Discovery of Prenyltransferase-Guided Hydroxyphenylacetic Acid Derivatives from Marine Fungus Penicillium sp. W21C371
Abstract
1. Introduction
2. Results
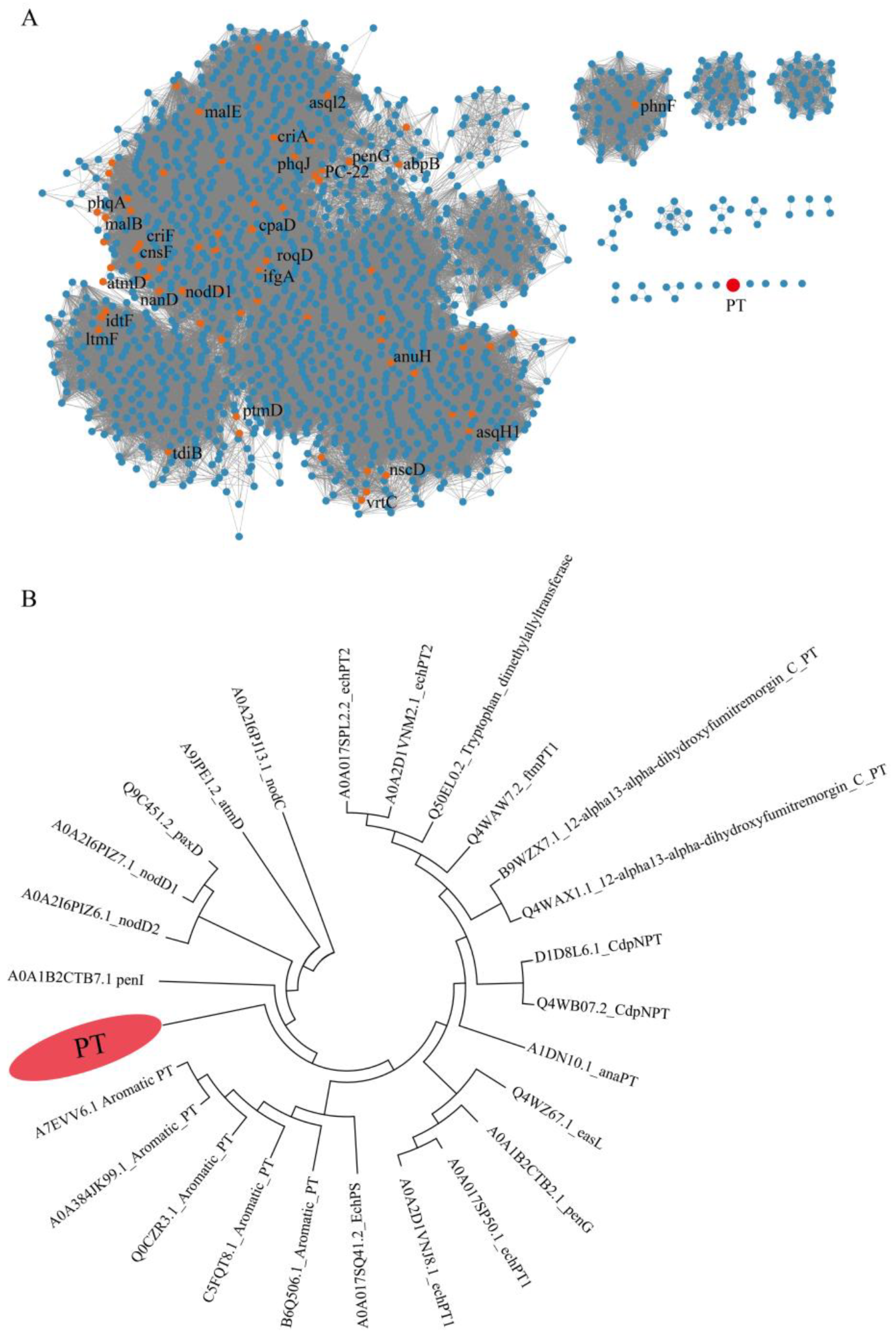
2.1. Bioinformatics Analysis and Phylogenetic Tree Construction of PTs
2.2. Structure Elucidation
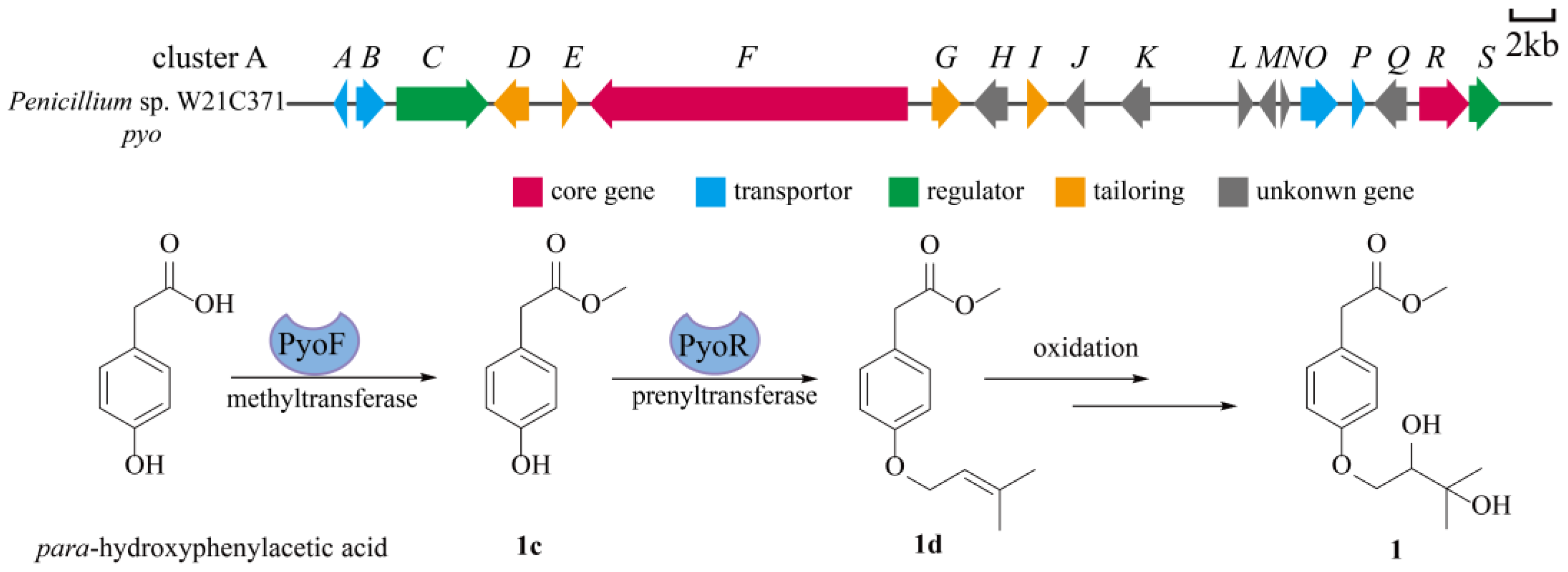
2.3. Plausible Biosynthetic Pathways of Peniprenydiol A
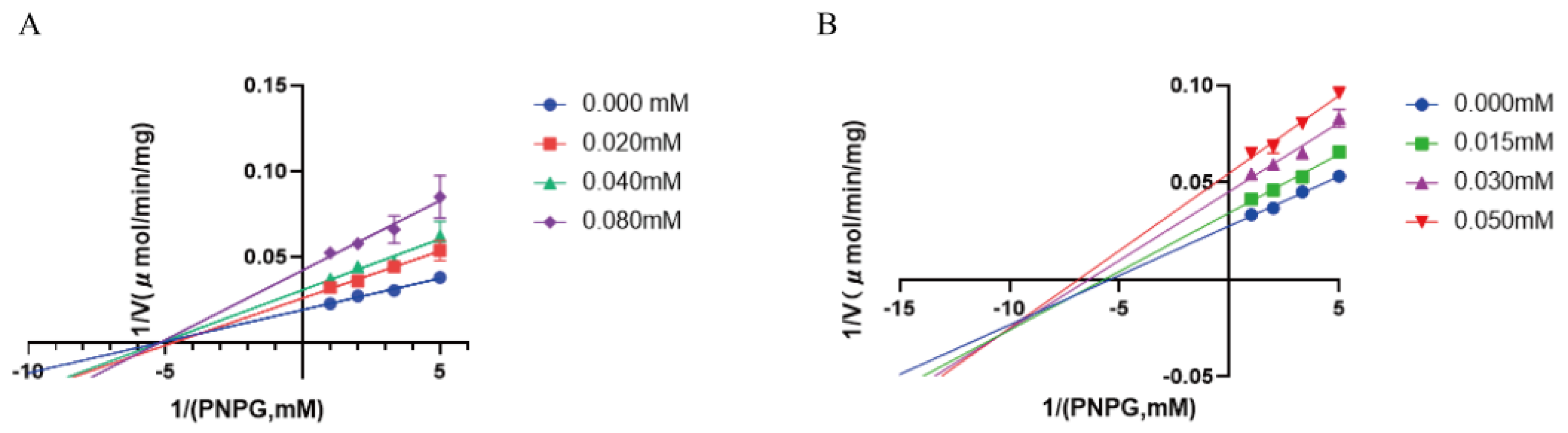
2.4. EcGUS Inhibition Assay
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Bioinformatics Analysis and Phylogenetic Tree Construction of PTs
3.4. Fermentation and Extraction
3.5. Isolation and Purification
3.6. Spectroscopic Data
3.7. Preparation of (R)- and (S)-MTPA Esters of 1b
3.8. Bioassay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Perrone, G.; Susca, A. Penicillium species and their associated mycotoxins. Methods Mol. Biol. 2017, 1542, 107–119. [Google Scholar]
- Petersen, C.; Sørensen, T.; Nielsen, M.R.; Sondergaard, T.E.; Sørensen, J.L.; Fitzpatrick, D.A.; Frisvad, J.C.; Nielsen, K.L. Comparative genomic study of the Penicillium genus elucidates a diverse pangenome and 15 lateral gene transfer events. IMA Fungus 2023, 14, 3. [Google Scholar] [CrossRef]
- Houbraken, J.; Frisvad, J.C.; Samson, R.A. Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus 2011, 2, 87–95. [Google Scholar] [CrossRef]
- Nielsen, J.C.; Grijseels, S.; Prigent, S.; Ji, B.; Dainat, J.; Nielsen, K.F.; Frisvad, J.C.; Workman, M.; Nielsen, J. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat. Microbiol. 2017, 2, 17044–17053. [Google Scholar] [CrossRef]
- Shu, W.S.; Huang, L.N. Microbial diversity in extreme environments. Nat. Rev. Microbiol. 2022, 20, 219–235. [Google Scholar] [CrossRef]
- König, G.M.; Kehraus, S.; Seibert, S.F.; Abdel-Lateff, A.; Müller, D. Natural products from marine organisms and their associated microbes. Chembiochem. 2006, 7, 229–238. [Google Scholar] [CrossRef]
- Ma, H.G.; Liu, Q.; Zhu, G.L.; Liu, H.S.; Zhu, W.M. Marine natural products sourced from marine-derived Penicillium fungi. J. Asian Nat. Prod. Res. 2016, 18, 92–115. [Google Scholar] [CrossRef]
- Liu, S.; Su, M.; Song, S.J.; Jung, J.H. Marine-derived Penicillium species as producers of cytotoxic metabolites. Mar. Drugs 2017, 15, 329. [Google Scholar] [CrossRef]
- Yang, X.; Liu, J.; Mei, J.; Jiang, R.; Tu, S.; Deng, H.; Liu, J.; Yang, S.; Li, J. Origins, structures, and bioactivities of secondary metabolites from marine-derived Penicillium fungi. Mini-Rev. Med. Chem. 2021, 21, 2000–2019. [Google Scholar] [CrossRef]
- Hu, X.Y.; Li, X.M.; Yang, S.Q.; Liu, H.; Meng, L.H.; Wang, B.G. Three new sesquiterpenoids from the algal-derived fungus Penicillium chermesinum EN-480. Mar. Drugs 2020, 18, 194. [Google Scholar] [CrossRef]
- Shah, M.; Sun, C.; Sun, Z.; Zhang, G.; Che, Q.; Gu, Q.; Zhu, T.; Li, D. Antibacterial polyketides from antarctica sponge-derived fungus Penicillium sp. HDN151272. Mar. Drugs 2020, 18, 71. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Hu, X.; Yue, Y.; Lu, X.; Wang, C.; Wei, B.; Zhang, H.; Wang, H. Medium-sized peptides from microbial sources with potential for antibacterial drug development. Nat. Prod. Rep. 2024; ahead of print. [Google Scholar] [CrossRef]
- Li, F.L.; Sun, W.G.; Zhang, S.T.; Gao, W.X.; Lin, S.; Yang, B.Y.; Chai, C.W.; Li, H.Q.; Wang, J.P.; Hu, Z.X.; et al. New cyclopiane diterpenes with anti-inflammatory activity from the sea sediment-derived fungus Penicillium sp. TJ403-2. Chin. Chem. Lett. 2020, 31, 197–201. [Google Scholar] [CrossRef]
- Dai, L.T.; Yang, L.; Kong, F.D.; Ma, Q.Y.; Xie, Q.Y.; Dai, H.F.; Yu, Z.F.; Zhao, Y.X. Cytotoxic indole-diterpenoids from the marine-derived fungus Penicillium sp. KFD28. Mar. Drugs 2021, 19, 613. [Google Scholar] [CrossRef]
- Pang, X.; Zhou, X.; Lin, X.; Yang, B.; Tian, X.; Wang, J.; Xu, S.; Liu, Y. Structurally various sorbicillinoids from the deep-sea sediment derived fungus Penicillium sp. SCSIO06871. Bioorg. Chem. 2021, 107, 104600. [Google Scholar] [CrossRef]
- Wang, P.; Jia, Y.; Wu, R.; Chen, Z.; Yan, R. Human gut bacterial β-glucuronidase inhibition: An emerging approach to manage medication therapy. Biochem. Pharmacol. 2021, 190, 114566. [Google Scholar] [CrossRef]
- Chen, S.; Yueh, M.F.; Bigo, C.; Barbier, O.; Wang, K.; Karin, M.; Nguyen, N.; Tukey, R.H. Intestinal glucuronidation protects against chemotherapy-induced toxicity by irinotecan (CPT-11). Proc. Natl. Acad. Sci. USA 2013, 110, 19143–19148. [Google Scholar] [CrossRef]
- Bauman, K.D.; Butler, K.S.; Moore, B.S.; Chekan, J.R. Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. 2021, 38, 2100–2129. [Google Scholar] [CrossRef]
- Chevrette, M.G.; Gavrilidou, A.; Mantri, S.; Selem-Mojica, N.; Ziemert, N.; Barona-Gómez, F. The confluence of big data and evolutionary genome mining for the discovery of natural products. Nat. Prod. Rep. 2021, 38, 2024–2040. [Google Scholar] [CrossRef]
- Fredimoses, M.; Zhou, X.; Ai, W.; Tian, X.; Yang, B.; Lin, X.; Xian, J.Y.; Liu, Y. Westerdijkin A, a new hydroxyphenylacetic acid derivative from deep sea fungus Aspergillus westerdijkiae SCSIO 05233. Nat. Prod. Res. 2015, 29, 158–162. [Google Scholar] [CrossRef]
- Pockrandt, D.; Ludwig, L.; Fan, A.; König, G.M.; Li, S.M. New insights into the biosynthesis of prenylated xanthones: Xptb from Aspergillus nidulans catalyses an O-prenylation of xanthones. ChemBioChem 2012, 13, 2764–2771. [Google Scholar] [CrossRef]
- Zeyhle, P.; Bauer, J.S.; Steimle, M.; Leipoldt, F.; Rösch, M.; Kalinowski, J.; Gross, H.; Heide, L. A membrane-bound prenyltransferase catalyzes the O-Prenylation of 1,6-dihydroxyphenazine in the marine bacterium Streptomyces sp. CNQ-509. ChemBioChem 2014, 15, 2385–2392. [Google Scholar] [CrossRef]
- Wang, F.; Fang, Y.; Zhang, M.; Lin, A.; Zhu, T.; Gu, Q.; Zhu, W. Six new ergosterols from the marine-derived fungus Rhizopus sp. Steroids 2008, 73, 19–26. [Google Scholar] [CrossRef]
- Fukuda, T.; Tomoda, H.; Omura, S. Citridones, new potentiators of antifungal miconazole activity, produced by Penicillium sp. FKH938. II. Structure elucidation. J. Antibiot. 2005, 58, 315–321. [Google Scholar] [CrossRef]
- Yaoita, Y.; Amemiya, K.; Ohnuma, H.; Furumura, K.; Masaki, A.; Matsuki, T.; Kikuchi, M. Sterol constituents from five edible mushrooms. Chem. Pharm. Bull. 1998, 46, 944–950. [Google Scholar] [CrossRef]
- Ohnuma, N.; Amemiya, K.; Kakuda, R.; Yaoita, Y.; Machida, K.; Kikuchi, M. Sterol constituents from two edible mushrooms, Lentinula edodes and Tricholoma matsutake. Chem. Pharm. Bull. 2000, 48, 749–751. [Google Scholar] [CrossRef]
- Lee, W.Y.; Park, Y.; Ahn, J.K.; Park, S. Cytotoxic activity of ergosta-4,6,8(14),22-tetraen-3-one from the sclerotia of Polyporus umbellatus. Bull. Korean Chem. Soc. 2005, 26, 1464–1466. [Google Scholar]
- Liu, X.H.; Miao, F.P.; Liang, X.R. Ergosteroid derivatives from an algicolous strain of Aspergillus ustus. Nat. Prod. Res. 2014, 28, 1182–1186. [Google Scholar] [CrossRef]
- Wang, X.H.; Hou, Y.Z.; Pan, X.H.; Wang, Q. Sterol compounds and their anti-complementary activities of Cordia dichotoma. Chem. Nat. Compd. 2020, 56, 759–760. [Google Scholar] [CrossRef]
- Amagata, T.; Tanaka, M.; Yamada, T.; Doi, M.; Minoura, K.; Ohishi, H.; Yamori, T.; Numata, A. Variation in cytostatic constituents of a sponge-derived Gymnascella dankaliensis by manipulating the carbon source. J. Nat. Prod. 2007, 70, 1731–1740. [Google Scholar] [CrossRef]
- Amagata, T.; Doi, M.; Tohgo, M.; Minoura, K.; Numata, A. Dankasterone, a new class of cytotoxic steroid produced by a Gymnascella species from a marine sponge. Chem. Commun. 1999, 30, 1321–1322. [Google Scholar] [CrossRef]
- Ghisalberti, E.; Hockless, D.; Rowland, C. Structural study of curvularin, a cell division inhibitor. Aust. J. Chem. 1993, 46, 571–577. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, H. Isolation of Secondary. Metabolites of 9F series marine fungi and their bioactivities against Pyricularia oryzae. Nat. Prod. Res. Dev. 2005, 17, 677–680. [Google Scholar]
- Kusano, M.; Nakagami, K.; Fujioka, S.; Kawano, T.; Shimada, A.; Kimura, Y. βγ-dehydrocurvularin and related compounds as nematicides of Pratylenchus penetrans from the fungus Aspergillus sp. Biosci. Biotechnol. Biochem. 2003, 67, 1413–1416. [Google Scholar] [CrossRef]
- Wu, Y.H.; Zhang, Z.H.; Zhong, Y.; Huang, J.J.; Li, X.X.; Jiang, J.Y.; Deng, Y.Y.; Zhang, L.H.; He, F. Sumalactones A–D, four new curvularin-type macrolides from a marine deep sea fungus Penicillium sumatrense. RSC Adv. 2017, 7, 40015–40019. [Google Scholar] [CrossRef]
- Yang, C.; Meng, Q.; Zhang, Y.; Hu, Y.; Xiao, S.J.; Zhang, Y.Q.; Ju, J.H.; Fu, S.B. Morelsins A–F, six sesquiterpenoids from the liquid culture of Morchella importuna. Tetrahedron 2020, 76, 131356. [Google Scholar] [CrossRef]
- Liu, D.Z.; Wang, F.; Liu, J.K. Sesquiterpenes from cultures of the basidiomycete Conocybe siliginea. J. Nat. Prod. 2007, 70, 1503–1506. [Google Scholar] [CrossRef]
- Wu, B.; Wu, L.J.; Zhang, L. Studies on the antibacterial chemical components of Senecio cannabifolius less (I). J. Shenyang Pharm. Univ. 2004, 21, 341–345. (In Chinese) [Google Scholar]
- Ward, L.C.; McCue, H.V.; Carnell, A.J. Carboxyl methyltransferases: Natural functions and potential applications in industrial biotechnology. ChemCatChem 2021, 13, 121–128. [Google Scholar] [CrossRef]
- Munakata, R.; Olry, A.; Takemura, T.; Tatsumi, K.; Ichino, T.; Villard, C.; Kageyama, J.; Kurata, T.; Nakayasu, M.; Jacob, F.; et al. Parallel evolution of UbiA superfamily proteins into aromatic O-prenyltransferases in plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2022294118. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Oberg, N.; Zallot, R.; John, A. EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme Function Initiative (EFI) web resource for genomic enzymology tools. J. Mol. Biol. 2023, 435, 168018. [Google Scholar] [CrossRef]
- Hoye, T.R.; Jeffrey, C.S.; Shao, F. Mosher ester analysis for the determination of absolute configuration of stereogenic (chiral) carbinol carbons. Nat. Protoc. 2007, 2, 2451–2458. [Google Scholar] [CrossRef]
- Zhou, T.S.; Wei, B.; He, M.; Li, Y.S.; Wang, Y.K.; Wang, S.J.; Chen, J.W.; Zhang, H.W.; Cui, Z.N.; Wang, H. Thiazolidin-2-cyanamides derivatives as novel potent Escherichia coli β-glucuronidase inhibitors and their structure–inhibitory activity relationships. J. Enzyme Inhib. Med. Chem. 2020, 35, 1736–1742. [Google Scholar] [CrossRef]
- Fukuda, T.; Hasegawa, Y.; Sakabe, Y.; Tomoda, H.; Omura, S. Citrinamides, new potentiators of antifungal miconazole activity, produced by Penicillium sp. FKI-1938. J. Antibiot. 2008, 61, 550–555. [Google Scholar] [CrossRef]
- Fukuda, T.; Shimoyama, K.; Nagamitsu, T.; Tomoda, H. Synthesis and biological activity of citridone A and its derivatives. J. Antibiot. 2014, 67, 445–450. [Google Scholar] [CrossRef]
- Zhang, Z.; Qiao, T.; Watanabe, K.; Tang, Y. Concise biosynthesis of phenylfuropyridones in fungi. Angew. Chem. Int. Ed. 2020, 59, 19889–19893. [Google Scholar] [CrossRef]
- Miyagawa, T.; Nagai, K.; Yamada, A.; Sugihara, Y.; Fukuda, T.; Fukuda, T.; Uchida, R.; Tomoda, H.; Omura, S.; Nagamitsu, T. Total synthesis of citridone A. Org. Lett. 2011, 13, 1158–1161. [Google Scholar] [CrossRef]
- Ermakova, E.; Zuev, Y. Effect of ergosterol on the fungal membrane properties. All-atom and coarse-grained molecular dynamics study. Chem. Phys. Lipids 2017, 209, 45–53. [Google Scholar] [CrossRef]
- Nes, W.D. Enzyme mechanisms for sterol C-methylations. Phytochemistry 2003, 64, 75–95. [Google Scholar] [CrossRef]


| Position | δC, Type | δH |
|---|---|---|
| 1 | 172.6, C | - |
| 2 | 40.4, CH2 | 3.56, s |
| 3 | 127.4, C | - |
| 4 | 131.1, CH | 7.19, d (8.4) |
| 4′ | 131.1, CH | 7.19, d (8.4) |
| 5 | 115.4, CH | 6.90, d (8.4) |
| 5′ | 115.4, CH | 6.90, d (8.4) |
| 6 | 159.2, C | - |
| 7 | 70.6, CH2 | 3.92, dd, (9.6, 7.8) |
| 4.26, dd, (9.6, 3.0) | ||
| 8 | 77.1, CH | 3.75, dd (7.8, 3.0) |
| 9 | 71.9, C | - |
| 10 | 26.7, CH3 | 1.22, s |
| 11 | 25.6, CH3 | 1.24, s |
| 12 | 51.9, CH3 | 3.62, s |
| Compound | IC50 (μM) |
|---|---|
| 2 | 44.60 ± 0.84 |
| 3 | 21.60 ± 0.76 |
| DSL | 47.94 ± 0.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Fan, Y.; Wang, C.; Tang, J.; Qiu, Y.; Xu, K.; Ding, Y.; Liu, Y.; Ying, Y.; Wang, H. Discovery of Prenyltransferase-Guided Hydroxyphenylacetic Acid Derivatives from Marine Fungus Penicillium sp. W21C371. Mar. Drugs 2024, 22, 296. https://doi.org/10.3390/md22070296
Wang C, Fan Y, Wang C, Tang J, Qiu Y, Xu K, Ding Y, Liu Y, Ying Y, Wang H. Discovery of Prenyltransferase-Guided Hydroxyphenylacetic Acid Derivatives from Marine Fungus Penicillium sp. W21C371. Marine Drugs. 2024; 22(7):296. https://doi.org/10.3390/md22070296
Chicago/Turabian StyleWang, Cancan, Ye Fan, Chenjie Wang, Jing Tang, Yixian Qiu, Keren Xu, Yingjia Ding, Ying Liu, Youmin Ying, and Hong Wang. 2024. "Discovery of Prenyltransferase-Guided Hydroxyphenylacetic Acid Derivatives from Marine Fungus Penicillium sp. W21C371" Marine Drugs 22, no. 7: 296. https://doi.org/10.3390/md22070296
APA StyleWang, C., Fan, Y., Wang, C., Tang, J., Qiu, Y., Xu, K., Ding, Y., Liu, Y., Ying, Y., & Wang, H. (2024). Discovery of Prenyltransferase-Guided Hydroxyphenylacetic Acid Derivatives from Marine Fungus Penicillium sp. W21C371. Marine Drugs, 22(7), 296. https://doi.org/10.3390/md22070296







