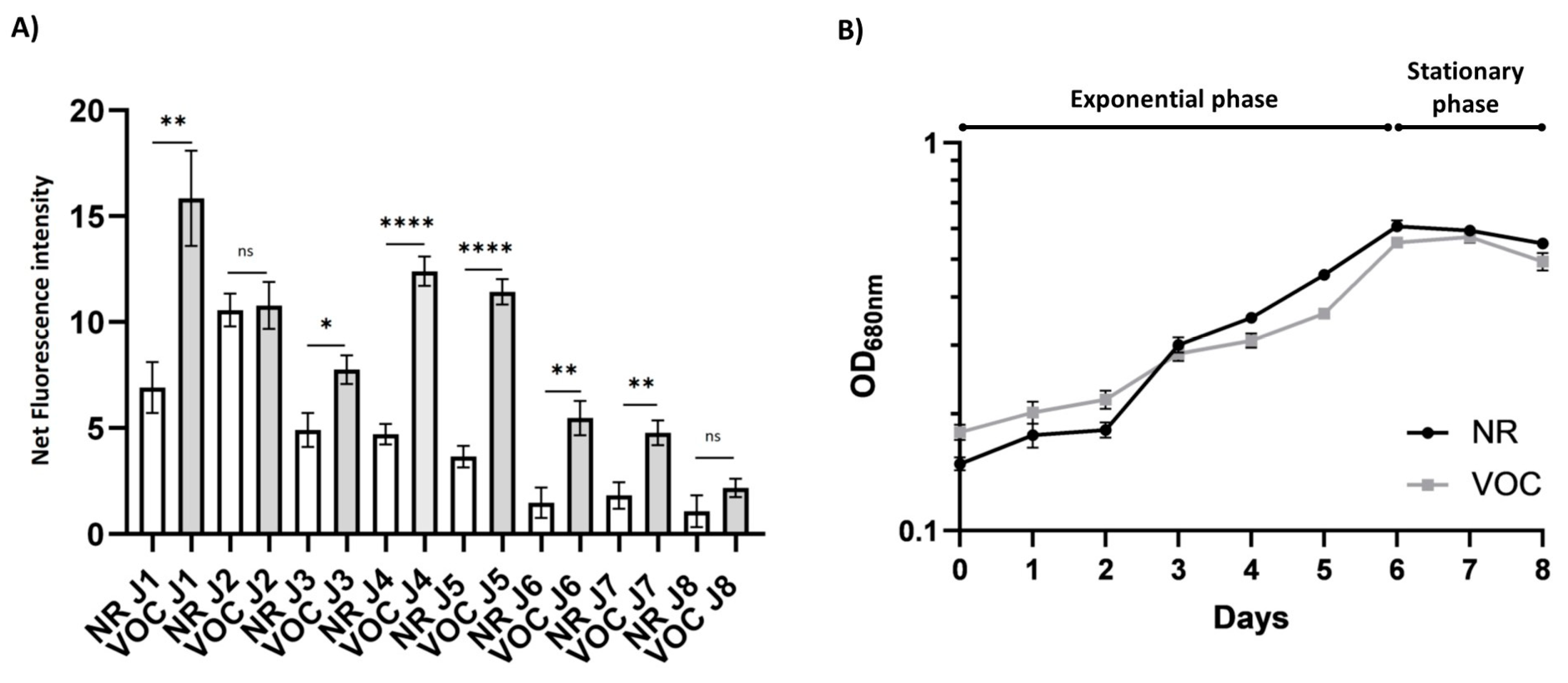
2.3. Characterization of the VOC Promoter
2.3.1. The Activity of the VOC Promoter Correlates with the Growth Phase of the Culture
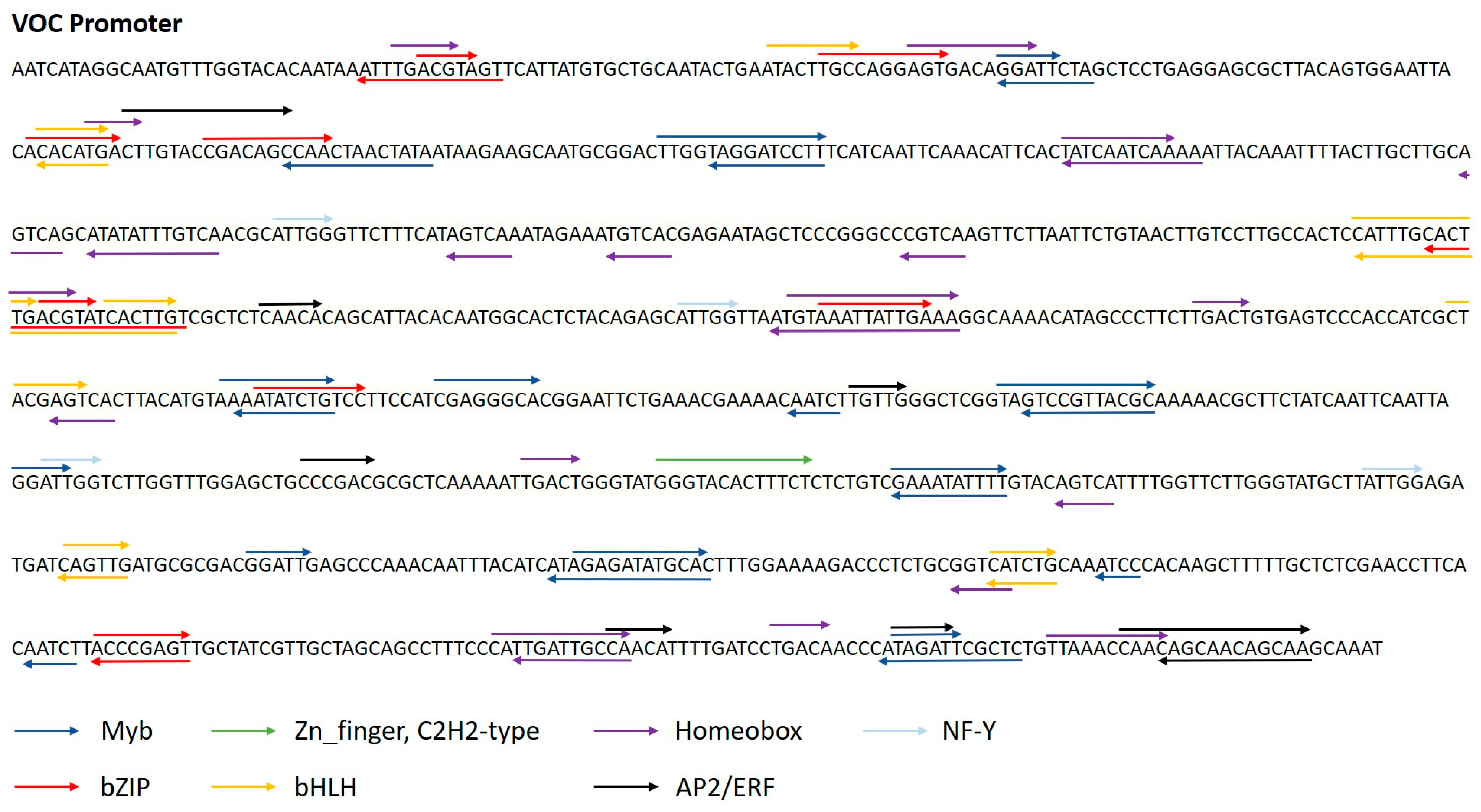
2.3.2. In Silico Analyses of the VOC Promoter
3. Materials and Methods
3.1. Cell Culture and Growth Conditions
3.2. Putative Regulatory Sequences Selection
3.3. Isolation of Putative Promoter Regions
3.4. Transformation Vectors Construction
3.5. Transformation of P. tricornutum by Bacterial Conjugation
3.6. Confirmation of Transgene Presence by PCR Amplification
3.7. Assessment of the Activity of Potential Promoter Sequences by eGFP Fluorescence Intensity Measure
3.8. Statistical Analyses
3.9. In Silico Analysis of Validated Promoter Sequences
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P. Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Falciatore, A.; Jaubert, M.; Bouly, J.-P.; Bailleul, B.; Mock, T. Diatom Molecular Research Comes of Age: Model Species for Studying Phytoplankton Biology and Diversity. Plant Cell 2020, 32, 547–572. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J. The Number, Speed, and Impact of Plastid Endosymbioses in Eukaryotic Evolution. Annu. Rev. Plant Biol. 2013, 64, 583–607. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum Genome Reveals the Evolutionary History of Diatom Genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, A.; Vieira, F.R.J.; Deton-Cabanillas, A.-F.; Veluchamy, A.; Cantrel, C.; Wang, G.; Vanormelingen, P.; Bowler, C.; Piganeau, G.; Hu, H.; et al. A Genomics Approach Reveals the Global Genetic Polymorphism, Structure, and Functional Diversity of Ten Accessions of the Marine Model Diatom Phaeodactylum tricornutum. ISME J. 2020, 14, 347–363. [Google Scholar] [CrossRef]
- Dorrell, R.G.; Villain, A.; Perez-Lamarque, B.; Audren de Kerdrel, G.; McCallum, G.; Watson, A.K.; Ait-Mohamed, O.; Alberti, A.; Corre, E.; Frischkorn, K.R.; et al. Phylogenomic Fingerprinting of Tempo and Functions of Horizontal Gene Transfer within Ochrophytes. Proc. Natl. Acad. Sci. USA 2021, 118, e2009974118. [Google Scholar] [CrossRef]
- Butler, T.; Kapoore, R.V.; Vaidyanathan, S. Phaeodactylum tricornutum: A Diatom Cell Factory. Trends Biotechnol. 2020, 38, 606–622. [Google Scholar] [CrossRef]
- Allen, A.E.; LaRoche, J.; Maheswari, U.; Lommer, M.; Schauer, N.; Lopez, P.J.; Finazzi, G.; Fernie, A.R.; Bowler, C. Whole-Cell Response of the Pennate Diatom Phaeodactylum tricornutum to Iron Starvation. Proc. Natl. Acad. Sci. USA 2008, 105, 10438–10443. [Google Scholar] [CrossRef]
- Gutenbrunner, S.A.; Thalhamer, J.; Schmid, A.-M.M. Proteinaceous and Immunochemical Distinctions Between the Oval and Fusiform Morphotypes of Phaeodactylum tricornutum (Bacillariophyceae). J. Phycol. 1994, 30, 129–136. [Google Scholar] [CrossRef]
- De Martino, A.; Meichenin, A.; Shi, J.; Pan, K.; Bowler, C. Genetic and Phenotypic Characterization of Phaeodactylum tricornutum (Bacillariophyceae) Accessions. J. Phycol. 2007, 43, 992–1009. [Google Scholar] [CrossRef]
- De Martino, A.; Bartual, A.; Willis, A.; Meichenin, A.; Villazán, B.; Maheswari, U.; Bowler, C. Physiological and Molecular Evidence That Environmental Changes Elicit Morphological Interconversion in the Model Diatom Phaeodactylum tricornutum. Protist 2011, 162, 462–481. [Google Scholar] [CrossRef]
- Lewin, J.C.; Lewin, R.A.; Philpott, D.E. Observations on Phaeodactylum tricornutum. Microbiology 1958, 18, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Ovide, C.; Kiefer-Meyer, M.-C.; Bérard, C.; Vergne, N.; Lecroq, T.; Plasson, C.; Burel, C.; Bernard, S.; Driouich, A.; Lerouge, P.; et al. Comparative in Depth RNA Sequencing of P. tricornutum’s Morphotypes Reveals Specific Features of the Oval Morphotype. Sci. Rep. 2018, 8, 14340. [Google Scholar] [CrossRef] [PubMed]
- Galas, L.; Burel, C.; Schapman, D.; Ropitaux, M.; Bernard, S.; Bénard, M.; Bardor, M. Comparative Structural and Functional Analyses of the Fusiform, Oval, and Triradiate Morphotypes of Phaeodactylum tricornutum Pt3 Strain. Front. Plant Sci. 2021, 12, 638181. [Google Scholar] [CrossRef] [PubMed]
- Borowitzka, M.A.; Volcani, B.E. The Polymorphic Diatom Phaeodactylum tricornutum: Ultrastructure of Its Morphotypes. J. Phycol. 1978, 14, 10–21. [Google Scholar] [CrossRef]
- Hempel, F.; Bozarth, A.S.; Lindenkamp, N.; Klingl, A.; Zauner, S.; Linne, U.; Steinbüchel, A.; Maier, U.G. Microalgae as Bioreactors for Bioplastic Production. Microb. Cell Factories 2011, 10, 81. [Google Scholar] [CrossRef]
- D’Adamo, S.; Schiano di Visconte, G.; Lowe, G.; Szaub-Newton, J.; Beacham, T.; Landels, A.; Allen, M.J.; Spicer, A.; Matthijs, M. Engineering the Unicellular Alga Phaeodactylum tricornutum for High-Value Plant Triterpenoid Production. Plant Biotechnol. J. 2019, 17, 75–87. [Google Scholar] [CrossRef]
- Fabris, M.; George, J.; Kuzhiumparambil, U.; Lawson, C.A.; Jaramillo-Madrid, A.C.; Abbriano, R.M.; Vickers, C.E.; Ralph, P. Extrachromosomal Genetic Engineering of the Marine Diatom Phaeodactylum tricornutum Enables the Heterologous Production of Monoterpenoids. ACS Synth. Biol. 2020, 9, 598–612. [Google Scholar] [CrossRef]
- Vavitsas, K.; Fabris, M.; Vickers, C.E. Terpenoid Metabolic Engineering in Photosynthetic Microorganisms. Genes 2018, 9, 520. [Google Scholar] [CrossRef]
- George, J.; Kahlke, T.; Abbriano, R.M.; Kuzhiumparambil, U.; Ralph, P.J.; Fabris, M. Metabolic Engineering Strategies in Diatoms Reveal Unique Phenotypes and Genetic Configurations With Implications for Algal Genetics and Synthetic Biology. Front. Bioeng. Biotechnol. 2020, 8, 513. [Google Scholar] [CrossRef]
- Hempel, F.; Maurer, M.; Brockmann, B.; Mayer, C.; Biedenkopf, N.; Kelterbaum, A.; Becker, S.; Maier, U.G. From Hybridomas to a Robust Microalgal-Based Production Platform: Molecular Design of a Diatom Secreting Monoclonal Antibodies Directed Against the Marburg Virus Nucleoprotein. Microb. Cell Factories 2017, 16, 131. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Lau, J.; Klingl, A.; Maier, U.G. Algae as Protein Factories: Expression of a Human Antibody and the Respective Antigen in the Diatom Phaeodactylum tricornutum. PLoS ONE 2011, 6, e28424. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Maier, U.G. An Engineered Diatom Acting like a Plasma Cell Secreting Human IgG Antibodies with High Efficiency. Microb. Cell Factories 2012, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Slattery, S.S.; Giguere, D.J.; Stuckless, E.E.; Shrestha, A.; Briere, L.-A.K.; Galbraith, A.; Reaume, S.; Boyko, X.; Say, H.H.; Browne, T.S.; et al. Phosphate-Regulated Expression of the SARS-CoV-2 Receptor-Binding Domain in the Diatom Phaeodactylum tricornutum for Pandemic Diagnostics. Sci. Rep. 2022, 12, 7010. [Google Scholar] [CrossRef]
- Kunert, R.; Reinhart, D. Advances in Recombinant Antibody Manufacturing. Appl. Microbiol. Biotechnol. 2016, 100, 3451–3461. [Google Scholar] [CrossRef]
- Vaucheret, H.; Béclin, C.; Elmayan, T.; Feuerbach, F.; Godon, C.; Morel, J.B.; Mourrain, P.; Palauqui, J.C.; Vernhettes, S. Transgene-Induced Gene Silencing in Plants. Plant J. 1998, 16, 651–659. [Google Scholar] [CrossRef]
- De Wilde, C.; Van Houdt, H.; De Buck, S.; Angenon, G.; De Jaeger, G.; Depicker, A. Plants as Bioreactors for Protein Production: Avoiding the Problem of Transgene Silencing. Plant Mol. Biol. 2000, 43, 347–359. [Google Scholar] [CrossRef]
- Windhagauer, M.; Abbriano, R.M.; Ashworth, J.; Barolo, L.; Jaramillo-Madrid, A.C.; Pernice, M.; Doblin, M.A. Characterisation of Novel Regulatory Sequences Compatible with Modular Assembly in the Diatom Phaeodactylum tricornutum. Algal Res. 2021, 53, 102159. [Google Scholar] [CrossRef]
- Slattery, S.S.; Diamond, A.; Wang, H.; Therrien, J.A.; Lant, J.T.; Jazey, T.; Lee, K.; Klassen, Z.; Desgagné-Penix, I.; Karas, B.J.; et al. An Expanded Plasmid-Based Genetic Toolbox Enables Cas9 Genome Editing and Stable Maintenance of Synthetic Pathways in Phaeodactylum tricornutum. ACS Synth. Biol. 2018, 7, 328–338. [Google Scholar] [CrossRef]
- Adler-Agnon (Shemesh), Z.; Leu, S.; Zarka, A.; Boussiba, S.; Khozin-Goldberg, I. Novel Promoters for Constitutive and Inducible Expression of Transgenes in the Diatom Phaeodactylum tricornutum Under Varied Nitrate Availability. J. Appl. Phycol. 2018, 30, 2763–2772. [Google Scholar] [CrossRef]
- Chu, L.; Ewe, D.; Río Bártulos, C.; Kroth, P.G.; Gruber, A. Rapid Induction of GFP Expression by the Nitrate Reductase Promoter in the Diatom Phaeodactylum tricornutum. PeerJ 2016, 4, e2344. [Google Scholar] [CrossRef] [PubMed]
- Erdene-Ochir, E.; Shin, B.-K.; Huda, M.N.; Kim, D.H.; Lee, E.H.; Song, D.-G.; Kim, Y.-M.; Kim, S.M.; Pan, C.-H. Cloning of a Novel Endogenous Promoter for Foreign Gene Expression in Phaeodactylum tricornutum. Appl. Biol. Chem. 2016, 59, 861–867. [Google Scholar] [CrossRef]
- Erdene-Ochir, E.; Shin, B.-K.; Kwon, B.; Jung, C.; Pan, C.-H. Identification and Characterisation of the Novel Endogenous Promoter HASP1 and Its Signal Peptide from Phaeodactylum tricornutum. Sci. Rep. 2019, 9, 9941. [Google Scholar] [CrossRef]
- Erdene-Ochir, E.; Shin, B.-K.; Huda, M.N.; Lee, E.H.; Song, D.-G.; Jung, C.; Pan, C.-H. Characterization of Endogenous Promoters of GapC1 and GS for Recombinant Protein Expression in Phaeodactylum tricornutum. MicrobiologyOpen 2021, 10, e1239. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-Y.; Yen, S.-C.; Kuo, P.-C.; Chung, C.-Y.; Yeh, K.-L.; Huang, C.-H.; Chang, J.; Lin, H.-J. Alkaline Phosphatase Promoter as an Efficient Driving Element for Exogenic Recombinant in the Marine Diatom Phaeodactylum tricornutum. Algal Res. 2017, 23, 58–65. [Google Scholar] [CrossRef]
- Tanaka, A.; Ohno, N.; Nakajima, K.; Matsuda, Y. Light and CO2/CAMP Signal Cross Talk on the Promoter Elements of Chloroplastic β-Carbonic Anhydrase Genes in the Marine Diatom Phaeodactylum tricornutum. Plant Physiol. 2016, 170, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, Z.; Leu, S.; Khozin-Goldberg, I.; Didi-Cohen, S.; Zarka, A.; Boussiba, S. Inducible Expression of Haematococcus Oil Globule Protein in the Diatom Phaeodactylum tricornutum: Association with Lipid Droplets and Enhancement of TAG Accumulation under Nitrogen Starvation. Algal Res. 2016, 18, 321–331. [Google Scholar] [CrossRef]
- Apt, K.E.; Kroth-Pancic, P.G.; Grossman, A.R. Stable Nuclear Transformation of the Diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 1996, 252, 572–579. [Google Scholar]
- Falciatore, A.; Casotti, R.; Leblanc, C.; Abrescia, C.; Bowler, C. Transformation of Nonselectable Reporter Genes in Marine Diatoms. Mar. Biotechnol. 1999, 1, 239–251. [Google Scholar] [CrossRef]
- Seo, S.; Jeon, H.; Hwang, S.; Jin, E.; Chang, K.S. Development of a New Constitutive Expression System for the Transformation of the Diatom Phaeodactylum tricornutum. Algal Res. 2015, 11, 50–54. [Google Scholar] [CrossRef]
- Zou, L.-G.; Chen, J.-W.; Zheng, D.-L.; Balamurugan, S.; Li, D.-W.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. High-Efficiency Promoter-Driven Coordinated Regulation of Multiple Metabolic Nodes Elevates Lipid Accumulation in the Model Microalga Phaeodactylum tricornutum. Microb. Cell Factories 2018, 17, 54. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.-G.; Balamurugan, S.; Zhou, T.-B.; Chen, J.-W.; Li, D.-W.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Potentiation of Concurrent Expression of Lipogenic Genes by Novel Strong Promoters in the Oleaginous Microalga Phaeodactylum tricornutum. Biotechnol. Bioeng. 2019, 116, 3006–3015. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Kadono, T.; Kira, N.; Suzuki, K.; Iwata, O.; Ohnishi, K.; Yamaguchi, H.; Adachi, M. Development of Endogenous Promoters That Drive High-Level Expression of Introduced Genes in the Model Diatom Phaeodactylum tricornutum. Mar. Genom. 2018, 42, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Harada, H.; Nakatsuma, D.; Ishida, M.; Matsuda, Y. Regulation of the Expression of Intracellular β-Carbonic Anhydrase in Response to CO2 and Light in the Marine Diatom Phaeodactylum tricornutum. Plant Physiol. 2005, 139, 1041–1050. [Google Scholar] [CrossRef]
- Garza, E.A.; Bielinski, V.A.; Espinoza, J.L.; Orlandi, K.; Alfaro, J.R.; Bolt, T.M.; Beeri, K.; Weyman, P.D.; Dupont, C.L. Validating a Promoter Library for Application in Plasmid-Based Diatom Genetic Engineering. ACS Synth. Biol. 2023, 12, 3215–3228. [Google Scholar] [CrossRef]
- Kassaw, T.K.; Paton, A.J.; Peers, G. Episome-Based Gene Expression Modulation Platform in the Model Diatom Phaeodactylum tricornutum. ACS Synth. Biol. 2022, 11, 191–204. [Google Scholar] [CrossRef]
- Karas, B.J.; Diner, R.E.; Lefebvre, S.C.; McQuaid, J.; Phillips, A.P.R.; Noddings, C.M.; Brunson, J.K.; Valas, R.E.; Deerinck, T.J.; Jablanovic, J.; et al. Designer Diatom Episomes Delivered by Bacterial Conjugation. Nat. Commun. 2015, 6, 6925. [Google Scholar] [CrossRef]
- Diner, R.E.; Bielinski, V.A.; Dupont, C.L.; Allen, A.E.; Weyman, P.D. Refinement of the Diatom Episome Maintenance Sequence and Improvement of Conjugation-Based DNA Delivery Methods. Front. Bioeng. Biotechnol. 2016, 4, 65. [Google Scholar] [CrossRef]
- Rayko, E.; Maumus, F.; Maheswari, U.; Jabbari, K.; Bowler, C. Transcription Factor Families Inferred from Genome Sequences of Photosynthetic Stramenopiles. New Phytol. 2010, 188, 52–66. [Google Scholar] [CrossRef]
- Buitrago-Flórez, F.J.; Restrepo, S.; Riaño-Pachón, D.M. Identification of Transcription Factor Genes and Their Correlation with the High Diversity of Stramenopiles. PLoS ONE 2014, 9, e111841. [Google Scholar] [CrossRef]
- Matthijs, M.; Fabris, M.; Obata, T.; Foubert, I.; Franco-Zorrilla, J.M.; Solano, R.; Fernie, A.R.; Vyverman, W.; Goossens, A. The Transcription Factor BZIP14 Regulates the TCA Cycle in the Diatom Phaeodactylum tricornutum. EMBO J. 2017, 36, 1559–1576. [Google Scholar] [CrossRef]
- Thiriet-Rupert, S.; Carrier, G.; Chénais, B.; Trottier, C.; Bougaran, G.; Cadoret, J.-P.; Schoefs, B.; Saint-Jean, B. Transcription Factors in Microalgae: Genome-Wide Prediction and Comparative Analysis. BMC Genom. 2016, 17, 282. [Google Scholar] [CrossRef]
- Annunziata, R.; Ritter, A.; Fortunato, A.E.; Manzotti, A.; Cheminant-Navarro, S.; Agier, N.; Huysman, M.J.J.; Winge, P.; Bones, A.M.; Bouget, F.-Y.; et al. BHLH-PAS Protein RITMO1 Regulates Diel Biological Rhythms in the Marine Diatom Phaeodactylum tricornutum. Proc. Natl. Acad. Sci. USA 2019, 116, 13137–13142. [Google Scholar] [CrossRef] [PubMed]
- Montsant, A.; Jabbari, K.; Maheswari, U.; Bowler, C. Comparative Genomics of the Pennate Diatom Phaeodactylum tricornutum. Plant Physiol. 2005, 137, 500–513. [Google Scholar] [CrossRef]
- Huang, A.; He, L.; Wang, G. Identification and Characterization of MicroRNAs from Phaeodactylum tricornutum by High-Throughput Sequencing and Bioinformatics Analysis. BMC Genom. 2011, 12, 337. [Google Scholar] [CrossRef] [PubMed]
- Rogato, A.; Richard, H.; Sarazin, A.; Voss, B.; Cheminant Navarro, S.; Champeimont, R.; Navarro, L.; Carbone, A.; Hess, W.R.; Falciatore, A. The Diversity of Small Non-Coding RNAs in the Diatom Phaeodactylum tricornutum. BMC Genom. 2014, 15, 698. [Google Scholar] [CrossRef] [PubMed]
- Cruz de Carvalho, M.H.; Sun, H.-X.; Bowler, C.; Chua, N.-H. Noncoding and Coding Transcriptome Responses of a Marine Diatom to Phosphate Fluctuations. New Phytol. 2016, 210, 497–510. [Google Scholar] [CrossRef]
- Rastogi, A.; Maheswari, U.; Dorrell, R.G.; Vieira, F.R.J.; Maumus, F.; Kustka, A.; McCarthy, J.; Allen, A.E.; Kersey, P.; Bowler, C.; et al. Integrative Analysis of Large Scale Transcriptome Data Draws a Comprehensive Landscape of Phaeodactylum tricornutum Genome and Evolutionary Origin of Diatoms. Sci. Rep. 2018, 8, 4834. [Google Scholar] [CrossRef]
- Hoguin, A.; Rastogi, A.; Bowler, C.; Tirichine, L. Genome-Wide Analysis of Allele-Specific Expression of Genes in the Model Diatom Phaeodactylum tricornutum. Sci. Rep. 2021, 11, 2954. [Google Scholar] [CrossRef]
- König, S.; Eisenhut, M.; Bräutigam, A.; Kurz, S.; Weber, A.P.M.; Büchel, C. The Influence of a Cryptochrome on the Gene Expression Profile in the Diatom Phaeodactylum tricornutum under Blue Light and in Darkness. Plant Cell Physiol. 2017, 58, 1914–1923. [Google Scholar] [CrossRef]
- Zhao, X.; Rastogi, A.; Deton Cabanillas, A.F.; Ait Mohamed, O.; Cantrel, C.; Lombard, B.; Murik, O.; Genovesio, A.; Bowler, C.; Bouyer, D.; et al. Genome Wide Natural Variation of H3K27me3 Selectively Marks Genes Predicted to Be Important for Cell Differentiation in Phaeodactylum tricornutum. New Phytol. 2021, 229, 3208–3220. [Google Scholar] [CrossRef] [PubMed]
- Remmers, I.M.; D’Adamo, S.; Martens, D.E.; de Vos, R.C.H.; Mumm, R.; America, A.H.P.; Cordewener, J.H.G.; Bakker, L.V.; Peters, S.A.; Wijffels, R.H.; et al. Orchestration of Transcriptome, Proteome and Metabolome in the Diatom Phaeodactylum tricornutum during Nitrogen Limitation. Algal Res. 2018, 35, 33–49. [Google Scholar] [CrossRef]
- Yang, Z.-K.; Ma, Y.-H.; Zheng, J.-W.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Proteomics to Reveal Metabolic Network Shifts towards Lipid Accumulation Following Nitrogen Deprivation in the Diatom Phaeodactylum tricornutum. J. Appl. Phycol. 2014, 26, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lin, X.; Liu, X.; Zhang, J.; Ge, F. Genome Annotation of a Model Diatom Phaeodactylum tricornutum Using an Integrated Proteogenomic Pipeline. Mol. Plant 2018, 11, 1292–1307. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.-Y.; Yang, Z.-K.; Zheng, J.-W.; Xie, Y.; Li, D.-W.; Murugan, S.B.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Examination of Metabolic Responses to Phosphorus Limitation via Proteomic Analyses in the Marine Diatom Phaeodactylum tricornutum. Sci. Rep. 2015, 5, 10373. [Google Scholar] [CrossRef]
- Bai, X.; Song, H.; Lavoie, M.; Zhu, K.; Su, Y.; Ye, H.; Chen, S.; Fu, Z.; Qian, H. Proteomic Analyses Bring New Insights into the Effect of a Dark Stress on Lipid Biosynthesis in Phaeodactylum tricornutum. Sci. Rep. 2016, 6, 25494. [Google Scholar] [CrossRef]
- Longworth, J.; Wu, D.; Huete-Ortega, M.; Wright, P.C.; Vaidyanathan, S. Proteome Response of Phaeodactylum tricornutum, during Lipid Accumulation Induced by Nitrogen Depletion. Algal Res. 2016, 18, 213–224. [Google Scholar] [CrossRef]
- Poirier, I.; Pallud, M.; Kuhn, L.; Hammann, P.; Demortière, A.; Jamali, A.; Chicher, J.; Caplat, C.; Gallon, R.K.; Bertrand, M. Toxicological Effects of CdSe Nanocrystals on the Marine Diatom Phaeodactylum tricornutum: The First Mass Spectrometry-Based Proteomic Approach. Ecotoxicol. Environ. Saf. 2018, 152, 78–90. [Google Scholar] [CrossRef]
- Chuberre, C.; Chan, P.; Walet-Balieu, M.-L.; Thiébert, F.; Burel, C.; Hardouin, J.; Gügi, B.; Bardor, M. Comparative Proteomic Analysis of the Diatom Phaeodactylum tricornutum Reveals New Insights into Intra- and Extra-Cellular Protein Contents of Its Oval, Fusiform, and Triradiate Morphotypes. Front. Plant Sci. 2022, 13, 673113. [Google Scholar] [CrossRef]
- Zaslavskaia, L.A.; Lippmeier, J.C.; Kroth, P.G.; Grossman, A.R.; Apt, K.E. Transformation of the Diatom Phaeodactylum tricornutum (Bacillariophyceae) with a Variety of Selectable Marker and Reporter Genes. J. Phycol. 2000, 36, 379–386. [Google Scholar] [CrossRef]
- Russo, M.T.; Annunziata, R.; Sanges, R.; Ferrante, M.I.; Falciatore, A. The Upstream Regulatory Sequence of the Light Harvesting Complex Lhcf2 Gene of the Marine Diatom Phaeodactylum tricornutum Enhances Transcription in an Orientation- and Distance-Independent Fashion. Mar. Genom. 2015, 24 Pt 1, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Toustou, C.; Boulogne, I.; Gonzalez, A.-A.; Bardor, M. Comparative RNA-Seq of Ten Phaeodactylum tricornutum Accessions: Unravelling Criteria for Robust Strain Selection from a Bioproduction Point of View. Mar. Drugs 2024, 22, 353. [Google Scholar] [CrossRef] [PubMed]
- Pavani, P.; Kumar, K.; Rani, A.; Venkatesu, P.; Lee, M.-J. The Influence of Sodium Phosphate Buffer on the Stability of Various Proteins: Insights into Protein-Buffer Interactions. J. Mol. Liq. 2021, 331, 115753. [Google Scholar] [CrossRef]
- Song, Z.; Lye, G.J.; Parker, B.M. Morphological and Biochemical Changes in Phaeodactylum tricornutum Triggered by Culture Media: Implications for Industrial Exploitation. Algal Res. 2020, 47, 101822. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Veluchamy, A.; Lin, X.; Maumus, F.; Rivarola, M.; Bhavsar, J.; Creasy, T.; O’Brien, K.; Sengamalay, N.A.; Tallon, L.J.; Smith, A.D.; et al. Insights into the Role of DNA Methylation in Diatoms by Genome-Wide Profiling in Phaeodactylum tricornutum. Nat. Commun. 2013, 4, 2091. [Google Scholar] [CrossRef]
- Li, M.Z.; Elledge, S.J. Harnessing Homologous Recombination in Vitro to Generate Recombinant DNA via SLIC. Nat. Methods 2007, 4, 251–256. [Google Scholar] [CrossRef]
- De Rybel, B.; van den Berg, W.; Lokerse, A.S.; Liao, C.-Y.; van Mourik, H.; Möller, B.; Llavata-Peris, C.I.; Weijers, D. A Versatile Set of Ligation-Independent Cloning Vectors for Functional Studies in Plants. Plant Physiol. 2011, 156, 1292–1299. [Google Scholar] [CrossRef]
- Jeong, J.-Y.; Yim, H.-S.; Ryu, J.-Y.; Lee, H.S.; Lee, J.-H.; Seen, D.-S.; Kang, S.G. One-Step Sequence- and Ligation-Independent Cloning as a Rapid and Versatile Cloning Method for Functional Genomics Studies. Appl. Environ. Microbiol. 2012, 78, 5440–5443. [Google Scholar] [CrossRef]
- Debit, A.; Charton, F.; Pierre-Elies, P.; Bowler, C.; Cruz de Carvalho, H. Differential Expression Patterns of Long Noncoding RNAs in a Pleiomorphic Diatom and Relation to Hyposalinity. Sci. Rep. 2023, 13, 2440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Burel, C.; Plasson, C.; Kiefer-Meyer, M.-C.; Ovide, C.; Gügi, B.; Wan, C.; Teo, G.; Mak, A.; Song, Z.; et al. Characterization of a GDP-Fucose Transporter and a Fucosyltransferase Involved in the Fucosylation of Glycoproteins in the Diatom Phaeodactylum tricornutum. Front. Plant Sci. 2019, 10, 610. [Google Scholar] [CrossRef]
- Truong, T.Q.; Park, Y.J.; Koo, S.Y.; Choi, J.-H.; Enkhbayar, A.; Song, D.-G.; Kim, S.M. Interdependence of Fucoxanthin Biosynthesis and Fucoxanthin-Chlorophyll a/c Binding Proteins in Phaeodactylum tricornutum Under Different Light Intensities. J. Appl. Phycol. 2023, 35, 25–42. [Google Scholar] [CrossRef]
- Mustafiz, A.; Singh, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Genome-Wide Analysis of Rice and Arabidopsis Identifies Two Glyoxalase Genes That Are Highly Expressed in Abiotic Stresses. Funct. Integr. Genom. 2011, 11, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.-N.; Yang, C.-W.; Wu, N.-Y.; Wang, H.-T.; Tseng, K.-C.; Chiu, Y.-H.; Lee, T.-Y.; Chang, W.-C. PlantPAN 4.0: Updated Database for Identifying Conserved Non-Coding Sequences and Exploring Dynamic Transcriptional Regulation in Plant Promoters. Nucleic Acids Res. 2024, 52, D1569–D1578. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Wang, W.; Fang, H.; Aslam, M.; Du, H.; Chen, J.; Luo, H.; Chen, W.; Liu, X. MYB Gene Family in the Diatom Phaeodactylum tricornutum Revealing Their Potential Functions in the Adaption to Nitrogen Deficiency and Diurnal Cycle. J. Phycol. 2022, 58, 121–132. [Google Scholar] [CrossRef]
- Liu, W.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W.-X. BHLH122 Is Important for Drought and Osmotic Stress Resistance in Arabidopsis and in the Repression of ABA Catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Y.; Sui, N. Transcriptional Regulation of BHLH during Plant Response to Stress. Biochem. Biophys. Res. Commun. 2018, 503, 397–401. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chow, C.-N.; Zheng, H.-Q.; Wu, N.-Y.; Chien, C.-H.; Huang, H.-D.; Lee, T.-Y.; Chiang-Hsieh, Y.-F.; Hou, P.-F.; Yang, T.-Y.; Chang, W.-C. PlantPAN 2.0: An Update of Plant Promoter Analysis Navigator for Reconstructing Transcriptional Regulatory Networks in Plants. Nucleic Acids Res. 2016, 44, D1154–D1160. [Google Scholar] [CrossRef] [PubMed]
| Gene ID | Protein Annotation (Blast2GO) | Promoter Name | Chromosomal Location | Promoter Predicted Length 1 | Promoter Final Length 2 |
|---|---|---|---|---|---|
| Phatr3_J39391 | predicted protein | P39391 | 19:379218–380555 | 896 bp | 896 bp |
| Phatr3_J33266 | solute carrier family 34 | PSCF34 | 3:462819–464704 | 1741 bp | 1701 bp |
| Phatr3_J37038 | syringomycin synthesis regulator | PSSR | 12:20667–21773 | 1890 bp | 1246 bp |
| Phatr3_J46468 | predicted protein | P46468 | 10:427467–428684 | 817 bp | 828 bp |
| Phatr3_Jdraft1668 | taurine catabolism family | PTC | bd_31x35:110713–112101 | 560 bp | 559 bp |
| Phatr3_J40651 | predicted protein | P40651 | 25:73019–73327 | 756 bp | 756 bp |
| Phatr3_J42538 | major intrinsic | PMI | 1:361483–362842 | 1089 bp | 1092 bp |
| Phatr3_J48356 | DUF11 domain-containing | PDUF11 | 17:217814–220228 | 986 bp | 985 bp |
| Phatr3_J33783 | predicted protein | P33783 | 4:340826–341837 | 747 bp | 744 bp |
| Phatr3_J41599 | predicted protein | P41599 | 32:49730–50980 | 1087 bp | 1076 bp |
| Phatr3_J36794 | predicted protein | P36794 | 11:381859–382908 | 1491 bp | 3 |
| Phatr3_J50361 | predicted protein | P50361 | 29:82531–83523 | 755 bp | 754 bp |
| Phatr3_J43621 | DUF2711 family | PDUF2711 | 2:1005210–1005785 | 478 bp | 477 bp |
| Phatr3_J34976 | alkaline phosphatase | PAP1 | 6:788784–789464 | 681 bp | 681 bp |
| Phatr3_J43494 | predicted protein | P43494 | 2:626125–627632 | 1669 bp | 3 |
| Phatr3_J47869 | alkaline phosphatase | PAP2 | 15:296103–298532 | 1972 bp | 3 |
| Phatr3_EG02507 | RING-H2 finger ATL74-like | PRHFA | 14:535986–536588 | 830 bp | 830 bp |
| Phatr3_J8683 | peptide methionine sulfoxide reductase | PPMSR | 1:241098–242192 | 597 bp | 596 bp |
| Phatr3_Jdraft1443 | P-loop containing nucleoside triphosphate hydrolase | PPCNTH | bd_32x35:212637–213710 | 1138 bp | 1125 bp |
| Phatr3_J40433 | solute carrier family 34 | PSCF34-2 | 23:381101–383068 | 735 bp | 727 bp |
| Phatr3_J35771 | predicted protein | P35771 | 8:716893–717834 | 1215 bp | 3 |
| Phatr3_J35102 | predicted protein | P35102 | 7:63434–63919 | 535 bp | 535 bp |
| Phatr3_J8537 | type VI secretion system tip | PSST5 | 5:930561–935372 | 1542 bp | 1542 bp |
| Phatr3_J36570 | serine hydrolase | PSH | 10:784031–785449 | 717 bp | 725 bp |
| Phatr3_J50252 | glutathione S-transferase Mu 3 | PGSTMu3 | 28:107611–109050 | 1095 bp | 1098 bp |
| Phatr3_J49693 | glycerophosphodiester phosphodiesterase | PGPPD | 23:448289–450075 | 860 bp | 3 |
| Phatr3_J34085 | Vicinal Oxygen Chelate family | PVOC | 4:1061396–1062934 | 957 bp | 957 bp |
| Phatr3_J40539 | predicted protein | P40539 | 24:193701–194375 | 687 bp | 687 bp |
| Phatr3_J46933 | predicted protein | P46933 | 11:854712–855482 | 1767 bp | 1772 bp |
| Phatr3_J36444 | predicted protein | P36444 | 10:464796–465641 | 851 bp | 851 bp |
| Phatr3_J48164 | hemerythrin domain-containing | PHDC | 16:393379–394224 | 905 bp | 872 bp |
| Phatr3_EG01906 | predicted protein | PE1906 | 26:436989–438052 | 479 bp | 472 bp |
| Phatr3_EG02422 | P-loop containing nucleoside triphosphate hydrolase | PPCNTH-2 | bd_32x35:126016–129603 | 953 bp | 953 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toustou, C.; Plasson, C.; Kiefer-Meyer, M.-C.; Bardor, M. Characterization of the VOC Promoter That Is Active Under Low-Salinity Conditions in the Diatom Phaeodactylum tricornutum. Mar. Drugs 2025, 23, 185. https://doi.org/10.3390/md23050185
Toustou C, Plasson C, Kiefer-Meyer M-C, Bardor M. Characterization of the VOC Promoter That Is Active Under Low-Salinity Conditions in the Diatom Phaeodactylum tricornutum. Marine Drugs. 2025; 23(5):185. https://doi.org/10.3390/md23050185
Chicago/Turabian StyleToustou, Charlotte, Carole Plasson, Marie-Christine Kiefer-Meyer, and Muriel Bardor. 2025. "Characterization of the VOC Promoter That Is Active Under Low-Salinity Conditions in the Diatom Phaeodactylum tricornutum" Marine Drugs 23, no. 5: 185. https://doi.org/10.3390/md23050185
APA StyleToustou, C., Plasson, C., Kiefer-Meyer, M.-C., & Bardor, M. (2025). Characterization of the VOC Promoter That Is Active Under Low-Salinity Conditions in the Diatom Phaeodactylum tricornutum. Marine Drugs, 23(5), 185. https://doi.org/10.3390/md23050185