Genome Mining-Guided Discovery of Two New Depsides from Talaromyces sp. HDN1820200
Abstract
1. Introduction
2. Results
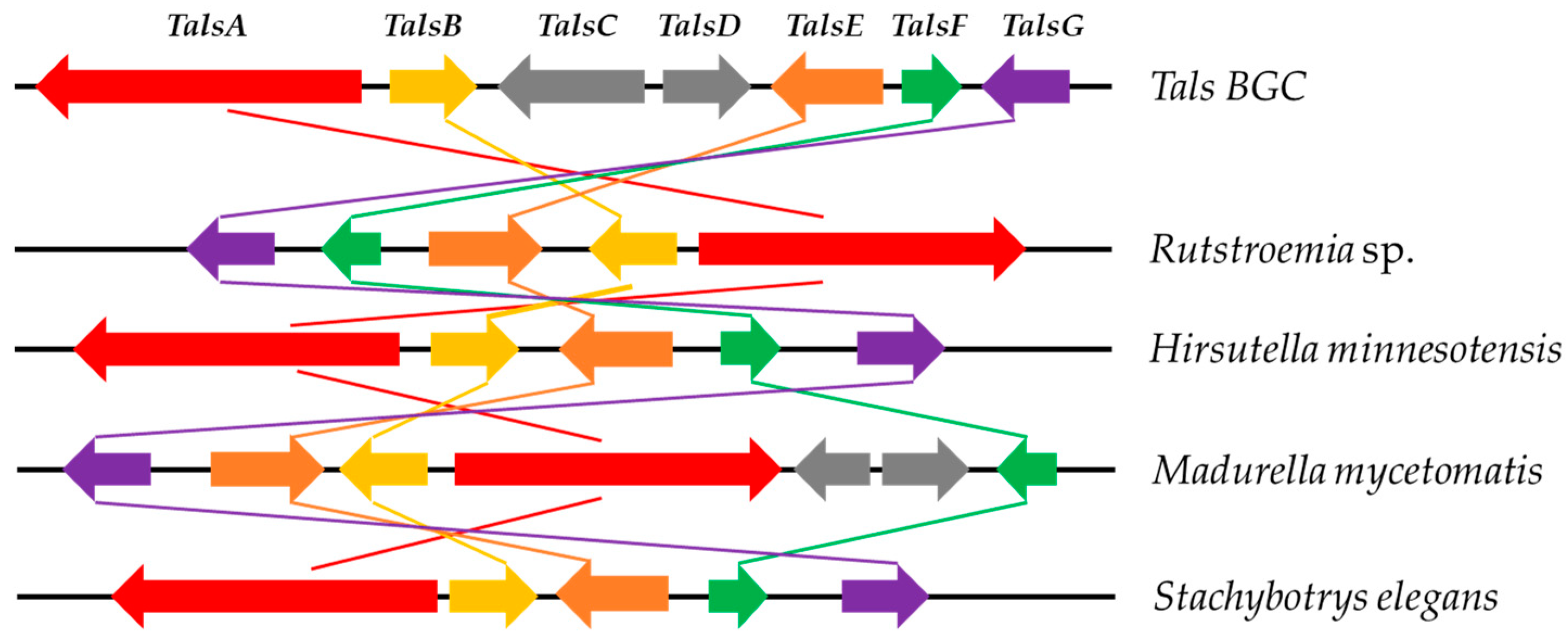
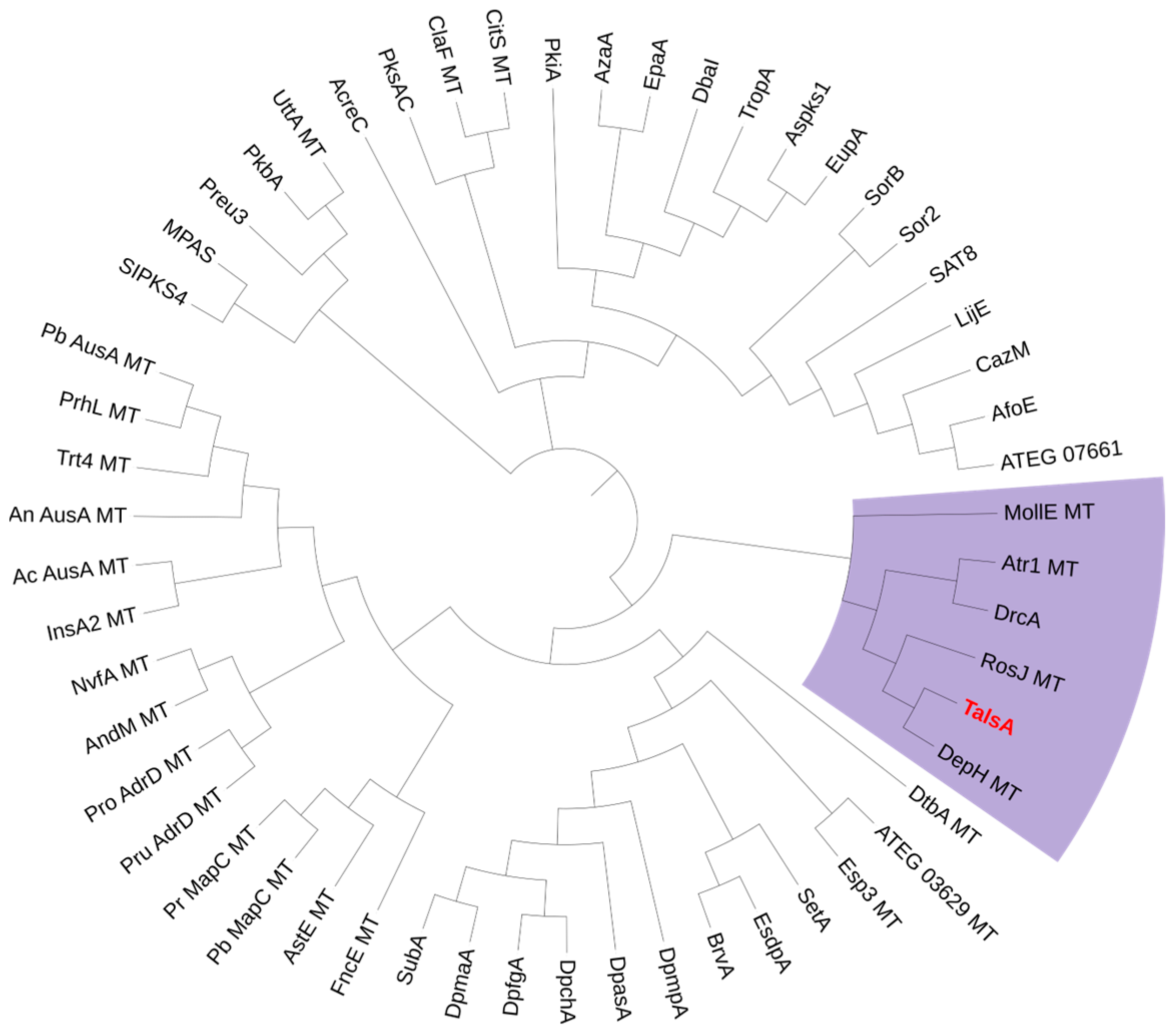
2.1. Bioinformatic Analysis of the TalsA and Tals BGC in Talaromyces sp. HDN1820200
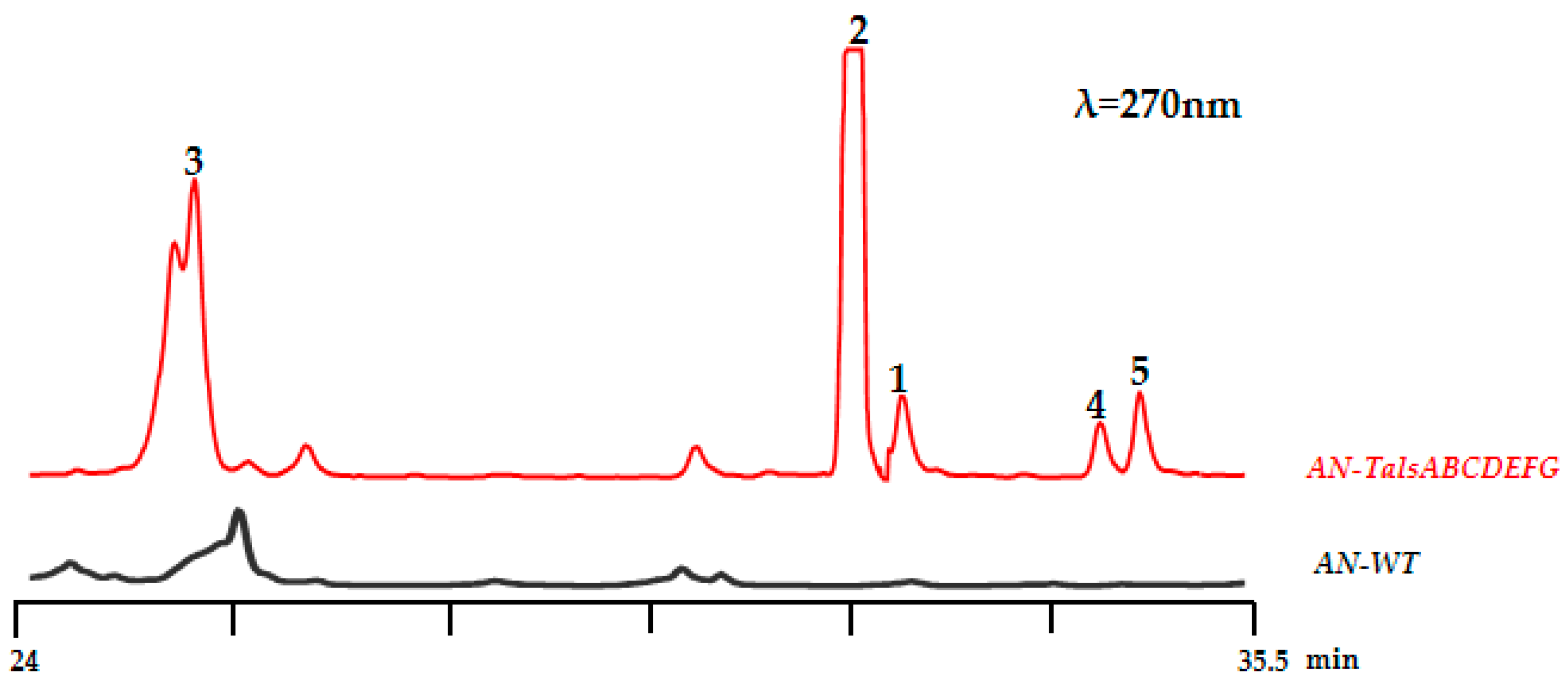
2.2. Heterologous Expression of the Tals Gene Cluster
2.3. Structure Elucidation of Compounds 1–5
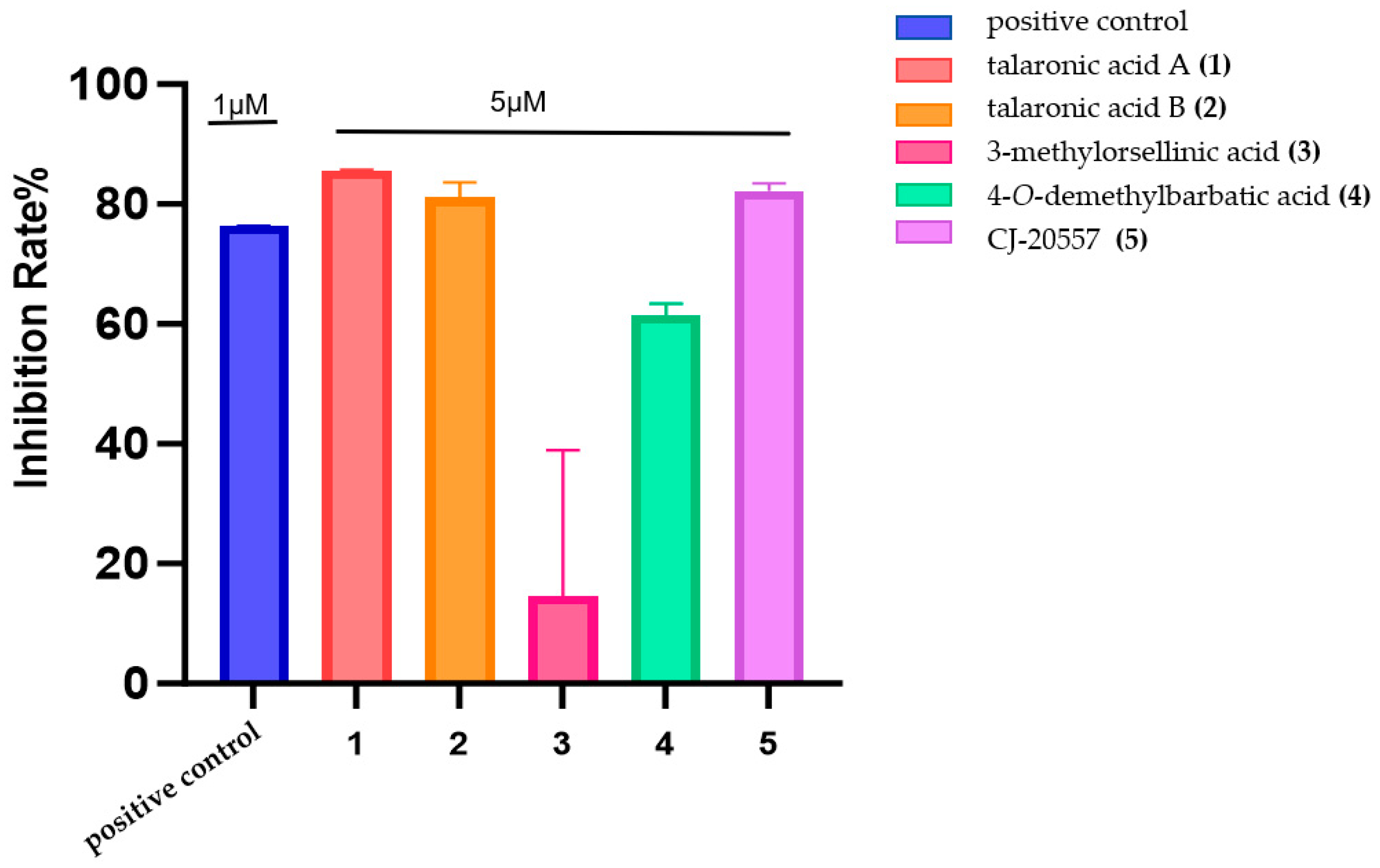
2.4. Testing of Anti-Inflammatory and Cytotoxic Activity
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Materials and Culture Conditions
3.3. Sequence Analysis of the TalsA Gene
3.4. Gene Cloning, Plasmid Construction, and Genetic Manipulation
3.5. Transformation of A. nidulans A1145
3.6. Fermentation and HPLC/LC-MS Analyses
3.7. Extraction, Isolation, and Purification
3.8. Anti-Inflammatory and Cytotoxicity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fischer, E. Synthesis of Depsides, Lichen-Substances and Tannins. J. Am. Chem. Soc. 1914, 36, 1170–1201. [Google Scholar] [CrossRef]
- Mohamed, G.A.J.M. Fungal Depsides—Naturally Inspiring Molecules: Biosynthesis, Structural Characterization, and Biological activities. Metabolites 2021, 11, 638. [Google Scholar] [CrossRef]
- Tao, H.; Abe, I. Enzymology and Biosynthesis of the Orsellinic Acid Derived Medicinal Meroterpenoids. Curr. Opin. Biotechnol. 2021, 69, 52–59. [Google Scholar] [CrossRef]
- Umezawa, H.; Shibamoto, N.; Naganawa, H.; Ayukawa, S.; Matsuzaki, M.; Takeuchi, T.; Kono, K.; Sakamoto, T. Isolation of Lecanoric Acid, an Inhibitor of Histidine Decarboxylase from a Fungus. J. Antibiot. 1974, 27, 587–596. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, W.-W.; Zheng, Q.-H.; Liu, Q.Y.; Zhong, P.; Hu, X.; Fang, Z.; Zhang, Q.J.C. Aculeatusquinones A—D, Novel Metabolites from the Marine-Derived Fungus Aspergillus aculeatus. Heterocycles 2013, 87, 861–868. [Google Scholar] [CrossRef]
- Chen, L.; Wei, X.; Matsuda, Y. Depside Bond Formation by the Starter-Unit Acyltransferase Domain of a Fungal Polyketide Synthase. J. Am. Chem. Soc. 2022, 144, 19225–19230. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, D.; Gao, S.; Cai, X.; Yao, M.; Xu, Y.; Gong, Y.; Zheng, K.; Mao, Y.; Yang, L.; et al. Didepside Formation by the Nonreducing Polyketide Synthase Preu6 of Preussia isomera Requires Interaction of Starter Acyl Transferase and Thioesterase Domains. Angew. Chem. Int. Ed. Engl. 2023, 62, e202214379. [Google Scholar] [CrossRef]
- Kim, W.; Liu, R.; Woo, S.; Kang, K.B.; Park, H.; Yu, Y.H.; Ha, H.H.; Oh, S.Y.; Yang, J.H.; Kim, H. Linking a Gene Cluster to Atranorin, a Major Cortical Substance of Lichens, through Genetic Dereplication and Heterologous Expression. MBio 2021, 12, e0111121. [Google Scholar] [CrossRef] [PubMed]
- Chooi, Y.-H.; Tang, Y. Navigating the Fungal Polyketide Chemical Space: From Genes to Molecules. J. Org. Chem. 2012, 77, 9933–9953. [Google Scholar] [CrossRef]
- Sakemi, S.; Hirai, H.; Ichiba, T.; Inagaki, T.; Kato, Y.; Kojima, N.; Nishida, H.; Parker, J.C.; Saito, T.; Tonai-Kachi, H.; et al. Thielavins as Glucose-6-Phosphatase (G6Pase) Inhibitors: Producing Strain, Fermentation, Isolation, Structural Elucidation and Biological Activities. J. Antibiot. 2002, 34, 941–951. [Google Scholar] [CrossRef]
- Sakai, K.; Kinoshita, H.; Nihira, T. Heterologous Expression System in Aspergillus oryzae for Fungal Biosynthetic Gene Clusters of Secondary Metabolites. Appl. Microbiol. Biotechnol. 2012, 93, 2011–2022. [Google Scholar] [CrossRef] [PubMed]
- Paluszczak, J.; Kleszcz, R.; Studzińska-Sroka, E.; Krajka-Kuźniak, V. Lichen-Derived Caperatic Acid and Physodic Acid Inhibit Wnt Signaling in Colorectal Cancer Cells. Mol. Cell. Biochem. 2018, 441, 109–124. [Google Scholar] [CrossRef]
- Reddy, R.G.; Veeraval, L.; Maitra, S.; Chollet-Krugler, M.; Tomasi, S.; Dévéhat, F.L.-L.; Boustie, J.; Chakravarty, S. Lichen-Derived Compounds Show Potential for Central Nervous System Therapeutics. Phytomedicine 2016, 23, 1527–1534. [Google Scholar] [CrossRef]
- Türk, H.; Yilmaz, M.; Tay, T.; Türk, A.; Kivanc, M. Antimicrobial Activity of Extracts of Chemical Races of the Lichen Pseudevernia furfuracea and their Physodic Acid, Chloroatranorin, Atranorin, and Olivetoric Acid Constituents. Z. Naturforschung C 2006, 61, 499–507. [Google Scholar] [CrossRef]
- Ma, K.; Bao, L.; Han, J.; Jin, T.; Yang, X.; Zhao, F.; Li, S.; Song, F.; Liu, M.; Liu, H. New Benzoate Derivatives and Hirsutane Type Sesquiterpenoids with Antimicrobial Activity and Cytotoxicity from the Solid-state Fermented Rice by the Medicinal Mushroom Stereum Hirsutum. Food Chem. 2014, 143, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Zhang, S.; Lu, X.; Ma, Y.; Fan, Y.; Shi, Y.; Dong, A.; Duan, B. Trivaric Acid, a Potent Depside Human Leukocyte Elastase Inhibitor. Biol. Pharm. Bull. 2012, 35, 2247–2251. [Google Scholar] [CrossRef] [PubMed]
- Kai, B.; Simon, S.; Katharina, S.; Rasmus, V.; Nadine, Z.; Yup, L.S.; Medema, M.H.; Tilmann, W.J.N.A.R. antiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, 81–87. [Google Scholar]
- Sudhir, K.; Glen, S.; Michael, L.; Christina, K.; Koichiro, T.J.M.B. Evolution. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar]
- Gilchrist, C.L.M.; Chooi, Y.H.J.B. Clinker & Clustermap.js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar]
- Mitchell, A.; Attwood, T.; Babbitt, P.C.; Finn, R. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S.C.; Medema, M.H.; Charkoudian, L.K.J.T.F.J. The Minimum Information About A Biosynthetic Gene Cluster Standard as a Means of Organizing Bioinformatic Data. FASEB J. 2018, 32, 547.2. [Google Scholar] [CrossRef]
- Nielsen, M.L.; Albertsen, L.; Lettier, G.L.; Nielsen, J.B.; Mortensen, U.H. Efficient PCR-based Gene Targeting with a Recyclable Marker for Aspergillus nidulans. Fungal Genet. Biol. 2006, 43, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, L.; Shen, M.; Liu, J.; Li, Y.; Xu, S.; Chen, L.; Shi, G.; Ding, Z. Establishment of an Efficient Polyethylene Glycol (PEG)-Mediated Transformation System in Pleurotus eryngii var. ferulae Using Comprehensive Optimization and Multiple Endogenous Promoters. J. Fungi 2022, 8, 186. [Google Scholar]
- Yee, D.A.; Tang, Y. Investigating Fungal Biosynthetic Pathways Using Heterologous Gene Expression: Aspergillus nidulans as a Heterologous Host. Methods Mol. Biol. 2022, 2489, 41–52. [Google Scholar] [PubMed]
- Mansbach, R.A.; Chakraborty, S.; Travers, T.; Gnanakaran, S.J.M.D. Graph-Directed Approach for Downselecting Toxins for Experimental Structure Determination. Mar. Drugs 2020, 18, 256. [Google Scholar] [CrossRef]
- Zhao, Q.; Lan, T.; Su, S.; Rao, Y. Induction of Apoptosis in MDA-MB-231 Breast Cancer Cells by a PARP1-Targeting PROTAC Small Molecule. Chem. Commun. 2019, 55, 369–372. [Google Scholar] [CrossRef]
- Demirel, D.; Ozkaya, F.C.; Ebrahim, W.; Sokullu, E.; Sahin, I.D. Aspergillus carneus Metabolite Averufanin Induced Cell Cycle Arrest and Apoptotic Cell Death on Cancer Cell Lines via Inducing DNA Damage. Sci. Rep. 2023, 13, 6460. [Google Scholar] [CrossRef]
- Preety, D.; Deepak, S.; Mradu, B.; Nalini, S. Evaluation of in Vitro Cytoprotective and Antioxidant Effects of Tinospora cordifolia in cultured HepG2 cells. J. Herb. Med. 2022, 31, 100529. [Google Scholar] [CrossRef]
- Ragab, A.E.; Badawy, E.T.; Aboukhatwa, S.M.; Kabbash, A.; El-Seoud, K.A.A. In Vitro Characterization of Inhibitors for Lung A549 and Leukemia K562 Cell Lines from Fungal Transformation of Arecoline Supported by In Silico Docking to M3-mAChR and ADME Prediction. Pharmaceuticals 2022, 15, 1171. [Google Scholar] [CrossRef]


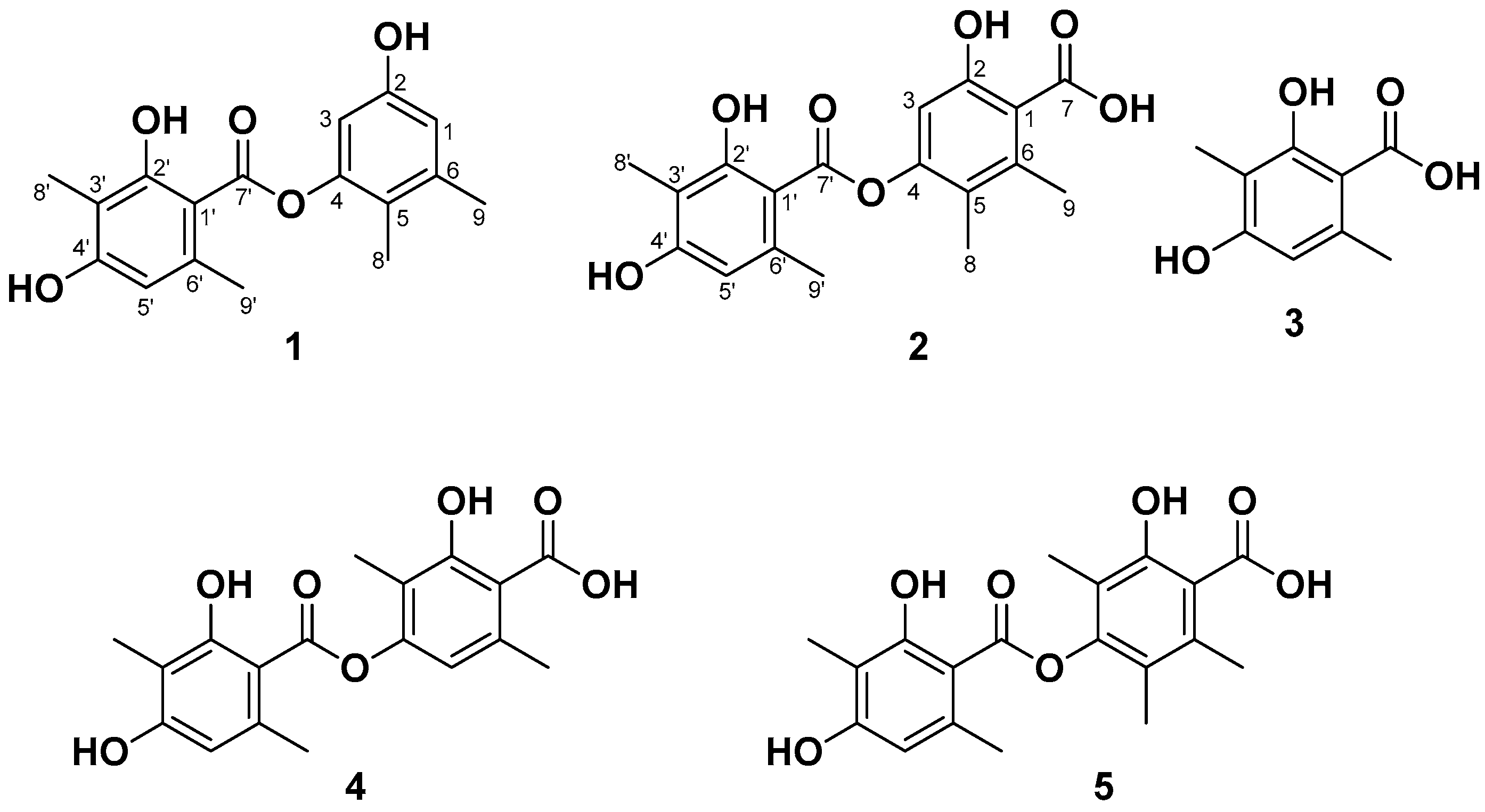

| NO. | 1 | 2 | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 106.7, C | 6.40, d (2.47) | 121.9, CH | - |
| 2 | 155.6, C | - | 152.7, C | - |
| 3 | 114.8, CH | 6.55, d (2.46) | 107.2, CH | 6.55, s |
| 4 | 149.9, C | - | 149.0, C | - |
| 5 | 118.3, C | - | 119.1, C | - |
| 6 | 138.6, C | - | 135.3, C | - |
| 7 | - | - | 169.6, C | - |
| 8-CH3 | 11.7, CH3 | 1.90, s | 11.9, CH3 | 1.94, s |
| 9-CH3 | 19.9, CH3 | 2.19, s | 16.9, CH3 | 2.19, s |
| 2-OH | - | 9.39, s | - | - |
| 1′ | 103.7, C | - | 103.8, C | - |
| 2′ | 162.6, C | - | 161.9, C | - |
| 3′ | 108.5, C | - | 108.6, C | - |
| 4′ | 160.5, C | - | 160.7, C | - |
| 5′ | 110.9, CH | 6.38, s | 110.1, CH | 6.39, s |
| 6′ | 139.0, C | - | 138.9, C | - |
| 7′ | 169.9, C | - | 169.0, C | - |
| 8′-CH3 | 8.1, CH3 | 1.96, s | 8.1, CH3 | 1.97, s |
| 9′-CH3 | 23.5, CH3 | 2.49, s | 23.7, CH3 | 2.48, s |
| 2′-OH | - | 11.37, s | - | 11.25, s |
| 4′-OH | - | 10.30, s | - | 10.36, s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Liu, L.; Huang, J.; Ren, X.; Zhang, G.; Che, Q.; Li, D.; Zhu, T. Genome Mining-Guided Discovery of Two New Depsides from Talaromyces sp. HDN1820200. Mar. Drugs 2025, 23, 130. https://doi.org/10.3390/md23030130
Zhang X, Liu L, Huang J, Ren X, Zhang G, Che Q, Li D, Zhu T. Genome Mining-Guided Discovery of Two New Depsides from Talaromyces sp. HDN1820200. Marine Drugs. 2025; 23(3):130. https://doi.org/10.3390/md23030130
Chicago/Turabian StyleZhang, Xiao, Luyang Liu, Jiani Huang, Xingtao Ren, Guojian Zhang, Qian Che, Dehai Li, and Tianjiao Zhu. 2025. "Genome Mining-Guided Discovery of Two New Depsides from Talaromyces sp. HDN1820200" Marine Drugs 23, no. 3: 130. https://doi.org/10.3390/md23030130
APA StyleZhang, X., Liu, L., Huang, J., Ren, X., Zhang, G., Che, Q., Li, D., & Zhu, T. (2025). Genome Mining-Guided Discovery of Two New Depsides from Talaromyces sp. HDN1820200. Marine Drugs, 23(3), 130. https://doi.org/10.3390/md23030130








