Voltage-Gated K+ Channel Modulation by Marine Toxins: Pharmacological Innovations and Therapeutic Opportunities
Abstract
1. Introduction
1.1. Voltage-Gated Ion Channels
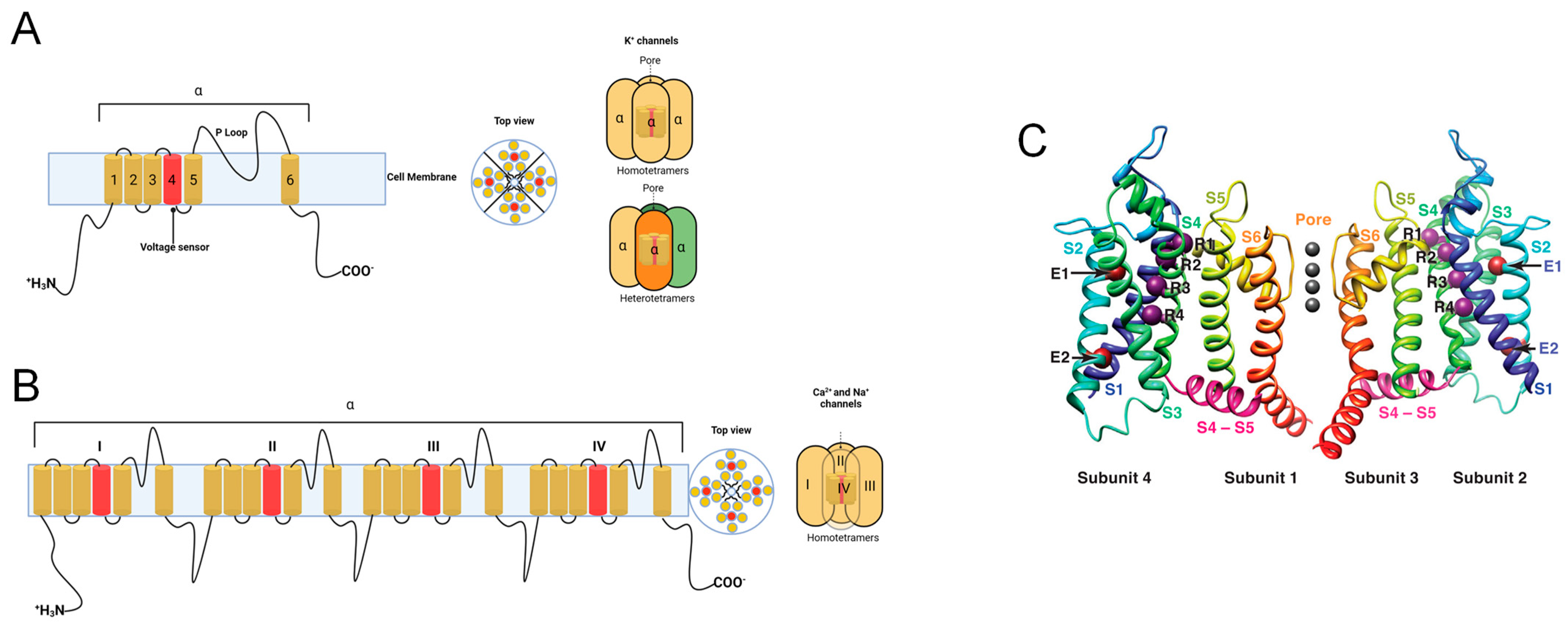

1.2. Voltage-Gated K+ Channels (VGKCs)
2. Kv Channel Blockers from Molluscan Peptides
2.1. Conotoxin
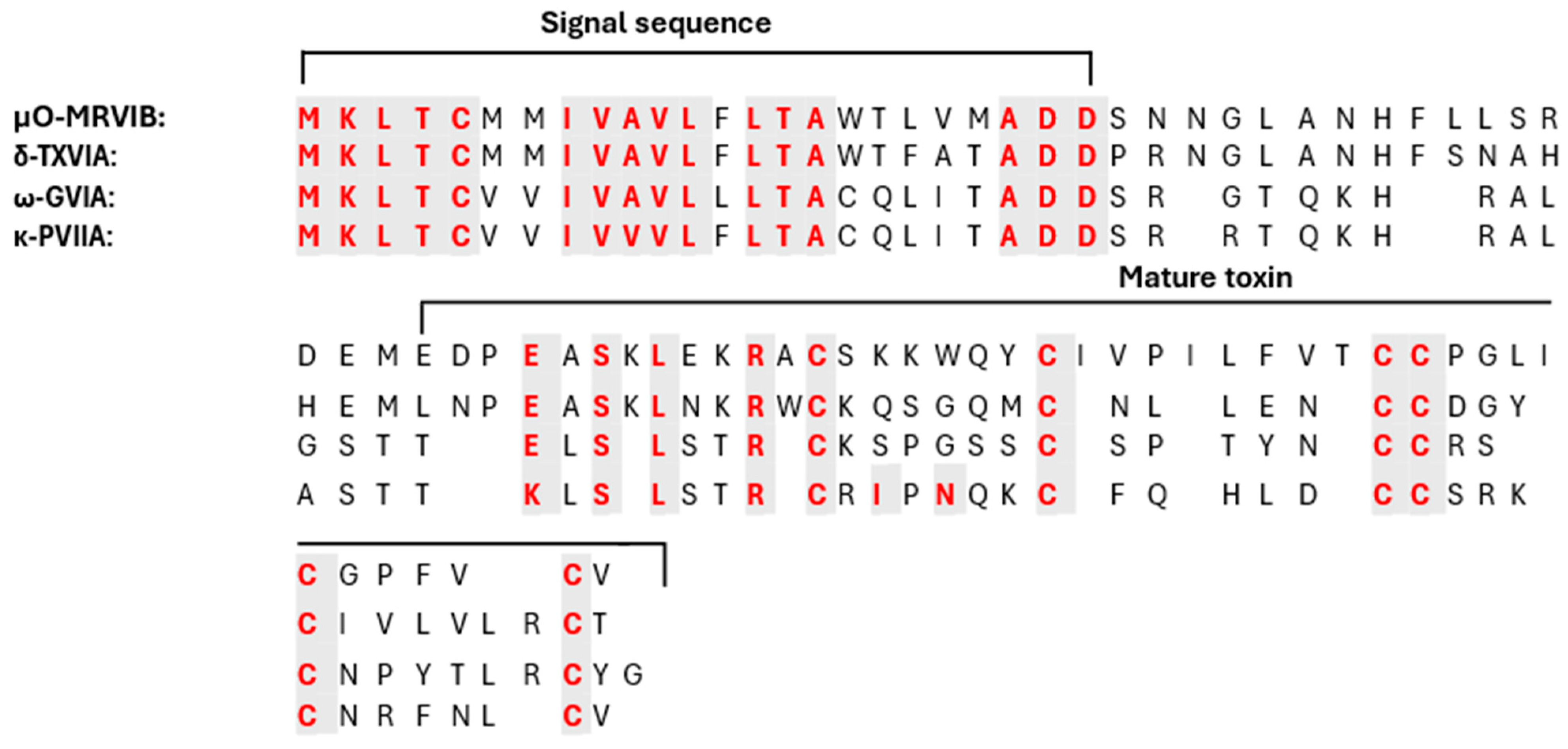
2.1.1. κ-Conotoxin PVIIA
2.1.2. κ-M-Conotoxins RIIIK and RIIIJ
2.1.3. Conkunitzin-S1
2.1.4. CPY or Tyrosine-Rich Conopeptides
2.1.5. Conotoxin Pl14a
2.1.6. Conotoxin sr11a
2.1.7. Other Conotoxins Modulating Kvs
2.2. Sea Anemones
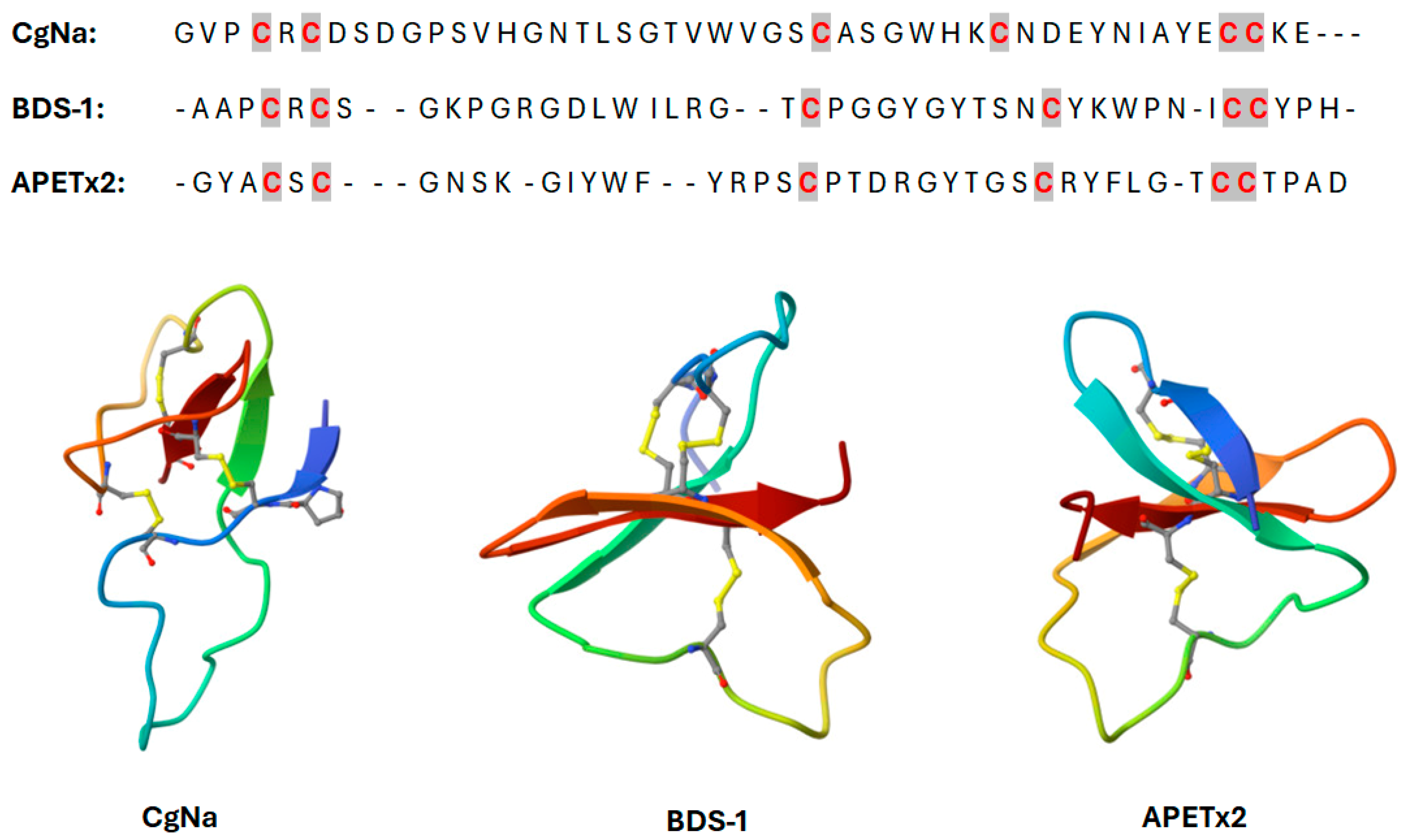
2.2.1. β-Defensin-like Peptides
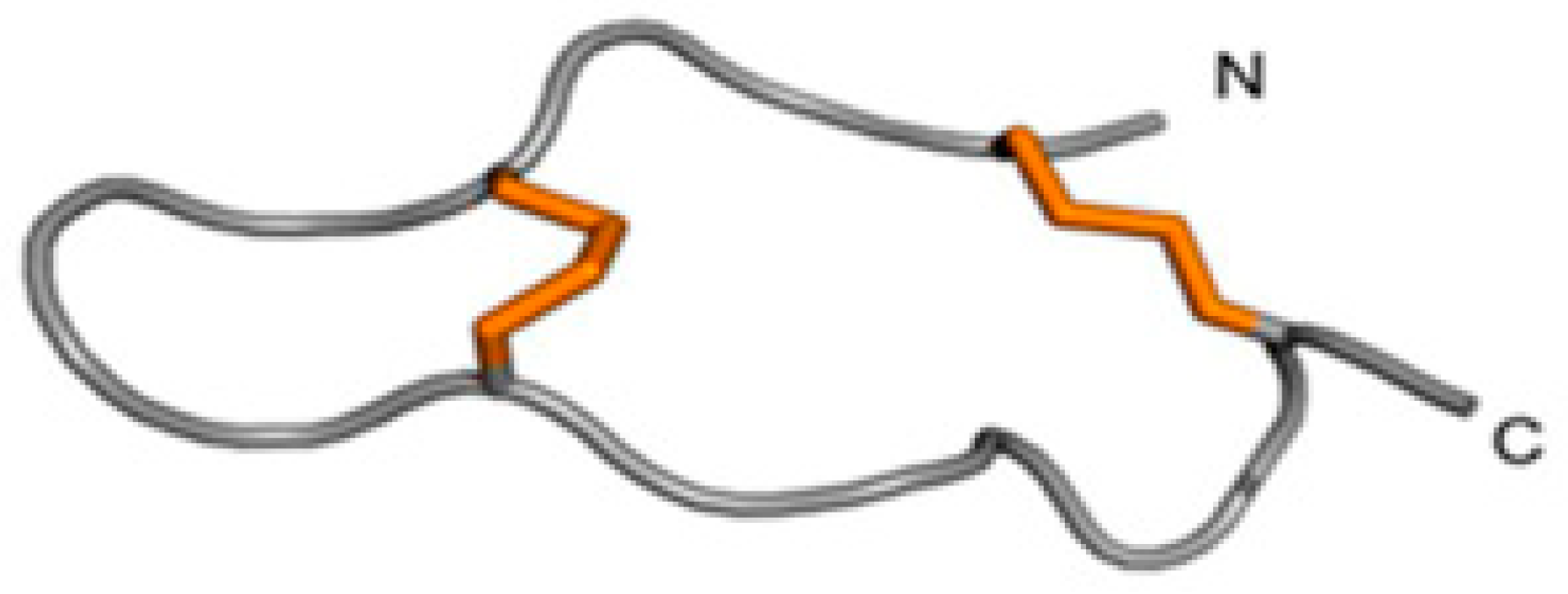
2.2.2. Inhibitor Cystine-Knot (ICK) Motif
2.2.3. Kunitz-Type Peptides
ShK Peptide
BgK Peptide
HmK Peptide
AETXk Peptide
AeK Peptide
Type-2 Kunitz Peptides
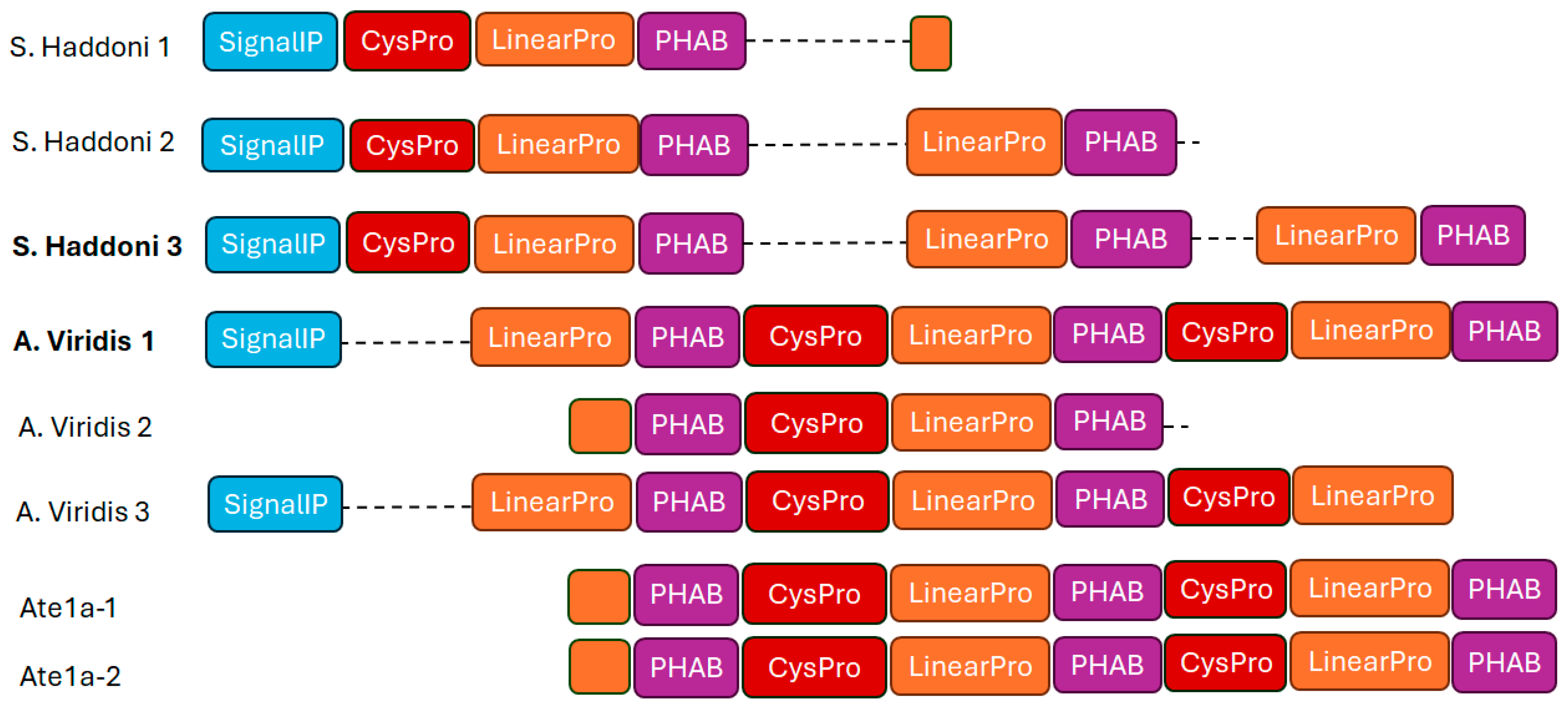
2.2.4. Proline-Hinged Asymmetric Β-Hairpin (PHAB)
2.2.5. kP-Crassipeptides
3. Marine Sponges
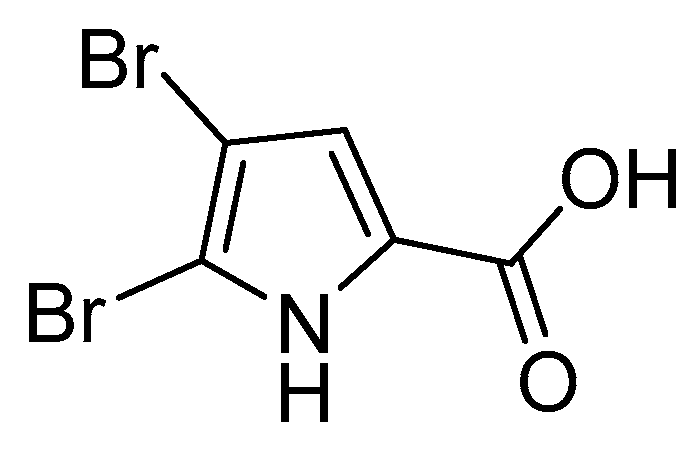
3.1. Pyrrole Alkaloids
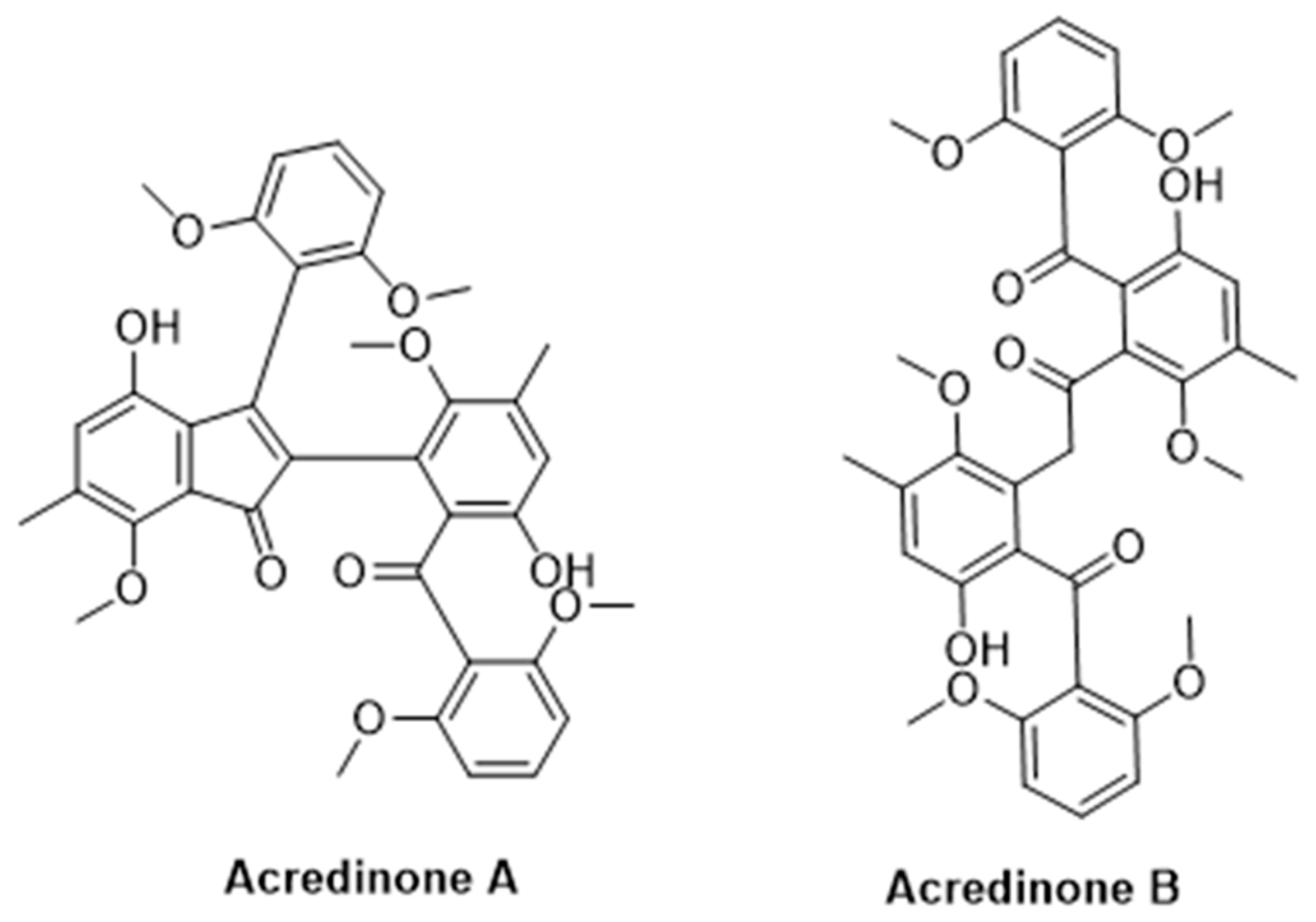
3.2. Secondary Metabolites
4. Additional Non-Peptide K+ Channels Modulators
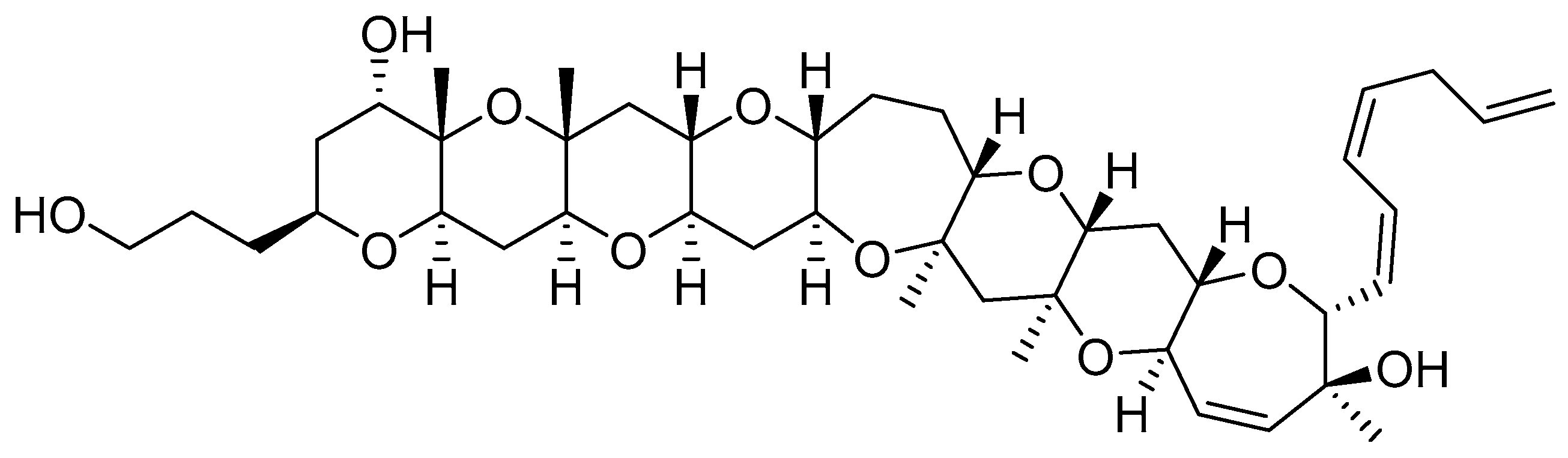
4.1. Gambierol
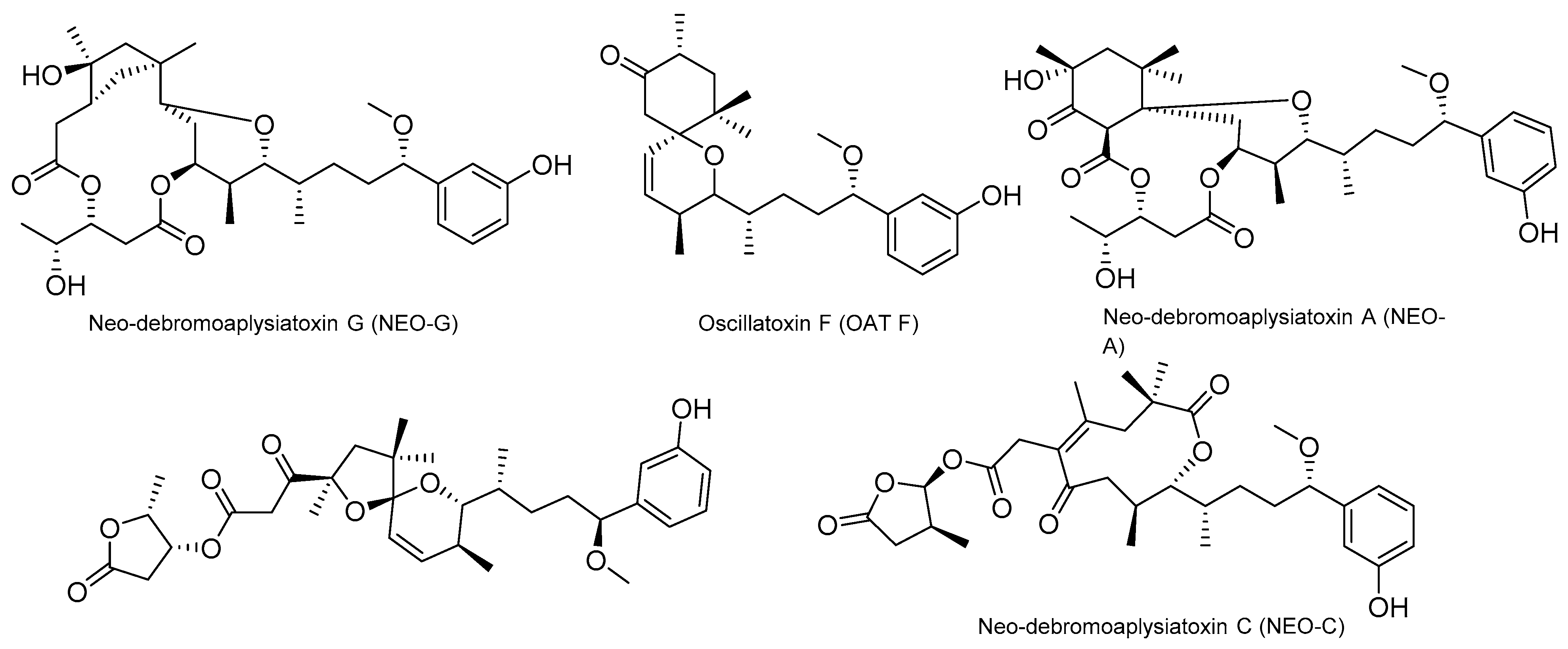
4.2. Aplysiatoxins
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Terlau, H.; Stühmer, W. Structure and Function of Voltage-Gated Ion Channels. Naturwissenschaften 1998, 85, 437–444. [Google Scholar] [CrossRef]
- Arias, H. Marine Toxins Targeting Ion Channels. Mar. Drugs 2006, 4, 37–69. [Google Scholar] [CrossRef]
- Yu, F.H.; Catterall, W.A. Overview of the voltage-gated sodium channel family. Genome Biol. 2003, 4, 207. [Google Scholar] [CrossRef]
- MacKinnon, R. Pore loops: An emerging theme in ion channel structure. Neuron 1995, 14, 889–892. [Google Scholar] [CrossRef][Green Version]
- Isacoff, E.Y.; Jan, Y.N.; Jan, L.Y. Evidence for the formation of heteromultimeric potassium channels in Xenopus oocytes. Nature 1990, 345, 530–534. [Google Scholar] [CrossRef]
- Ruppersberg, J.P.; Schröter, K.H.; Sakmann, B.; Stocker, M.; Sewing, S.; Pongs, O. Heteromultimeric channels formed by rat brain potassium-channel proteins. Nature 1990, 345, 535–537. [Google Scholar] [CrossRef]
- Catterall, W.A. Ion Channel Voltage Sensors: Structure, Function, and Pathophysiology. Neuron 2010, 67, 915–928. [Google Scholar] [CrossRef]
- Doyle, D.A.; Cabral, J.M.; Pfuetzner, R.A.; Kuo, A.; Gulbis, J.M.; Cohen, S.L.; Chait, B.T.; MacKinnon, R. The Structure of the Potassium Channel: Molecular Basis of K+ Conduction and Selectivity. Science 1998, 280, 69–77. [Google Scholar] [CrossRef]
- Kim, D.M.; Nimigean, C.M. Voltage-Gated Potassium Channels: A Structural Examination of Selectivity and Gating. Cold Spring Harb. Perspect. Biol. 2016, 8, a029231. [Google Scholar] [CrossRef] [PubMed]
- Sukomon, N.; Fan, C.; Nimigean, C.M. Ball-and-Chain Inactivation in Potassium Channels. Annu. Rev. Biophysics. 2023, 52, 91–111. [Google Scholar] [CrossRef]
- Kise, Y.; Kasuya, G.; Okamoto, H.H.; Yamanouchi, D.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Nakajo, K.; Nureki, O. Structural basis of gating modulation of Kv4 channel complexes. Nature 2021, 599, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.F.; Bae, C.; Stix, R.; Fernández-Mariño, A.I.; Huffer, K.; Chang, T.H.; Jiang, J.; Faraldo-Gómez, J.D.; Swartz, K.J. Structure of the Shaker Kv channel and mechanism of slow C-type inactivation. Sci. Adv. 2022, 8, eabm7814. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.T.; Tyagi, A.; Chandy, K.G.; Bhushan, S. Mechanisms Underlying C-type Inactivation in Kv Channels: Lessons From Structures of Human Kv1.3 and Fly Shaker-IR Channels. Front. Pharmacol. 2022, 13, 924289. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Mariño, A.I.; Tan, X.F.; Bae, C.; Huffer, K.; Jiang, J.; Swartz, K.J. Inactivation of the Kv2.1 channel through electromechanical coupling. Nature 2023, 622, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Miller, C. An overview of the potassium channel family. Genome Biol. 2000, 1, REVIEWS0004. [Google Scholar] [CrossRef] [PubMed]
- Harding, S.D.; Armstrong, J.F.; Faccenda, E.; Southan, C.; Alexander, S.P.H.; Davenport, A.P.; Spedding, M.; Davies, J.A. The IUPHAR/BPS Guide to PHARMACOLOGY in 2024. Nucleic Acids Res. 2024, 52, D1438–D1449. [Google Scholar] [CrossRef] [PubMed]
- Rudy, B. Diversity and ubiquity of K channels. Neuroscience 1988, 25, 729–749. [Google Scholar] [CrossRef]
- Lewis, R.S.; Cahalan, M.D. Potassium and Calcium Channels in Lymphocytes. Annu. Rev. Immunol. 1995, 13, 623–653. [Google Scholar] [CrossRef] [PubMed]
- Gutman, G.A.; Chandy, K.G.; Grissmer, S.; Lazdunski, M.; McKinnon, D.; Pardo, L.A.; Robertson, G.A.; Rudy, B.; Sanguinetti, M.C.; Stühmer, W.; et al. International Union of Pharmacology. LIII. Nomenclature and Molecular Relationships of Voltage-Gated Potassium Channels. Pharmacol. Rev. 2005, 57, 473–508. [Google Scholar] [CrossRef]
- Filosa, J.A.; Bonev, A.D.; Straub, S.V.; Meredith, A.L.; Wilkerson, M.K.; Aldrich, R.W.; Nelson, M.T. Local potassium signaling couples neuronal activity to vasodilation in the brain. Nat. Neurosci. 2006, 9, 1397–1403. [Google Scholar] [CrossRef]
- Honma, T.; Shiomi, K. Peptide Toxins in Sea Anemones: Structural and Functional Aspects. Mar. Biotechnol. 2006, 8, 1–10. [Google Scholar] [CrossRef]
- Long, S.B.; Campbell, E.B.; MacKinnon, R. Crystal Structure of a Mammalian Voltage-Dependent Shaker Family K+ Channel. Science 2005, 309, 897–903. [Google Scholar] [CrossRef]
- Shen, N.V.; Pfaffinger, P.J. Molecular recognition and assembly sequences involved in the subfamily-specific assembly of voltage-gated K+ channel subunit proteins. Neuron 1995, 14, 625–633. [Google Scholar] [CrossRef]
- Roux, B. Ion Conduction and Selectivity in K+ Channels. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 153–171. [Google Scholar] [CrossRef]
- Tian, C.; Zhu, R.; Zhu, L.; Qiu, T.; Cao, Z.; Kang, T. Potassium Channels: Structures, Diseases, and Modulators. Chem. Biol. Drug Des. 2013, 83, 1–26. [Google Scholar] [CrossRef]
- Musella, S.; Carotenuto, L.; Iraci, N.; Baroli, G.; Ciaglia, T.; Nappi, P.; Basilicata, M.G.; Salviati, E.; Barrese, V.; Vestuto, V.; et al. Beyond Retigabine: Design, Synthesis, and Pharmacological Characterization of a Potent and Chemically Stable Neuronal Kv7 Channel Activator with Anticonvulsant Activity. J. Med. Chem. 2022, 65, 11340–11364. [Google Scholar] [CrossRef]
- Ostacolo, C.; Miceli, F.; Di Sarno, V.; Nappi, P.; Iraci, N.; Soldovieri, M.V.; Ciaglia, T.; Ambrosino, P.; Vestuto, V.; Lauritano, A.; et al. Synthesis and Pharmacological Characterization of Conformationally Restricted Retigabine Analogues as Novel Neuronal Kv7 Channel Activators. J. Med. Chem. 2019, 63, 163–185. [Google Scholar] [CrossRef]
- Pichon, Y.; Prime, L.; Benquet, P.; Tiaho, F. Some aspects of the physiological role of ion channels in the nervous system. Eur. Biophys. J. 2004, 33, 211–226. [Google Scholar] [CrossRef]
- Lebbe, E.; Peigneur, S.; Wijesekara, I.; Tytgat, J. Conotoxins Targeting Nicotinic Acetylcholine Receptors: An Overview. Mar. Drugs 2014, 12, 2970–3004. [Google Scholar] [CrossRef]
- Shiomi, K. Novel peptide toxins recently isolated from sea anemones. Toxicon 2009, 54, 1112–1118. [Google Scholar] [CrossRef]
- Norton, R.S.; Olivera, B.M. Conotoxins down under. Toxicon 2006, 48, 780–798. [Google Scholar] [CrossRef]
- Dave, K.; Lahiry, A. Conotoxins: Review and Docking Studies to determine potentials of Conotoxin as an Anticancer Drug Molecule. Curr. Top. Med. Chem. 2012, 12, 845–851. [Google Scholar] [CrossRef]
- Prashanth, J.R.; Brust, A.; Jin, A.-H.; Alewood, P.F.; Dutertre, S.; Lewis, R.J. Cone snail venomics: From novel biology to novel therapeutics. Future Med. Chem. 2014, 6, 1659–1675. [Google Scholar] [CrossRef]
- Gao, B.; Peng, C.; Yang, J.; Yi, Y.; Zhang, J.; Shi, Q. Cone Snails: A Big Store of Conotoxins for Novel Drug Discovery. Toxins 2017, 9, 397. [Google Scholar] [CrossRef]
- Dao, F.-Y.; Yang, H.; Su, Z.-D.; Yang, W.; Wu, Y.; Hui, D.; Chen, W.; Tang, H.; Lin, H. Recent Advances in Conotoxin Classification by Using Machine Learning Methods. Molecules 2017, 22, 1057. [Google Scholar] [CrossRef]
- Hopkins, C.; Grilley, M.; Miller, C.; Shon, K.-J.; Cruz, L.J.; Gray, W.R.; Dykert, J.; Rivier, J.; Yoshikami, D.; Olivera, B.M. A New Family of Conus Peptides Targeted to the Nicotinic Acetylcholine Receptor. J. Biol. Chem. 1995, 270, 22361–22367. [Google Scholar] [CrossRef]
- Shon, K.-J.; Olivera, B.M.; Watkins, M.; Jacobsen, R.B.; Gray, W.R.; Floresca, C.Z.; Cruz, L.J.; Hillyard, D.R.; Brink, A.; Terlau, H.; et al. μ-Conotoxin PIIIA, a New Peptide for Discriminating among Tetrodotoxin-Sensitive Na Channel Subtypes. J. Neurosci. 1998, 18, 4473–4481. [Google Scholar] [CrossRef]
- Pruneau, D.; Angus, J.A. ω-Conotoxin GVIA is a potent inhibitor of sympathetic neurogenic responses in rat small mesenteric arteries. Br. J. Pharmacol. 2012, 100, 180–184. [Google Scholar] [CrossRef]
- Naranjo, D.; Díaz-Franulic, I. Binding of κ-Conotoxin-PVIIA to Open and Closed Shaker K-Channels Are Differentially Affected by the Ionic Strength. Mar. Drugs 2020, 18, 533. [Google Scholar] [CrossRef]
- Terlau, H.; Boccaccio, A.; Olivera, B.M.; Conti, F. The Block of Shaker K+ Channels by κ-Conotoxin Pviia Is State Dependent. J. Gen. Physiol. 1999, 114, 125–140. [Google Scholar] [CrossRef]
- Shon, K.-J.; Stocker, M.; Terlau, H.; Stühmer, W.; Jacobsen, R.; Walker, C.; Grilley, M.; Watkins, M.; Hillyard, D.R.; Gray, W.R.; et al. κ-Conotoxin Pviia Is a Peptide Inhibiting theShaker K+ Channel. J. Biol. Chem. 1998, 273, 33–38. [Google Scholar] [CrossRef]
- Terlau, H.; Olivera, B.M. ConusVenoms: A Rich Source of Novel Ion Channel-Targeted Peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Santos, A.D.; Olivera, B.M. Conus Peptides Targeted to Specific Nicotinic Acetylcholine Receptor Subtypes. Annu. Rev. Biochem. 1999, 68, 59–88. [Google Scholar] [CrossRef]
- Craig, A.G.; Bandyopadhyay, P.; Olivera, B.M. Post-translationally modified neuropeptides from Conus venoms. Eur. J. Biochem. 2001, 264, 271–275. [Google Scholar] [CrossRef]
- Jacobsen, R.B.; Koch, E.D.; Lange-Malecki, B.; Stocker, M.; Verhey, J.; Van Wagoner, R.M.; Vyazovkina, A.; Olivera, B.M.; Terlau, H. Single Amino Acid Substitutions in κ-Conotoxin PVIIA Disrupt Interaction with the Shaker K+ Channel. J. Biol. Chem. 2000, 275, 24639–24644. [Google Scholar] [CrossRef]
- Adams, M.E.; Olivera, B.M. Neurotoxins: Overview of an emerging research technology. Trends Neurosci. 1994, 17, 151–155. [Google Scholar] [CrossRef]
- Terlau, H.; Shon, K.J.; Grilley, M.; Stocker, M.; Stühmer, W.; Olivera, B.M. Strategy for rapid immobilization of prey by a fish-hunting marine snail. Nature 1996, 381, 148–151. [Google Scholar] [CrossRef]
- Sudarslal, S.; Singaravadivelan, G.; Ramasamy, P.; Ananda, K.; Sarma, S.P.; Sikdar, S.K.; Krishnan, K.S.; Balaram, P. A novel 13 residue acyclic peptide from the marine snail, Conus monile, targets potassium channels. Biochem. Biophys. Res. Commun. 2004, 317, 682–688. [Google Scholar] [CrossRef]
- Kauferstein, S.; Huys, I.; Lamthanh, H.; Stöcklin, R.; Sotto, F.; Menez, A.; Tytgat, J.; Mebs, D. A novel conotoxin inhibiting vertebrate voltage-sensitive potassium channels. Toxicon 2003, 42, 43–52. [Google Scholar] [CrossRef]
- Kauferstein, S.; Huys, I.; Kuch, U.; Melaun, C.; Tytgat, J.; Mebs, D. Novel conopeptides of the I-superfamily occur in several clades of cone snails. Toxicon 2004, 44, 539–548. [Google Scholar] [CrossRef]
- Chen, P.; Dendorfer, A.; Finol-Urdaneta, R.K.; Terlau, H.; Olivera, B.M. Biochemical Characterization of κM-RIIIJ, a Kv1.2 Channel Blocker. J. Biol. Chem. 2010, 285, 14882–14889. [Google Scholar] [CrossRef] [PubMed]
- Verdier, L.; Al-Sabi, A.; Rivier, J.E.F.; Olivera, B.M.; Terlau, H.; Carlomagno, T. Identification of a Novel Pharmacophore for Peptide Toxins Interacting with K+ Channels. J. Biol. Chem. 2005, 280, 21246–21255. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, S.; Finol-Urdaneta, R.K.; Köpfer, D.; Markushina, A.; Song, J.; French, R.J.; Kopec, W.; de Groot, B.L.; Giacobassi, M.J.; Leavitt, L.S.; et al. Conotoxin κM-RIIIJ, a tool targeting asymmetric heteromeric Kv1 channels. Proc. Natl. Acad. Sci. USA 2019, 116, 1059–1064. [Google Scholar] [CrossRef]
- Saikia, C.; Dym, O.; Altman-Gueta, H.; Gordon, D.; Reuveny, E.; Karbat, I. A Molecular Lid Mechanism of K+ Channel Blocker Action Revealed by a Cone Peptide. J. Mol. Biol. 2021, 433, 166957. [Google Scholar] [CrossRef] [PubMed]
- Karbat, I.; Altman-Gueta, H.; Fine, S.; Szanto, T.; Hamer-Rogotner, S.; Dym, O.; Frolow, F.; Gordon, D.; Panyi, G.; Gurevitz, M.; et al. Pore-modulating toxins exploit inherent slow inactivation to block K+ channels. Proc. Natl. Acad. Sci. USA 2019, 116, 18700–18709. [Google Scholar] [CrossRef] [PubMed]
- Imperial, J.S.; Chen, P.; Sporning, A.; Terlau, H.; Daly, N.L.; Craik, D.J.; Alewood, P.F.; Olivera, B.M. Tyrosine-rich conopeptides affect voltage-gated K+ channels. J. Biol. Chem. 2008, 283, 23026–23032. [Google Scholar] [CrossRef] [PubMed]
- Imperial, J.S.; Bansal, P.S.; Alewood, P.F.; Daly, N.L.; Craik, D.J.; Sporning, A.; Terlau, H.; López-Vera, E.; Bandyopadhyay, P.K.; Olivera, B.M. A novel conotoxin inhibitor of Kv1.6 channel and nAChR subtypes defines a new superfamily of conotoxins. Biochemistry 2006, 45, 8331–8340. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, M.B.; Pérez-Reyes, L.I.; López, Z.; de la Cotera, E.P.H.; Falcón, A.; Ayala, C.; Galván, M.; Salvador, C.; Escobar, L.I. Peptide sr11a from Conus spurius is a novel peptide blocker for Kv1 potassium channels. Peptides 2010, 31, 1287–1291. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, M.B.; López-Vera, E.; de la Cotera, E.P.H.; Falcón, A.; Olivera, B.M.; Maillo, M. I-conotoxins in vermivorous species of the West Atlantic: Peptide sr11a from Conus spurius. Peptides 2007, 28, 18–23. [Google Scholar] [CrossRef]
- Scanlon, M.J.; Naranjo, D.; Thomas, L.; Alewood, P.F.; Lewis, R.J.; Craik, D.J. Solution structure and proposed binding mechanism of a novel potassium channel toxin κ-conotoxin PVIIA. Structure 1997, 5, 1585–1597. [Google Scholar] [CrossRef]
- Koch, E.D.; Olivera, B.M.; Terlau, H.; Conti, F. The binding of kappa-Conotoxin PVIIA and fast C-type inactivation of Shaker K+ channels are mutually exclusive. Biophys. J. 2004, 86, 191–209. [Google Scholar] [CrossRef]
- Savarin, P.; Guenneugues, M.; Gilquin, B.; Lamthanh, H.; Gasparini, S.; Zinn-Justin, S.; Ménez, A. Three-Dimensional Structure of κ-Conotoxin PVIIA, a Novel Potassium Channel-Blocking Toxin from Cone Snails. Biochemistry 1998, 37, 5407–5416. [Google Scholar] [CrossRef] [PubMed]
- García, E.; Scanlon, M.; Naranjo, D. A Marine Snail Neurotoxin Shares with Scorpion Toxins a Convergent Mechanism of Blockade on the Pore of Voltage-Gated K Channels. J. Gen. Physiol. 1999, 114, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Dauplais, M.; Gilquin, B.; Possani, L.D.; Gurrola-Briones, G.; Roumestand, C.; Menez, A. Determination of the Three-Dimensional Solution Structure of Noxiustoxin: Analysis of Structural Differences with Related Short-Chain Scorpion Toxins. Biochemistry 2002, 34, 16563–16573. [Google Scholar] [CrossRef] [PubMed]
- Dauplais, M.; Lecoq, A.; Song, J.; Cotton, J.; Jamin, N.; Gilquin, B.; Roumestand, C.; Vita, C.; de Medeiros, C.C.; Rowan, E.G.; et al. On the Convergent Evolution of Animal Toxins. J. Biol. Chem. 1997, 272, 4302–4309. [Google Scholar] [CrossRef] [PubMed]
- Imredy, J.P.; MacKinnon, R. Energetic and structural interactions between δ-dendrotoxin and a voltage-gated potassium channel 1 1Edited by G. von Heijne. J. Mol. Biol. 2000, 296, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Stampe, P.; Kolmakova-Partensky, L.; Miller, C. Intimations of potassium channel structure from a complete functional map of the molecular surface of charybdotoxin. Biochemistry 2002, 33, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Wang, K.; Dai, X.; Cao, Y.; Liu, S.; Jiang, H.; Fan, C.; Wu, W.; Chen, J. The Role of Individual Disulfide Bonds of μ-Conotoxin GIIIA in the Inhibition of NaV1.4. Mar. Drugs 2016, 14, 213. [Google Scholar] [CrossRef]
- Ferber, M.; Sporning, A.; Jeserich, G.; DeLaCruz, R.; Watkins, M.; Olivera, B.M.; Terlau, H. A Novel Conus Peptide Ligand for K+Channels. J. Biol. Chem. 2003, 278, 2177–2183. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Jeserich, G. Molecular structure and expression of shaker type potassium channels in glial cells of trout CNS. J. Neurosci. Res. 1998, 51, 284–292. [Google Scholar] [CrossRef]
- Ferber, M.; Al-Sabi, A.; Stocker, M.; Olivera, B.M.; Terlau, H. Identification of a mammalian target of κM-conotoxin RIIIK. Toxicon 2004, 43, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Bayrhuber, M.; Vijayan, V.; Ferber, M.; Graf, R.; Korukottu, J.; Imperial, J.; Garrett, J.E.; Olivera, B.M.; Terlau, H.; Zweckstetter, M.; et al. Conkunitzin-S1 Is the First Member of a New Kunitz-type Neurotoxin Family. J. Biol. Chem. 2005, 280, 23766–23770. [Google Scholar] [CrossRef] [PubMed]
- Finol-Urdaneta, R.K.; Remedi, M.S.; Raasch, W.; Becker, S.; Clark, R.B.; Strüver, N.; Pavlov, E.; Nichols, C.G.; French, R.J.; Terlau, H. Block of Kv1.7 potassium currents increases glucose-stimulated insulin secretion. EMBO Mol. Med. 2012, 4, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Finol-Urdaneta, R.K.; Belovanovic, A.; Micic-Vicovac, M.; Kinsella, G.K.; McArthur, J.R.; Al-Sabi, A. Marine Toxins Targeting Kv1 Channels: Pharmacological Tools and Therapeutic Scaffolds. Mar. Drugs 2020, 18, 173. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus Venom Peptide Pharmacology. Pharmacol. Rev. 2012, 64, 259–298. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.S.; Guerrero, A.J.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Nakamura, F.; Fusetani, N. Marine Pharmacology in 2016–2017: Marine Compounds with Antibacterial, Antidiabetic, Antifungal, Anti-Inflammatory, Antiprotozoal, Antituberculosis and Antiviral Activities; Affecting the Immune and Nervous Systems, and Other Miscellaneous Mechanisms of Action. Mar. Drugs 2021, 19, 49. [Google Scholar] [CrossRef]
- Jakubowski, J.A.; Kelley, W.P.; Sweedler, J.V.; Gilly, W.F.; Schulz, J.R. Intraspecific variation of venom injected by fish-hunting Conus snails. J. Exp. Biol. 2005, 208, 2873–2883. [Google Scholar] [CrossRef] [PubMed]
- Madio, B.; King, G.F.; Undheim, E.A.B. Sea Anemone Toxins: A Structural Overview. Mar. Drugs 2019, 17, 325. [Google Scholar] [CrossRef]
- Chintiroglou, C.; Koukouras, A. The feeding habits of three Mediterranean sea anemone species, Anemonia viridis (Forskål),Actinia equina (Linnaeus) andCereus pedunculatus (Pennant). Helgoländer Meeresunters. 1992, 46, 53–68. [Google Scholar] [CrossRef]
- Logashina, Y.A.; Solstad, R.G.; Mineev, K.S.; Korolkova, Y.V.; Mosharova, I.V.; Dyachenko, I.A.; Palikov, V.A.; Palikova, Y.A.; Murashev, A.N.; Arseniev, A.S.; et al. New Disulfide-Stabilized Fold Provides Sea Anemone Peptide to Exhibit Both Antimicrobial and TRPA1 Potentiating Properties. Toxins 2017, 9, 154. [Google Scholar] [CrossRef]
- Nevalainen, T.J.; Peuravuori, H.J.; Quinn, R.J.; Llewellyn, L.E.; Benzie, J.A.H.; Fenner, P.J.; Winkel, K.D. Phospholipase A2 in Cnidaria. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 139, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Maček, P. Cytolytic peptide and protein toxins from sea anemones (Anthozoa: Actiniaria). Toxicon 2002, 40, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.S. Structure and structure-function relationships of sea anemone proteins that interact with the sodium channel. Toxicon 1991, 29, 1051–1084. [Google Scholar] [CrossRef] [PubMed]
- Jouiaei, M.; Yanagihara, A.; Madio, B.; Nevalainen, T.; Alewood, P.; Fry, B. Ancient Venom Systems: A Review on Cnidaria Toxins. Toxins 2015, 7, 2251–2271. [Google Scholar] [CrossRef] [PubMed]
- Rees, B.; Bilwes, A. Three-dimensional structures of neurotoxins and cardiotoxins. Chem. Res. Toxicol. 2002, 6, 385–406. [Google Scholar] [CrossRef] [PubMed]
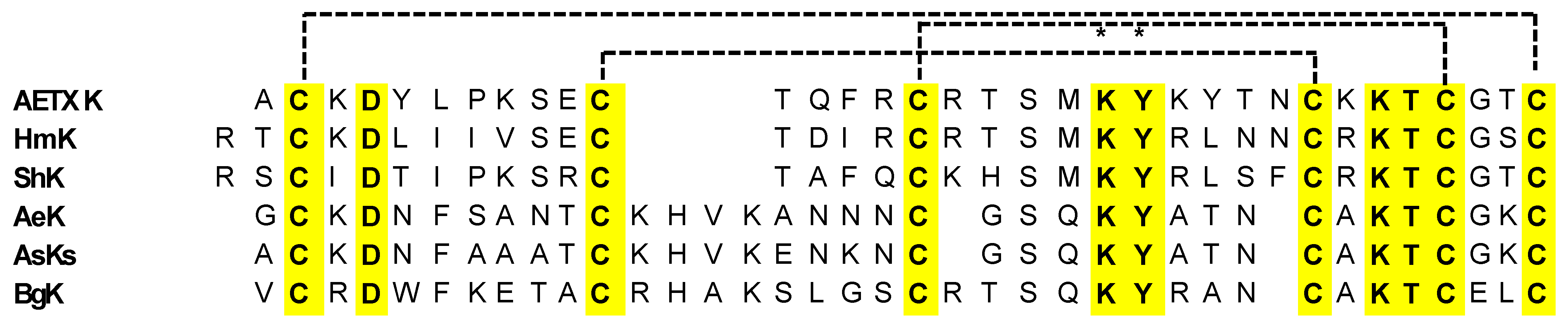
- Pennington, M.W.; Byrnes, M.E.; Zaydenberg, I.; Khaytin, I.; De Chastonay, J.; Krafte, D.S.; Hill, R.; Mahnir, V.M.; Volberg, W.A.; Gorczyca, W.; et al. Chemical synthesis and characterization of ShK toxin: A potent potassium channel inhibitor from a sea anemone. Int. J. Pept. Protein Res. 2009, 46, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Diochot, S.; Schweitz, H.; Béress, L.; Lazdunski, M. Sea Anemone Peptides with a Specific Blocking Activity against the Fast Inactivating Potassium Channel Kv3.4. J. Biol. Chem. 1998, 273, 6744–6749. [Google Scholar] [CrossRef] [PubMed]
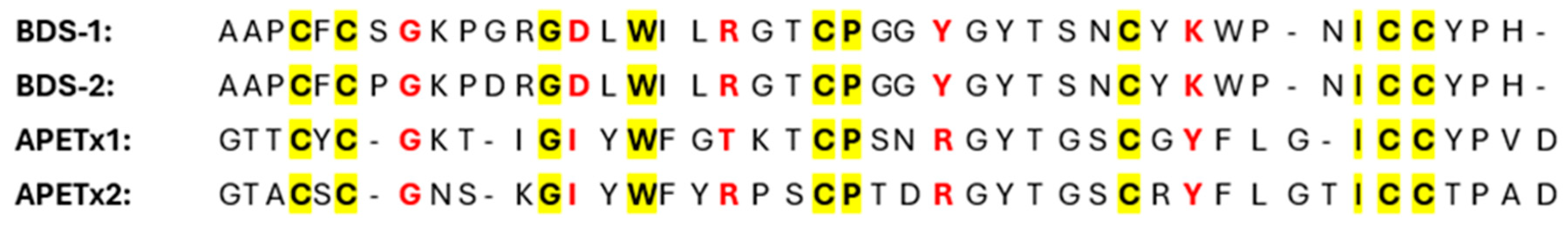
- Yeung, S.Y.M.; Thompson, D.; Wang, Z.; Fedida, D.; Robertson, B. Modulation of Kv3 Subfamily Potassium Currents by the Sea Anemone Toxin BDS: Significance for CNS and Biophysical Studies. J. Neurosci. 2005, 25, 8735–8745. [Google Scholar] [CrossRef] [PubMed]
- Diochot, S.; Loret, E.; Bruhn, T.; Béress, L.; Lazdunski, M. APETx1, a New Toxin from the Sea Anemone Anthopleura elegantissima, Blocks Voltage-Gated HumanEther-a-go-go–Related Gene Potassium Channels. Mol. Pharmacol. 2003, 64, 59–69. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, X.S.; Diochot, S.; Lazdunski, M.; Tseng, G.N. APETx1 from Sea Anemone Anthopleura elegantissima Is a Gating Modifier Peptide Toxin of the Human Ether-a-go-go- Related Potassium Channel. Mol. Pharmacol. 2007, 72, 259–268. [Google Scholar] [CrossRef]
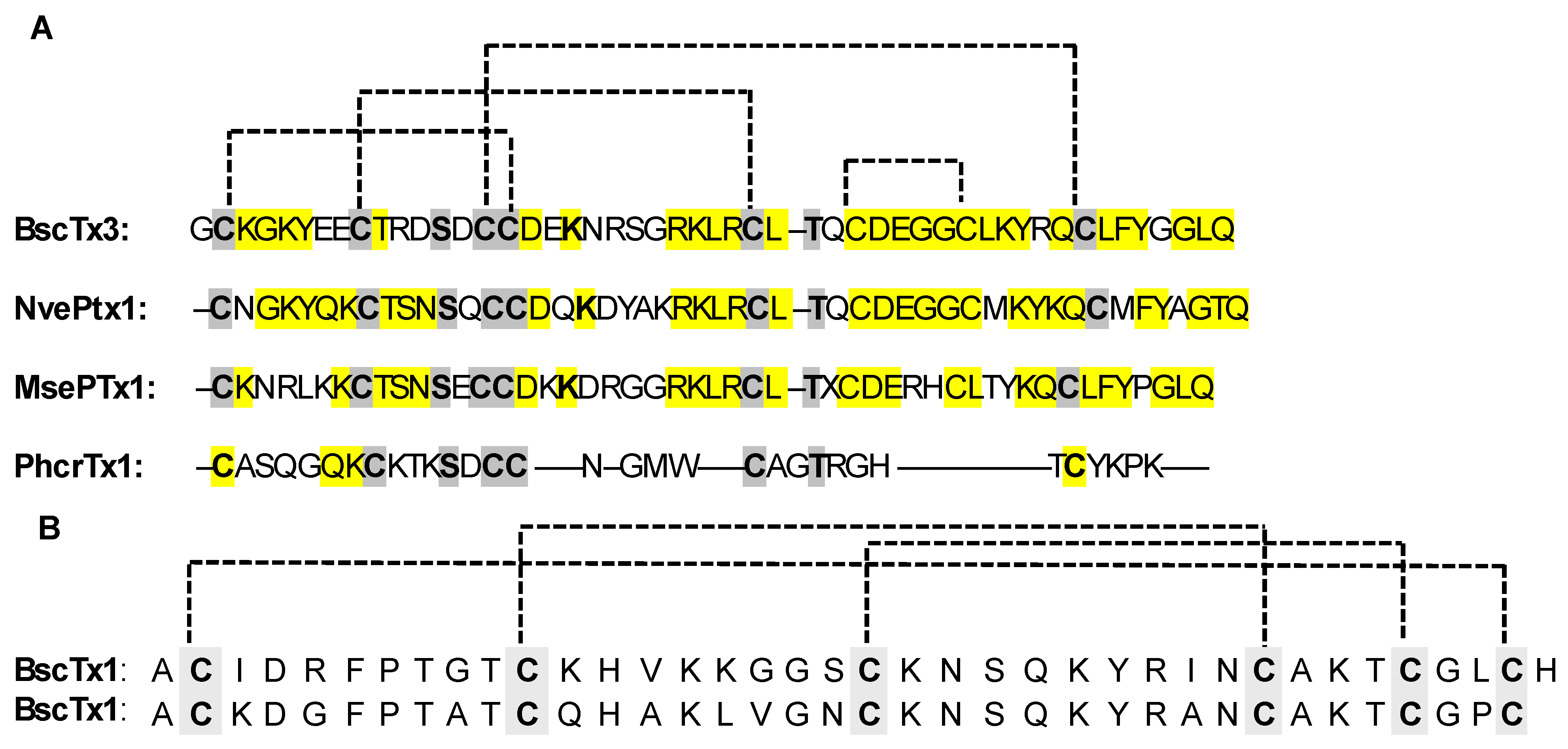
- Orts, D.J.; Peigneur, S.; Madio, B.; Cassoli, J.S.; Montandon, G.G.; Pimenta, A.M.; Bicudo, J.E.; Freitas, J.C.; Zaharenko, A.J.; Tytgat, J. Biochemical and electrophysiological characterization of two sea anemone type 1 potassium toxins from a geographically distant population of Bunodosoma caissarum. Mar. Drugs 2013, 11, 655–679. [Google Scholar] [CrossRef] [PubMed]
- Orts, D.J.B.; Moran, Y.; Cologna, C.T.; Peigneur, S.; Madio, B.; Praher, D.; Quinton, L.; De Pauw, E.; Bicudo, J.E.P.W.; Tytgat, J.; et al. BcsTx3 is a founder of a novel sea anemone toxin family of potassium channel blocker. FEBS J. 2013, 280, 4839–4852. [Google Scholar] [CrossRef] [PubMed]
- Schweitz, H.; Bruhn, T.; Guillemare, E.; Moinier, D.; Lancelin, J.M.; Béress, L.; Lazdunski, M. Kalicludines and kaliseptine. Two different classes of sea anemone toxins for voltage sensitive K+ channels. J. Biol. Chem. 1995, 270, 25121–25126. [Google Scholar] [CrossRef] [PubMed]
- Peigneur, S.; Billen, B.; Derua, R.; Waelkens, E.; Debaveye, S.; Béress, L.; Tytgat, J. A bifunctional sea anemone peptide with Kunitz type protease and potassium channel inhibiting properties. Biochem. Pharmacol. 2011, 82, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Madio, B.; Peigneur, S.; Chin, Y.K.Y.; Hamilton, B.R.; Henriques, S.T.; Smith, J.J.; Cristofori-Armstrong, B.; Dekan, Z.; Boughton, B.A.; Alewood, P.F.; et al. PHAB toxins: A unique family of predatory sea anemone toxins evolving via intra-gene concerted evolution defines a new peptide fold. Cell. Mol. Life Sci. 2018, 75, 4511–4524. [Google Scholar] [CrossRef] [PubMed]
- Orts, D.J.B.; Peigneur, S.; Silva-Gonçalves, L.C.; Arcisio-Miranda, M.; PWBicudo, J.E.; Tytgat, J. AbeTx1 Is a Novel Sea Anemone Toxin with a Dual Mechanism of Action on Shaker-Type K+ Channels Activation. Mar. Drugs 2018, 16, 360. [Google Scholar] [CrossRef] [PubMed]
- Racapé, J.; Lecoq, A.; Romi-Lebrun, R.; Liu, J.; Kohler, M.; Garcia, M.L.; Ménez, A.; Gasparini, S. Characterization of a novel radiolabeled peptide selective for a subpopulation of voltage-gated potassium channels in mammalian brain. J. Biol. Chem. 2002, 277, 3886–3893. [Google Scholar] [CrossRef] [PubMed]
- Zidar, N.; Žula, A.; Tomašič, T.; Rogers, M.; Kirby, R.W.; Tytgat, J.; Peigneur, S.; Kikelj, D.; Ilaš, J.; Mašič, L.P. Clathrodin, hymenidin and oroidin, and their synthetic analogues as inhibitors of the voltage-gated potassium channels. Eur. J. Med. Chem. 2017, 139, 232–341. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Honma, T.; Nagai, H.; Ishida, M.; Nagashima, Y.; Shiomi, K. Isolation and cDNA cloning of a potassium channel peptide toxin from the sea anemone Anemonia erythraea. Toxicon 2006, 48, 536–542. [Google Scholar] [CrossRef]
- Minagawa, S.; Ishida, M.; Nagashima, Y.; Shiomi, K. Primary structure of a potassium channel toxin from the sea anemone Actinia equina. FEBS Lett. 1998, 427, 149–151. [Google Scholar] [CrossRef]
- Smith, J.J.; Blumenthal, K.M. Site-3 sea anemone toxins: Molecular probes of gating mechanisms in voltage-dependent sodium channels. Toxicon 2007, 49, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Fu, J.; Yuan, L.; Liao, Y.; Li, M.; Li, X.; Yi, B.; Zhang, J.; Gao, B. Diversity analysis of sea anemone peptide toxins in different tissues of Heteractis crispa based on transcriptomics. Sci. Rep. 2024, 14, 7684. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Jo, S.; Bean, B.P. Modulation of neuronal sodium channels by the sea anemone peptide BDS-I. J. Neurophysiol. 2012, 107, 3155–3167. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Huang, J.; Yuan, X.; Peng, B.; Liu, W.; Han, S.; He, X. Toxins Targeting the KV1.3 Channel: Potential Immunomodulators for Autoimmune Diseases. Toxins 2015, 7, 1749–1764. [Google Scholar] [CrossRef] [PubMed]
- Fusetani, N.; Kem, W. (Eds.) Marine Toxins as Research Tools; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Pannaccione, A.; Piccialli, I.; Ciccone, R. The new KV3.4 inhibitor BDS-I [1–8] as a potential pharmacological opportunity in Alzheimer’s disease therapy. Neural Regen. Res. 2020, 15, 1255–1256. [Google Scholar] [CrossRef] [PubMed]
- Baranauskas, G.; Tkatch, T.; Nagata, K.; Yeh, J.Z.; Surmeier, D.J. Kv3.4 subunits enhance the repolarizing efficiency of Kv3.1 channels in fast-spiking neurons. Nat. Neurosci. 2003, 6, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Angulo, E.; Noé, V.; Casadó, V.; Mallol, J.; Gomez-Isla, T.; Lluis, C.; Ferrer, I.; Ciudad, C.J.; Franco, R. Up-regulation of the Kv3.4 potassium channel subunit in early stages of Alzheimer’s disease. J. Neurochem. 2004, 91, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Iraci, N.; Carotenuto, L.; Ciaglia, T.; Belperio, G.; Di Matteo, F.; Mosca, I.; Carleo, G.; Basilicata, G.M.; Ambrosino, P.; Turcio, R.; et al. In silico assisted identification, synthesis and in vitro pharmacological characterization of potent and selective blockers of the epilepsy-associated KCNT1 channel. J. Med. Chem. 2024, 67, 9124–9149. [Google Scholar] [CrossRef] [PubMed]
- Gustina, A.S.; Trudeau, M.C. hERG potassium channel gating is mediated by N- and C-terminal region interactions. J. Gen. Physiol. 2011, 137, 315–325. [Google Scholar] [CrossRef]
- Chagot, B.; Escoubas, P.; Diochot, S.; Bernard, C.; Lazdunski, M.; Darbon, H. Solution structure of APETx2, a specific peptide inhibitor of ASIC3 proton-gated channels. Protein Sci. 2009, 14, 2003–2010. [Google Scholar] [CrossRef]
- Matsumura, K.; Shimomura, T.; Kubo, Y.; Takayuki, O.; Kobayashi, N.; Imai, S.; Yanase, N.; Akimoto, M.; Fukuda, M.; Yokogawa, M.; et al. Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization. BMC Mol. Cell. Biol. 2021, 22, 3. [Google Scholar] [CrossRef]
- Torres, A.M.; Kuchel, P.W. The β-defensin-fold family of polypeptides. Toxicon 2004, 44, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Pignataro, G.; Li, M.; Chang, S.; Simon, R. Acid-sensing ion channels (ASICs) as pharmacological targets for neurodegenerative diseases. Curr. Opin. Pharmacol. 2008, 8, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.B.; Mobli, M.; King, G.F. Toxin structures as evolutionary tools: Using conserved 3D folds to study the evolution of rapidly evolving peptides. BioEssays 2016, 38, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.A.; Salceda, E.; Garateix, A.G.; Zaharenko, A.J.; Peigneur, S.; López, O.; Pons, T.; Richardson, M.; Díaz, M.; Hernández, Y.; et al. A novel sea anemone peptide that inhibits acid-sensing ion channels. Peptides 2014, 53, 3–12. [Google Scholar] [CrossRef]
- Ranasinghe, S.; McManus, D.P. Structure and function of invertebrate Kunitz serine protease inhibitors. Dev. Comp. Immunol. 2013, 39, 219–227. [Google Scholar] [CrossRef]
- Rauer, H.; Pennington, M.; Cahalan, M.; Chandy, K.G. Structural Conservation of the Pores of Calcium-activated and Voltage-gated Potassium Channels Determined by a Sea Anemone Toxin. J. Biol. Chem. 1999, 274, 21885–21892. [Google Scholar] [CrossRef]
- Dy, C.Y.; Buczek, P.; Imperial, J.S.; Bulaj, G.; Horvath, M.P. Structure of conkunitzin-S1, a neurotoxin and Kunitz-fold disulfide variant from cone snail. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 980–990. [Google Scholar] [CrossRef]
- Lintermans, L.L.; Stegeman, C.A.; Muñoz-Elías, E.J.; Tarcha, E.J.; Iadonato, S.P.; Rutgers, A.; Heeringa, P.; Abdulahad, W.H. Kv1.3 blockade by ShK186 modulates CD4+ effector memory T-cell activity of patients with granulomatosis with polyangiitis. Rheumatology 2024, 63, 198–208. [Google Scholar] [CrossRef]
- Tarcha, E.J.; Chi, V.; Muñoz-Elías, E.J.; Bailey, D.; Londono, L.M.; Upadhyay, S.K.; Norton, K.; Banks, A.; Tjong, I.; Nguyen, H.; et al. Durable Pharmacological Responses from the Peptide ShK-186, a Specific Kv1.3 Channel Inhibitor That Suppresses T Cell Mediators of Autoimmune Disease. J. Pharmacol. Exp. Ther. 2012, 342, 642–653. [Google Scholar] [CrossRef]
- Tyagi, A.; Ahmed, T.; Jian, S.; Bajaj, S.; Ong, S.T.; Goay, S.S.M.; Zhao, Y.; Vorobyov, I.; Tian, C.; Chandy, K.G.; et al. Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug. Proc. Natl. Acad. Sci. USA 2022, 119, e2113536119. [Google Scholar] [CrossRef]
- Tarcha, E.J.; Olsen, C.M.; Probst, P.; Peckham, D.; Muñoz-Elías, E.J.; Kruger, J.G.; Iadonato, S.P. Safety and pharmacodynamics of dalazatide, a Kv1.3 channel inhibitor, in the treatment of plaque psoriasis: A randomized phase 1b trial. PLoS ONE 2017, 12, e0180762. [Google Scholar] [CrossRef]
- Chang, S.C.; Huq, R.; Chhabra, S.; Beeton, C.; Pennington, M.W.; Smith, B.J.; Norton, R.S. N-Terminally extended analogues of the K⁺ channel toxin from Stichodactyla helianthus as potent and selective blockers of the voltage-gated potassium channel Kv1.3. FEBS J. 2015, 282, 2247–2259. [Google Scholar] [CrossRef]
- Pennington, M.W.; Harunur Rashid, M.; Tajhya, R.B.; Beeton, C.; Kuyucak, S.; Norton, R.S. A C-terminally amidated analogue of ShK is a potent and selective blocker of the voltage-gated potassium channel Kv1.3. FEBS Lett. 2012, 586, 3996–4001. [Google Scholar] [CrossRef]
- Pennington, M.W.; Beeton, C.; Galea, C.A.; Smith, B.J.; Chi, V.; Monaghan, K.P.; Garcia, A.; Rangaraju, S.; Giuffrida, A.; Plank, D.; et al. Engineering a stable and selective peptide blocker of the Kv1.3 channel in T lymphocytes. Mol. Pharmacol. 2009, 75, 762–773. [Google Scholar] [CrossRef]
- Rashid, M.H.; Heinzelmann, G.; Huq, R.; Tajhya, R.B.; Chang, S.C.; Chhabra, S.; Pennington, M.W.; Beeton, C.; Norton, R.S.; Kuyucak, S. A potent and selective peptide blocker of the Kv1.3 channel: Prediction from free-energy simulations and experimental confirmation. PLoS ONE 2013, 8, e78712. [Google Scholar] [CrossRef]
- Rashid, M.H.; Kuyucak, S. Affinity and selectivity of ShK toxin for the Kv1 potassium channels from free energy simulations. J. Phys. Chem. B 2012, 116, 4812–4822. [Google Scholar] [CrossRef]
- Selvakumar, P.; Fernández-Mariño, A.I.; Khanra, N.; He, C.; Paquette, A.J.; Wang, B.; Huang, R.; Smider, V.V.; Rice, W.J.; Swartz, K.J.; et al. Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators. Nat. Commun. 2022, 13, 3854. [Google Scholar] [CrossRef]
- Chandy, K.G.; Sanches, K.; Norton, R.S. Structure of the voltage-gated potassium channel KV1.3: Insights into the inactivated conformation and binding to therapeutic leads. Channels 2023, 17, 2253104. [Google Scholar] [CrossRef]
- Cotton, J.; Crest, M.; Bouet, F.; Alessandri, N.; Gola, M.; Forest, E.; Karlsson, E.; Castañeda, O.; Harvey, A.L.; Vita, C.; et al. A potassium-channel toxin from the sea anemone Bunodosoma granulifera, an inhibitor for Kv1 channels. Revision of the amino acid sequence, disulfide-bridge assignment, chemical synthesis, and biological activity. Eur. J. Biochem. 1997, 244, 192–202. [Google Scholar] [CrossRef]
- Beraud, E.; Viola, A.; Regaya, I.; Confort-Gouny, S.; Siaud, P.; Ibarrola, D.; Le Fur, Y.; Barbaria, J.; Pellissier, J.F.; Sabatier, J.M.; et al. Block of neural Kv1.1 potassium channels for neuroinflammatory disease therapy. Ann. Neurol. 2006, 60, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Gilquin, B.; Braud, S.; Eriksson, M.A.; Roux, B.; Bailey, T.D.; Priest, B.T.; Garcia, M.L.; Ménez, A.; Gasparini, S. A variable residue in the pore of Kv1 channels is critical for the high affinity of blockers from sea anemones and scorpions. J. Biol. Chem. 2005, 280, 27093–27102. [Google Scholar] [CrossRef] [PubMed]
- Gendeh, G.S.; Young, L.C.; de Medeiros, C.L.; Jeyaseelan, K.; Harvey, A.L.; Chung, M.C. A new potassium channel toxin from the sea anemone Heteractis magnifica: Isolation, cDNA cloning, and functional expression. Biochemistry 1997, 36, 11461–11471. [Google Scholar] [CrossRef] [PubMed]
- Sanches, K.; Prypoten, V.; Chandy, K.G.; Chalmers, D.K.; Norton, R.S. Interaction of the Inhibitory Peptides ShK and HmK with the Voltage-Gated Potassium Channel KV1.3: Role of Conformational Dynamics. J. Chem. Inf. Model. 2023, 63, 3043–3053. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, R.; Peigneur, S.; Pons, T.; Alvarez, C.; González, L.; Chávez, M.; Tytgat, J. The Kunitz-Type Protein ShPI-1 Inhibits Serine Proteases and Voltage-Gated Potassium Channels. Toxins 2016, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L. Recent studies on dendrotoxins and potassium ion channels. Gen. Pharmacol. Vasc. Syst. 1997, 28, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Honma, T.; Kawahata, S.; Ishida, M.; Nagai, H.; Nagashima, Y.; Shiomi, K. Novel peptide toxins from the sea anemone Stichodactyla haddoni. Peptides 2008, 29, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, O.; Harvey, A.L. Discovery and characterization of cnidarian peptide toxins that affect neuronal potassium ion channels. Toxicon 2009, 54, 1119–1124. [Google Scholar] [CrossRef] [PubMed]
- Imperial, J.S.; Cabang, A.B.; Song, J.; Raghuraman, S.; Gajewiak, J.; Watkins, M.; Showers-Corneli, P.; Fedosov, A.; Concepcion, G.P.; Terlau, H.; et al. A family of excitatory peptide toxins from venomous crassispirine snails: Using Constellation Pharmacology to assess bioactivity. Toxicon 2014, 89, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Cabang, A.B.; Imperial, J.S.; Gajewiak, J.; Watkins, M.; Corneli, P.S.; Olivera, B.M.; Concepcion, G.P. Characterization of a venom peptide from a crassispirid gastropod. Toxicon 2011, 58, 672–680. [Google Scholar] [CrossRef]
- Carbone, D.; De Franco, M.; Pecoraro, C.; Bassani, D.; Pavan, M.; Cascioferro, S.; Parrino, B.; Cirrincione, G.; Dall’Acqua, S.; Sut, S.; et al. Structural Manipulations of Marine Natural Products Inspire a New Library of 3-Amino-1,2,4-Triazine PDK Inhibitors Endowed with Antitumor Activity in Pancreatic Ductal Adenocarcinoma. Mar. Drugs 2023, 21, 288. [Google Scholar] [CrossRef] [PubMed]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine Sponge Derived Natural Products between 2001 and 2010: Trends and Opportunities for Discovery of Bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [PubMed]
- Carbone, D.; Vestuto, V.; Ferraro, M.R.; Ciaglia, T.; Pecoraro, C.; Sommella, E.; Cascioferro, S.; Salviati, E.; Novi, S.; Tecce, M.F.; et al. Metabolomics-assisted discovery of a new anticancer GLS-1 inhibitor chemotype from a nortopsentin-inspired library: From phenotype screening to target identification. Eur. J. Med. Chem. 2022, 234, 114233. [Google Scholar] [CrossRef] [PubMed]
- Varijakzhan, D.; Loh, J.Y.; Yap, W.S.; Yusoff, K.; Seboussi, R.; Lim, S.E.; Lai, K.S.; Chong, C.M. Bioactive Compounds from Marine Sponges: Fundamentals and Applications. Mar. Drugs 2021, 19, 246. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Manconi, R.; Perino, E.; Pronzato, R. A new species of Agelas from the Zanzibar Archipelago, western Indian Ocean (Porifera, Demospongiae). ZooKeys 2016, 553, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Forenza, S.; Minale, L.; Riccio, R.; Fattorusso, E. New bromo-pyrrole derivatives from the sponge Agelas oroides. J. Chem. Soc. D Chem. Commun. 1971, 18, 1129–1130. [Google Scholar] [CrossRef]
- Morales, J.J.; Rodríguez, A.D. The Structure of Clathrodin, a Novel Alkaloid Isolated from the Caribbean Sea Sponge Agelas clathrodes. J. Nat. Prod. 2004, 54, 629–631. [Google Scholar] [CrossRef]
- Chu, M.-J.; Li, M.; Ma, H.; Li, P.-L.; Li, G.-Q. Secondary metabolites from marine sponges of the genus Agelas: A comprehensive update insight on structural diversity and bioactivity. RSC Adv. 2022, 12, 7789–7820. [Google Scholar] [CrossRef]
- Suciati; Fraser, J. A.; Lambert, L.K.; Pierens, G.K.; Bernhardt, P.V.; Garson, M.J. Secondary Metabolites of the Sponge-Derived Fungus Acremonium persicinum. J. Nat. Prod. 2013, 76, 1432–1440. [Google Scholar] [CrossRef]
- Kim, H.; Yang, I.; Ryu, S.-Y.; Won, D.H.; Giri, A.G.; Wang, W.; Choi, H.; Chin, J.; Hahn, D.; Kim, E.; et al. Acredinones A and B, Voltage-Dependent Potassium Channel Inhibitors from the Sponge-Derived Fungus Acremonium sp. F9A015. J. Nat. Prod. 2015, 78, 363–367. [Google Scholar] [CrossRef]
- Cuypers, E.; Abdel-Mottaleb, Y.; Kopljar, I.; Rainier, J.D.; Raes, A.L.; Snyders, D.J.; Tytgat, J. Gambierol, a toxin produced by the dinoflagellate Gambierdiscus toxicus, is a potent blocker of voltage-gated potassium channels. Toxicon 2008, 51, 974–983. [Google Scholar] [CrossRef]
- Lewis, R.J. Ciguatera: Australian perspectives on a global problem. Toxicon 2006, 48, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, G.M.; Lewis, R.J. Ciguatoxins: Cyclic Polyether Modulators of Voltage-gated Iion Channel Function. Mar. Drugs 2006, 4, 82–118. [Google Scholar] [CrossRef]
- Birinyi-Strachan, L.C.; Gunning, S.J.; Lewis, R.J.; Nicholson, G.M. Block of voltage-gated potassium channels by Pacific ciguatoxin-1 contributes to increased neuronal excitability in rat sensory neurons. Toxicol. Appl. Pharmacol. 2005, 204, 175–186. [Google Scholar] [CrossRef]
- Louzao, M.C.; Cagide, E.; Vieytes, M.R.; Sasaki, M.; Fuwa, H.; Yasumoto, T.; Botana, L.M. The sodium channel of human excitable cells is a target for gambierol. Cell Physiol. Biochem. 2006, 17, 257–268. [Google Scholar] [CrossRef]
- Kopljar, I.; Labro, A.J.; Cuypers, E.; Johnson, H.W.; Rainier, J.D.; Tytgat, J.; Snyders, D.J. A polyether biotoxin binding site on the lipid-exposed face of the pore domain of Kv channels revealed by the marine toxin gambierol. Proc. Natl. Acad. Sci. USA 2009, 106, 9896–9901. [Google Scholar] [CrossRef]
- Xie, C.; Kessi, M.; Yin, F.; Peng, J. Roles of KCNA2 in Neurological Diseases: From Physiology to Pathology. Mol. Neurobiol. 2024. Epub ahead of print. [Google Scholar] [CrossRef]
- Zhang, H.H.; Zhang, X.K.; Si, R.R.; Shen, S.C.; Liang, T.T.; Fan, T.T.; Chen, W.; Xu, L.H.; Han, B.N. Chemical and Biological Study of Novel Aplysiatoxin Derivatives from the Marine Cyanobacterium Lyngbya sp. Toxins 2020, 12, 733. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.T.; Zhang, H.H.; Tang, Y.H.; Zhang, F.Z.; Han, B.N. Two New Neo-debromoaplysiatoxins-A Pair of Stereoisomers Exhibiting Potent Kv1.5 Ion Channel Inhibition Activities. Mar. Drugs 2019, 17, 652. [Google Scholar] [CrossRef]





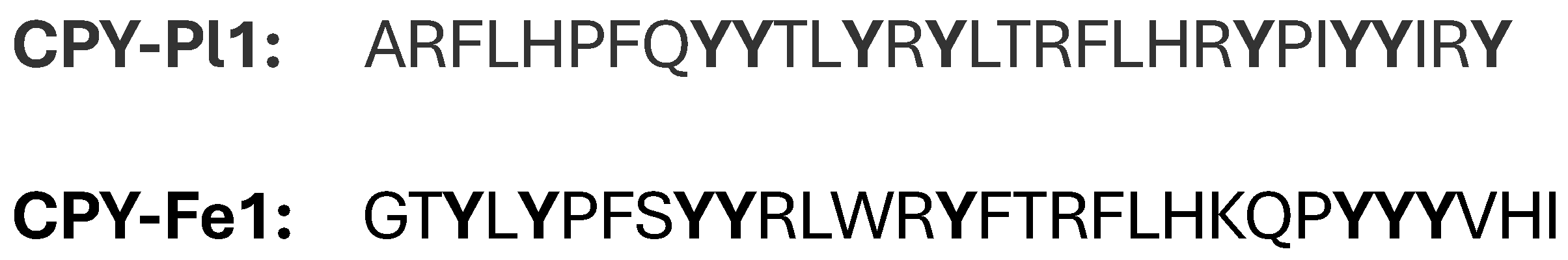





| Conopeptide | Target Channel | Potency or Efficacy | Reference(s) |
|---|---|---|---|
| κ-PVIIA | Fly Shaker | IC50= 60 nM | [39] |
| hKv1.1, hKv1.4 | IC50 = 1 µM | [47] | |
| Mo1659 | K+ currents in rat DRG | 29.6% current amplitude reduction at 200 nM | [48] |
| κ-BtX | KCa1.1 | 229% current amplitude increase at 10 nM (EC50= 0.7 nM) | [39] |
| ViTx | rKv1.1 | IC50= 1.59 μM | [49,50] |
| rKv1.3 | IC50 = 2.09 μM | [49,50] | |
| κA-SIVA | Fly Shaker | 54% current amplitude reduction at 100 nM | [50] |
| κM-RIIIK | hKv1.2 | IC50 = 352 nM | [51] |
| hKv1.2/1.7 | IC50 = 680 nM | [51] | |
| hKv1.2/1.5 and hKv1.2/1.1 | IC50 = 2.76 μM | [51] | |
| hKv1.2/1.1 | IC50 = 2.80 μM | [51] | |
| hKv1.2/1.6 | IC50 = 7.70 μM | [51] | |
| hTSha1 | IC50 = 20 nM (closed state) 60 nM at 0 mV | [52] | |
| κM-RIIIJ | hKv1.2 | IC50 = 33 nM | [53] |
| hKv1.2/1.1 | IC50 = 12 nM | [53] | |
| hKv1.2/1.3 | IC50 = 165 nM | [53] | |
| hKv1.2/1.4 | IC50 = 8.13 μM | [53] | |
| hKv1.2/1.5 | IC50 = 287 nM | [53] | |
| hKv1.2/1.6 | IC50 = 24 nM | [53] | |
| hKv1.2/1.7 | IC50 = 370 nM | [53] | |
| Conkunitzin-S1 | Fly Shaker | IC50 = 502 nM | [54] |
| hKv1.2 | IC50 = 3.4 μM | [55] | |
| hKv1.7 | IC50= 439 nM | [55] | |
| hKv1.2–1.7 | IC50 = 180 nM | [55] | |
| hKv1.7–1.2 | IC50 = 390 nM | [55] | |
| hKv1.7–1.2 | IC50 = 390 nM | [55] | |
| CPY-Pl1 | Mammalian Kv1.2 | IC50 = 2.0 μM | [56] |
| Mammalian Kv 1.6 | IC50 = 170 nM | [56] | |
| CPY-Fe1 | Mammalian Kv 1.6 | IC50 = 8.8 nM | [56] |
| pl14a | Mammalian Kv1.6 | IC50 = 1.5 μM | [57] |
| sr11a | rKv1.2 | IC50 = 640 nM | [58,59] |
| hKv1.6 | IC50 = 640 nM | [58,59] |
| Sea Anemones Peptide | Target Channel | Potency, Efficacy or Affinity | Reference(s) |
|---|---|---|---|
| BDS-I | hKv3.4 | IC50 = 47 nM | [87] |
| mKv3.1 | IC50 = 220 nM | [88] | |
| rKv3.2 | 48.1% current amplitude inhibition at 500 nM (+40 mV) | [88] | |
| BDS-II | hKv3.4 | IC50 = 56 nM | [87] |
| mKv3.1 | IC50 = 750 nM | [88] | |
| rKv3.2 | 52.5% current amplitude inhibition at 500 nM (+40 mV) | [88] | |
| APETx1 | hERG1 | IC50 = 34 nM | [89] |
| hERG3 | Equally responsive as hERG1 (determined by Kd) | [90] | |
| BcsTx1 | rKv1.1 | IC50 = 405 nM | [91] |
| rKv1.2 | IC50 = 30 pM | [91] | |
| hKv1.3 | IC50 = 74 nM | [91] | |
| rKv1.6 | IC50 = 1.31 nM | [91] | |
| Shaker IR | IC50 = 275 nM | [91] | |
| BcsTx2 | rKv1.1 | IC50 = 14.4 nM | [91] |
| rKv1.2 | IC50 = 80.4 nM | [91] | |
| hKv1.3 | IC50 = 13.1 nM | [91] | |
| rKv1.6 | IC50 = 7.76 nM | [91] | |
| Shaker IR | IC50 = 49.1 nM | [91] | |
| BcsTx3 | rKv1.2 | IC50 = 172.6 nM | [92] |
| rKv1.3 | IC50 = 1.0 μM | [92] | |
| rKv1.6 | IC50 = 2.2 μM | [92] | |
| Shaker IR | IC50 = 94.2 nM | [92] | |
| AsKC1 | rKv1.2 | IC50 = 2.8 µM | [93] |
| AsKC2 | rKv1.2 | IC50 = 1.1 µM | [93] |
| AsKC3 | rKv1.2 | IC50 = 1.3 µM | [93] |
| APEKTx1 | rKv1.1 | IC50 = 0.9 nM | [94] |
| SHTX-III | rKv1 | Ki = 650 nM | [95] |
| AbeTx1 | rKv1.1 | IC50 = 672 nM | [96] |
| rKv1.2 | IC50 = 167 nM | [96] | |
| rKv1.6 | IC50 = 116 nM | [96] | |
| Ate1a | rKv1.1 | IC50 = 33 nM | [95] |
| rKv1.2 | IC50 = 12 nM | [95] | |
| hKv1.3 | IC50 = 3.0 μM | [95] | |
| rKv1.6 | IC50 = 191 nM | [95] | |
| BgK | rKv1.1 | Ki = 34 pM | [97] |
| rKv1.2 | Ki = 66 pM | [97] | |
| rKv1.3 | Ki = 777 pM | [97] | |
| rKv1.6 | Ki = 13 pM | [97] | |
| ShK | mKv1.1 | Ki = 16 pM | [98] |
| rKv1.2 | Ki = 9 nM | [98] | |
| mKv1.3 | Ki = 11 pM | [98] | |
| hKv1.6 | Ki = 165 pM | [98] | |
| mKv1.7 | Ki = 11.5 nM | [98] | |
| hKCa4 | Ki = 28 nM | [98] | |
| HmK | hKv1.2 | Ki = 2.5 nM | [99] |
| hKv1.3 | Ki = 3.1 nM | [99] | |
| Fly Shaker | Ki = 1.0 nM | [99] | |
| AsKs | rKv1.2 | IC50 = 140 nM | [93] |
| AeK | rKv1 | IC50 = 22 nM | [100] |
| AETXk | hKv1.2 | Ki = 2.2 µM | [99] |
| hKv1.3 | Ki = 1.3 µM | [99] | |
| Fly Shaker | Ki = 445 nM | [99] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turcio, R.; Di Matteo, F.; Capolupo, I.; Ciaglia, T.; Musella, S.; Di Chio, C.; Stagno, C.; Campiglia, P.; Bertamino, A.; Ostacolo, C. Voltage-Gated K+ Channel Modulation by Marine Toxins: Pharmacological Innovations and Therapeutic Opportunities. Mar. Drugs 2024, 22, 350. https://doi.org/10.3390/md22080350
Turcio R, Di Matteo F, Capolupo I, Ciaglia T, Musella S, Di Chio C, Stagno C, Campiglia P, Bertamino A, Ostacolo C. Voltage-Gated K+ Channel Modulation by Marine Toxins: Pharmacological Innovations and Therapeutic Opportunities. Marine Drugs. 2024; 22(8):350. https://doi.org/10.3390/md22080350
Chicago/Turabian StyleTurcio, Rita, Francesca Di Matteo, Ilaria Capolupo, Tania Ciaglia, Simona Musella, Carla Di Chio, Claudio Stagno, Pietro Campiglia, Alessia Bertamino, and Carmine Ostacolo. 2024. "Voltage-Gated K+ Channel Modulation by Marine Toxins: Pharmacological Innovations and Therapeutic Opportunities" Marine Drugs 22, no. 8: 350. https://doi.org/10.3390/md22080350
APA StyleTurcio, R., Di Matteo, F., Capolupo, I., Ciaglia, T., Musella, S., Di Chio, C., Stagno, C., Campiglia, P., Bertamino, A., & Ostacolo, C. (2024). Voltage-Gated K+ Channel Modulation by Marine Toxins: Pharmacological Innovations and Therapeutic Opportunities. Marine Drugs, 22(8), 350. https://doi.org/10.3390/md22080350








