Understanding the Impact of Nitrogen Availability: A Limiting Factor for Enhancing Fucoxanthin Productivity in Microalgae Cultivation
Abstract
1. Introduction
2. Results
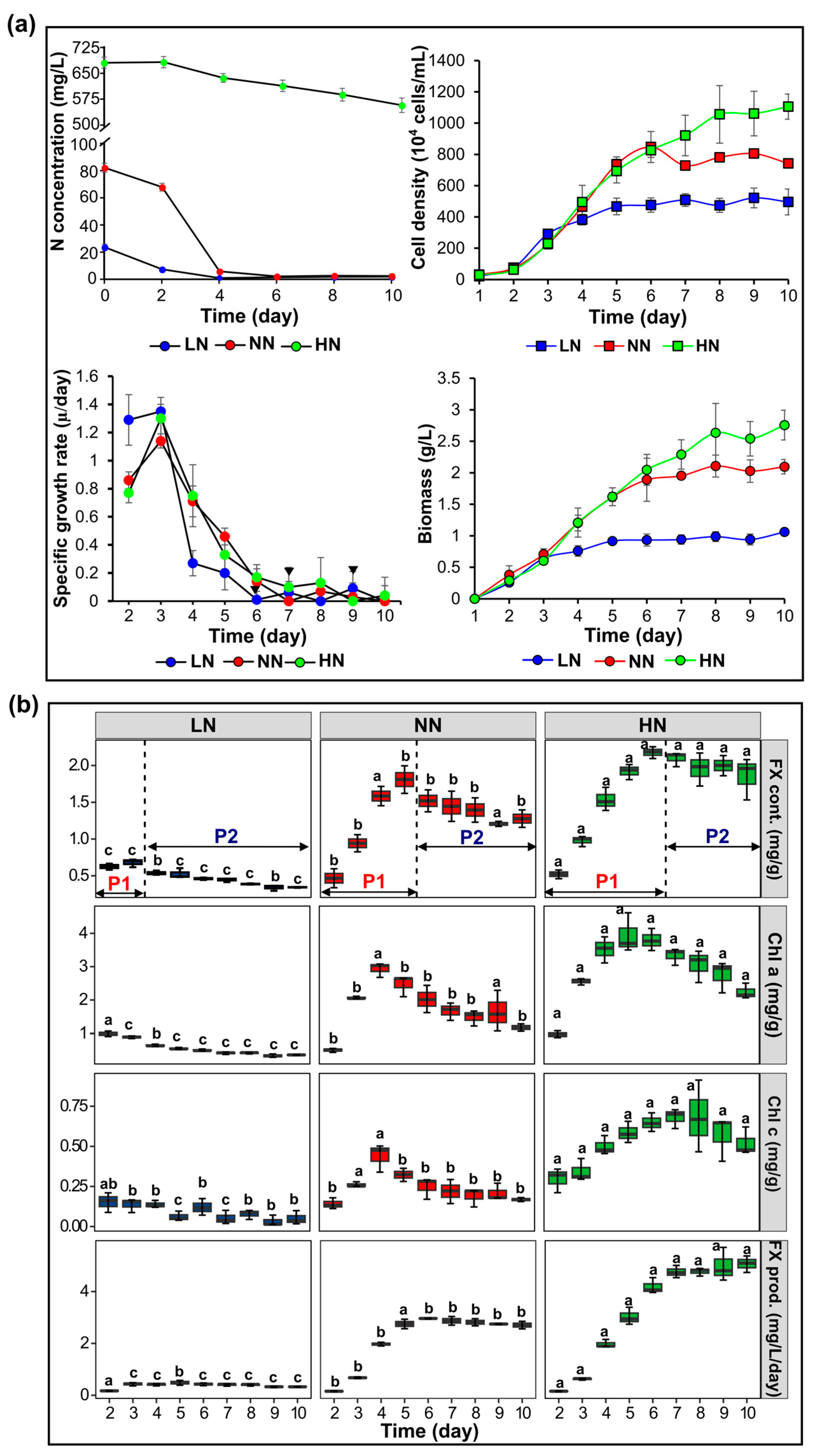
2.1. Effect of Nitrogen Availability on Various Physiological Characteristics of Phaeodactylum tricornutum
2.2. Semi-Quantification of FCP Complexes in the Thylakoid Membrane of Day 5 Samples
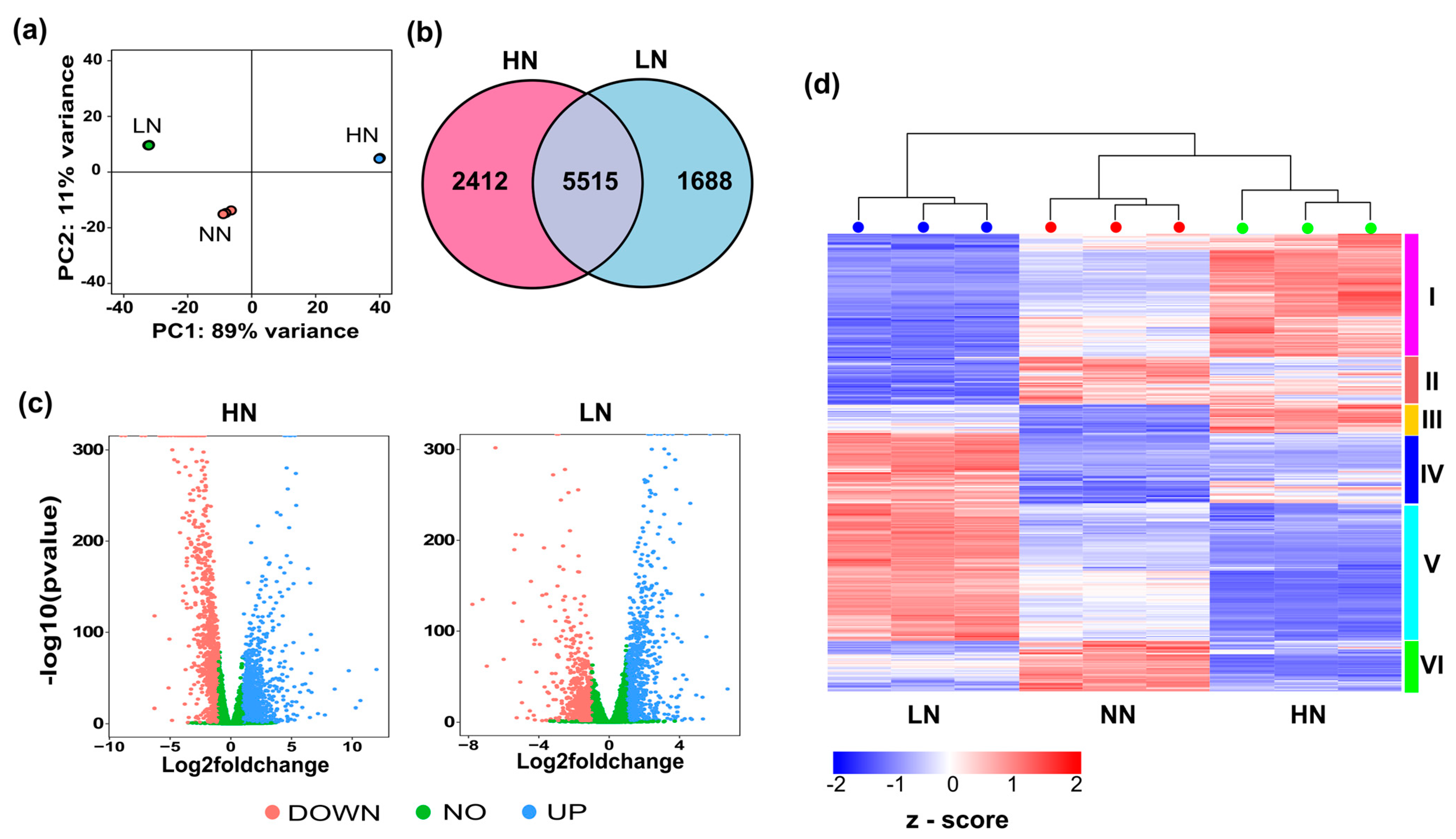
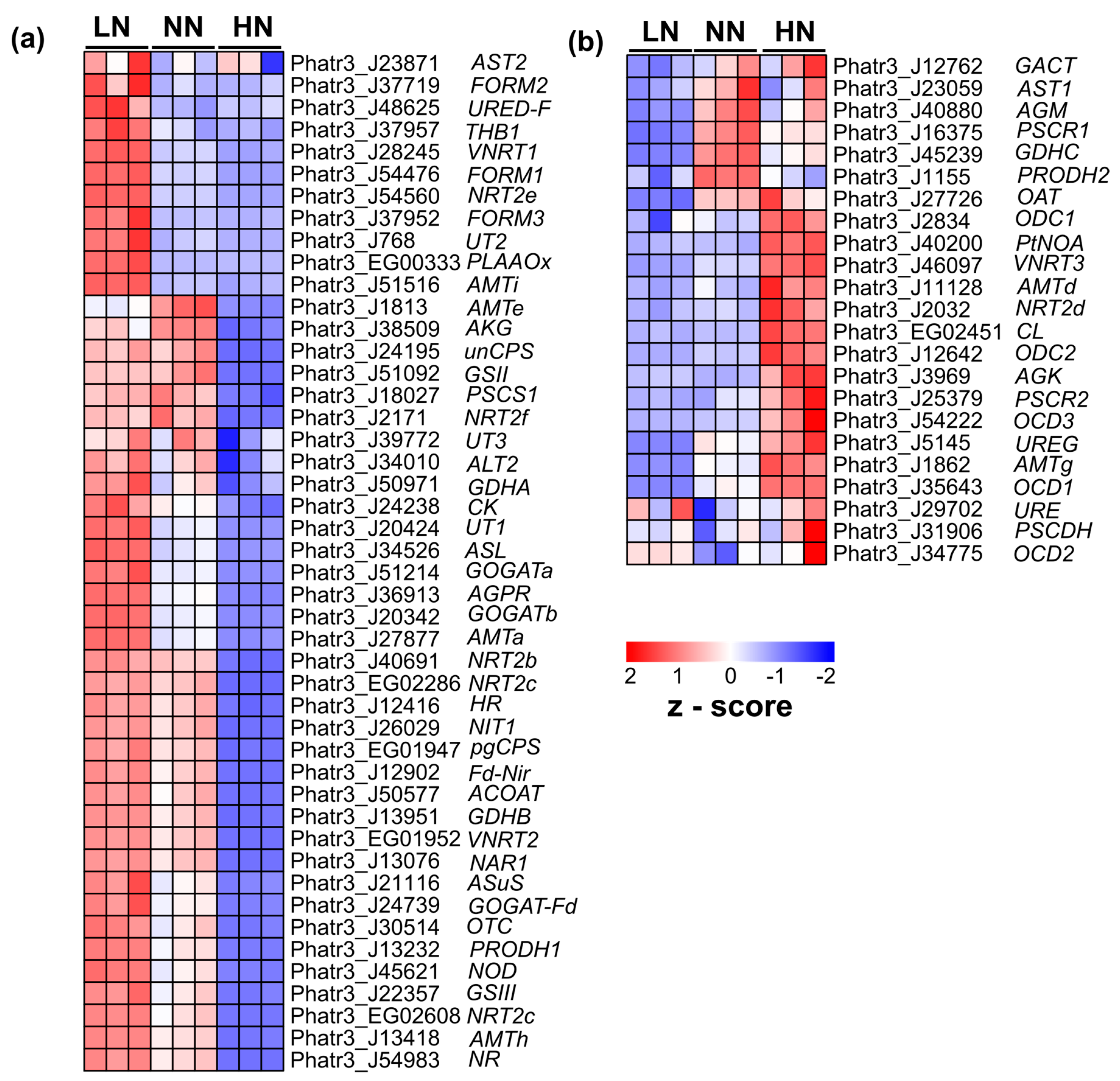
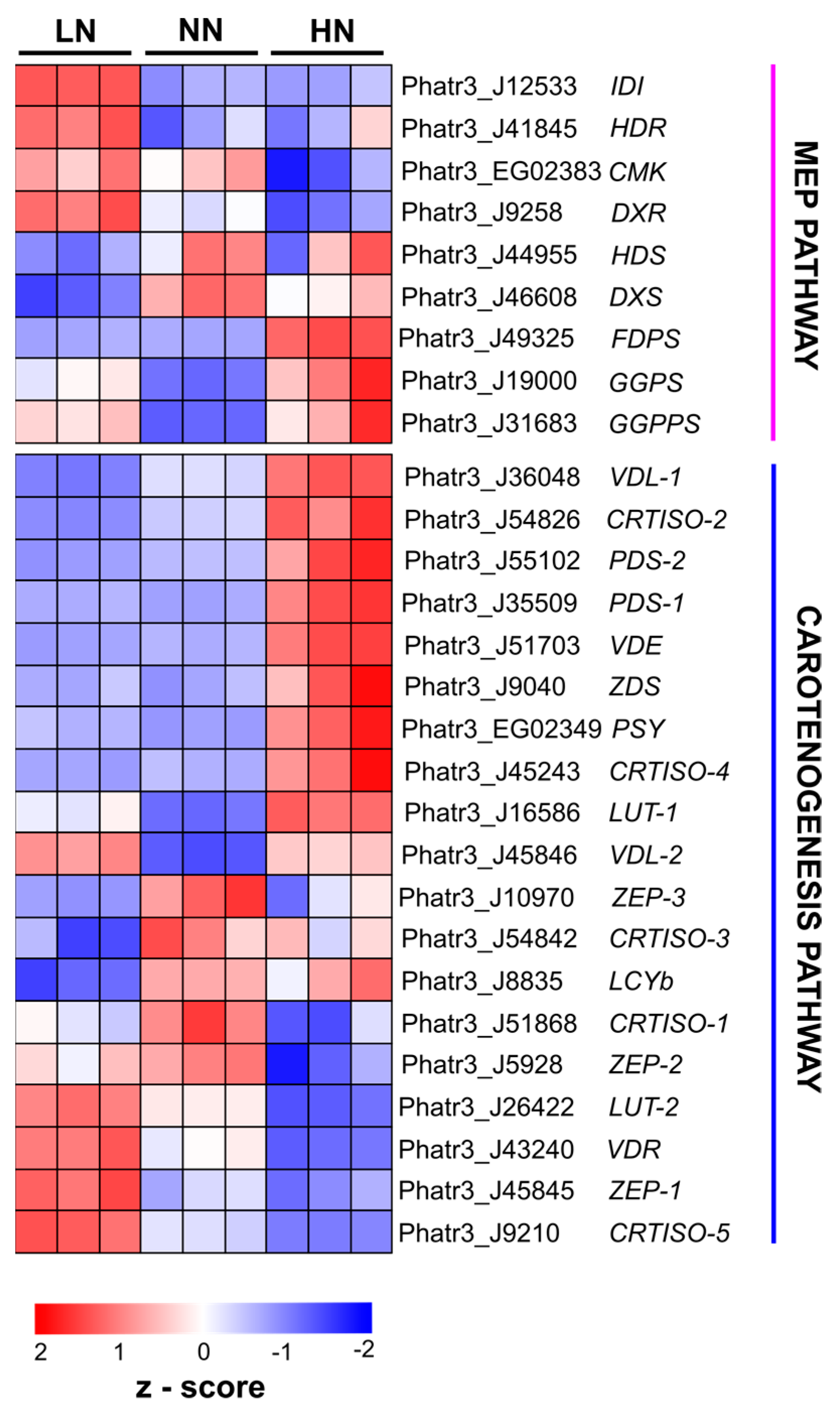
2.3. Transcriptome Analysis of Phaeodactylum tricornutum under Varying Nitrogen Conditions
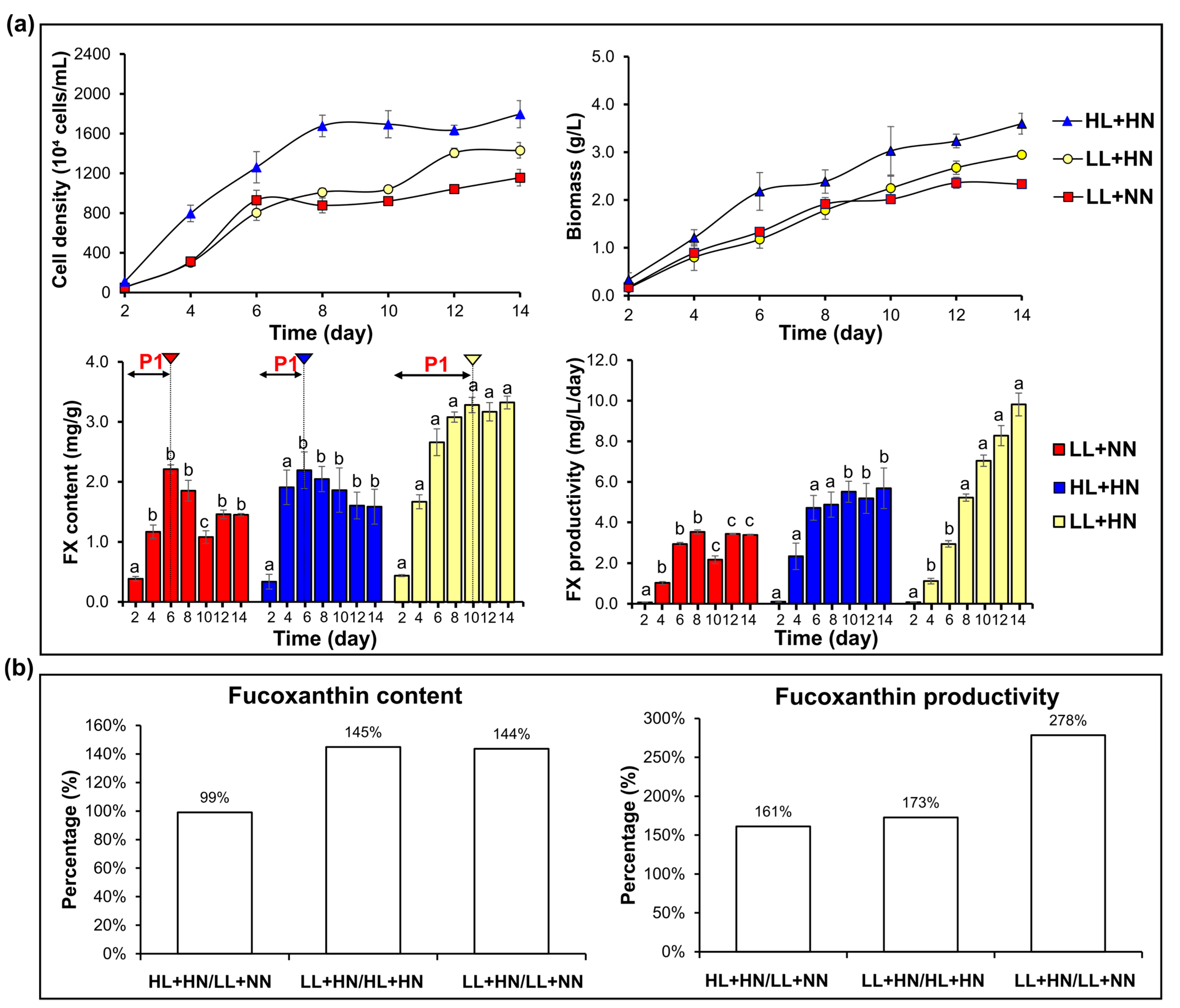
2.4. Combination of Light and Nitrogen Supply to Enhance Fucoxanthin Production
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. P. tricornutum Cultivation
4.3. Growth Determination and Biomass Production
4.4. Quantification of Primary Photosynthetic Pigments
4.4.1. Fucoxanthin Quantification with HPLC-UV-DAD Analysis
4.4.2. Quantification of Chlorophylls with a UV/Vis Spectrophotometer
4.5. Total Nitrogen Content in P. tricornutum Culture Medium
4.6. Semi-Quantification of Fucoxanthin–Chlorophyll a/c-Binding Proteins
4.6.1. Thylakoid Membrane Isolation
4.6.2. Proteomic Analysis with SDS-PAGE Coupled with LC-MS Analysis
4.7. RNA Sequencing Analysis of Day 5 Samples
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mohibbullah, M.; Haque, M.N.; Sohag, A.A.M.; Hossain, M.T.; Zahan, M.S.; Uddin, M.J.; Hannan, M.A.; Moon, I.S.; Choi, J.-S. A systematic review on marine algae-derived fucoxanthin: An update of pharmacological insights. Mar. Drugs 2022, 20, 279. [Google Scholar] [CrossRef]
- Sachindra, N.M.; Sato, E.; Maeda, H.; Hosokawa, M.; Niwano, Y.; Kohno, M.; Miyashita, K. Radical scavenging and singlet oxygen quenching activity of marine carotenoid fucoxanthin and its metabolites. J. Agric. Food Chem. 2007, 55, 8516–8522. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Shin, H.-Y.; Park, J.-H.; Koo, S.Y.; Kim, S.M.; Yang, S.-H. Fucoxanthin from microalgae Phaeodactylum tricornutum inhibits pro-inflammatory cytokines by regulating both NF-κB and NLRP3 inflammasome activation. Sci. Rep. 2021, 11, 543. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Mendonca, P.; Messeha, S.S.; Soliman, K.F.A. Anticancer effects of fucoxanthin through cell cycle arrest, apoptosis induction, and angiogenesis inhibition in triple-negative breast cancer cells. Molecules 2023, 28, 6536. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, S.; Yang, G.; Pan, K.; Wang, L.; Hu, Z. A review on the progress, challenges and prospects in commercializing microalgal fucoxanthin. Biotechnol. Adv. 2021, 53, 107865. [Google Scholar] [CrossRef] [PubMed]
- Pajot, A.; Hao Huynh, G.; Picot, L.; Marchal, L.; Nicolau, E. Fucoxanthin from algae to human, an extraordinary bioresource: Insights and advances in up and downstream processes. Mar. Drugs 2022, 20, 222. [Google Scholar] [CrossRef] [PubMed]
- Butler, T.; Kapoore, R.V.; Vaidyanathan, S. Phaeodactylum tricornutum: A diatom cell factory. Trends Biotechnol. 2020, 38, 606–622. [Google Scholar] [CrossRef] [PubMed]
- Gelzinis, A.; Butkus, V.; Songaila, E.; Augulis, R.; Gall, A.; Büchel, C.; Robert, B.; Abramavicius, D.; Zigmantas, D.; Valkunas, L. Mapping energy transfer channels in fucoxanthin–chlorophyll protein complex. Biochim. Biophys. Acta Bioenerg. 2015, 1847, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.; Philippidis, G.P. Fucoxanthin production from diatoms: Current advances and challenges. In Algae: Multifarious Applications for a Sustainable World; Mandotra, S.K., Upadhyay, A.K., Ahluwalia, A.S., Eds.; Springer: Singapore, 2021; pp. 227–242. [Google Scholar]
- Kadono, T.; Kira, N.; Suzuki, K.; Iwata, O.; Ohama, T.; Okada, S.; Nishimura, T.; Akakabe, M.; Tsuda, M.; Adachi, M. Effect of an introduced phytoene synthase gene expression on carotenoid biosynthesis in the marine diatom Phaeodactylum tricornutum. Mar. Drugs 2015, 13, 5334–5357. [Google Scholar] [CrossRef]
- Manfellotto, F.; Stella, G.R.; Falciatore, A.; Brunet, C.; Ferrante, M.I. Engineering the unicellular alga Phaeodactylum tricornutum for enhancing carotenoid production. Antioxidants 2020, 9, 757. [Google Scholar] [CrossRef]
- Eilers, U.; Bikoulis, A.; Breitenbach, J.; Büchel, C.; Sandmann, G. Limitations in the biosynthesis of fucoxanthin as targets for genetic engineering in Phaeodactylum tricornutum. J. Appl. Phycol. 2016, 28, 123–129. [Google Scholar] [CrossRef]
- Hao, T.-B.; Lu, Y.; Zhang, Z.-H.; Liu, S.-F.; Wang, X.; Yang, W.-D.; Balamurugan, S.; Li, H.-Y. Hyperaccumulation of fucoxanthin by enhancing methylerythritol phosphate pathway in Phaeodactylum tricornutum. Appl. Microbiol. Biotechnol. 2021, 105, 8783–8793. [Google Scholar] [CrossRef]
- Wang, W.; Yu, L.-J.; Xu, C.; Tomizaki, T.; Zhao, S.; Umena, Y.; Chen, X.; Qin, X.; Xin, Y.; Suga, M.; et al. Structural basis for blue-green light harvesting and energy dissipation in diatoms. Science 2019, 363, eaav0365. [Google Scholar] [CrossRef]
- Khaw, Y.S.; Yusoff, F.M.; Tan, H.T.; Noor Mazli, N.A.; Nazarudin, M.F.; Shaharuddin, N.A.; Omar, A.R.; Takahashi, K. Fucoxanthin production of microalgae under different culture factors: A systematic review. Mar. Drugs 2022, 20, 592. [Google Scholar] [CrossRef] [PubMed]
- McClure, D.D.; Luiz, A.; Gerber, B.; Barton, G.W.; Kavanagh, J.M. An investigation into the effect of culture conditions on fucoxanthin production using the marine microalgae Phaeodactylum tricornutum. Algal Res. 2018, 29, 41–48. [Google Scholar] [CrossRef]
- Truong, T.Q.; Park, Y.J.; Koo, S.Y.; Choi, J.-H.; Enkhbayar, A.; Song, D.-G.; Kim, S.M. Interdependence of fucoxanthin biosynthesis and fucoxanthin-chlorophyll a/c binding proteins in Phaeodactylum tricornutum under different light intensities. J. Appl. Phycol. 2023, 35, 25–42. [Google Scholar] [CrossRef]
- Yang, R.; Wei, D. Improving fucoxanthin production in mixotrophic culture of marine diatom Phaeodactylum tricornutum by LED light shift and nitrogen supplementation. Front. Bioeng. Biotechnol. 2020, 8, 820. [Google Scholar] [CrossRef]
- Sharma, A.K.; Nymark, M.; Flo, S.; Sparstad, T.; Bones, A.M.; Winge, P. Simultaneous knockout of multiple LHCF genes using single sgRNAs and engineering of a high-fidelity Cas9 for precise genome editing in marine algae. J. Plant Biotechnol. 2021, 19, 1658–1669. [Google Scholar] [CrossRef]
- Nymark, M.; Volpe, C.; Hafskjold, M.C.G.; Kirst, H.; Serif, M.; Vadstein, O.; Bones, A.M.; Melis, A.; Winge, P. Loss of ALBINO3b insertase results in truncated light-harvesting antenna in diatoms. Plant Physiol. 2019, 181, 1257–1276. [Google Scholar] [CrossRef]
- Yang, W.; Zhou, L.; Wang, J.; Wang, L.; Gao, S.; Wang, G. Knockout of a diatom cryptochrome by CRISPR/Cas9 causes an increase in light-harvesting protein levels and accumulation of fucoxanthin. Algal Res. 2022, 66, 102822. [Google Scholar] [CrossRef]
- Zarrinmehr, M.J.; Farhadian, O.; Heyrati, F.P.; Keramat, J.; Koutra, E.; Kornaros, M.; Daneshvar, E. Effect of nitrogen concentration on the growth rate and biochemical composition of the microalga, Isochrysis galbana. Egypt. J. Aquat. Res. 2020, 46, 153–158. [Google Scholar] [CrossRef]
- Pancha, I.; Chokshi, K.; George, B.; Ghosh, T.; Paliwal, C.; Maurya, R.; Mishra, S. Nitrogen stress triggered biochemical and morphological changes in the microalgae Scenedesmus sp. CCNM 1077. Bioresour. Technol. 2014, 156, 146–154. [Google Scholar] [CrossRef]
- Cao, Z.; Shen, X.; Wang, X.; Zhu, B.; Pan, K.; Li, Y. Effect of nitrogen concentration on the alkalophilic microalga Nitzschia sp. NW129-a promising feedstock for the integrated production of lipids and fucoxanthin in biorefinery. Front. Mar. Sci. 2022, 8, 830590. [Google Scholar] [CrossRef]
- Wu, Z.; Qiu, S.; Abbew, A.-W.; Chen, Z.; Liu, Y.; Zuo, J.; Ge, S. Evaluation of nitrogen source, concentration and feeding mode for co-production of fucoxanthin and fatty acids in Phaeodactylum tricornutum. Algal Res. 2022, 63, 102655. [Google Scholar] [CrossRef]
- Xia, S.; Gao, B.; Fu, J.; Xiong, J.; Zhang, C. Production of fucoxanthin, chrysolaminarin, and eicosapentaenoic acid by Odontella aurita under different nitrogen supply regimes. J. Biosci. Bioeng. 2018, 126, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Afonso, C.; Bragança, A.R.; Rebelo, B.A.; Serra, T.S.; Abranches, R. Optimal nitrate supplementation in Phaeodactylum tricornutum culture medium increases biomass and fucoxanthin production. Foods 2022, 11, 568. [Google Scholar] [CrossRef] [PubMed]
- Rui, X.; Amenorfenyo, D.K.; Peng, K.; Li, H.; Wang, L.; Huang, X.; Li, C.; Li, F. Effects of different nitrogen concentrations on co-production of fucoxanthin and fatty acids in Conticribra weissflogii. Mar. Drugs 2023, 21, 106. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Chen, L.; Cheng, W.; Liu, T. Combined production of fucoxanthin and EPA from two diatom strains Phaeodactylum tricornutum and Cylindrotheca fusiformis cultures. Bioprocess Biosyst. Eng. 2018, 41, 1061–1071. [Google Scholar] [CrossRef] [PubMed]
- Remmers, I.M.; D’Adamo, S.; Martens, D.E.; de Vos, R.C.H.; Mumm, R.; America, A.H.P.; Cordewener, J.H.G.; Bakker, L.V.; Peters, S.A.; Wijffels, R.H. Orchestration of transcriptome, proteome and metabolome in the diatom Phaeodactylum tricornutum during nitrogen limitation. Algal Res. 2018, 35, 33–49. [Google Scholar] [CrossRef]
- Alipanah, L.; Rohloff, J.; Winge, P.; Bones, A.M.; Brembu, T. Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Exp. Bot. 2015, 66, 6281–6296. [Google Scholar] [CrossRef] [PubMed]
- Sulochana, S.B.; Arumugam, M. Targeted metabolomic and biochemical changes during nitrogen stress mediated lipid accumulation in Scenedesmus quadricauda CASA CC202. Front. Bioeng. Biotechnol. 2020, 8, 585632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-M.; Chen, H.; He, C.-L.; Wang, Q. Nitrogen starvation induced oxidative stress in an oil-producing green alga Chlorella sorokiniana C3. PLoS ONE 2013, 8, e69225. [Google Scholar] [CrossRef] [PubMed]
- Yilancioglu, K.; Cokol, M.; Pastirmaci, I.; Erman, B.; Cetiner, S. Oxidative stress is a mediator for increased lipid accumulation in a newly isolated Dunaliella salina strain. PLoS ONE 2014, 9, e91957. [Google Scholar] [CrossRef] [PubMed]
- Benasla, A.; Hausler, R. A two-step cultivation strategy for high biomass production and lipid accumulation of raphidocelis subcapitata immobilized in alginate gel. Biomass 2021, 1, 94–104. [Google Scholar] [CrossRef]
- Nur, M.M.A.; Muizelaar, W.; Boelen, P.; Buma, A.G.J. Environmental and nutrient conditions influence fucoxanthin productivity of the marine diatom Phaeodactylum tricornutum grown on palm oil mill effluent. J. Appl. Phycol. 2018, 31, 111–122. [Google Scholar] [CrossRef]
- Villanova, V.; Fortunato, A.E.; Singh, D.; Bo, D.D.; Conte, M.; Obata, T.; Jouhet, J.; Fernie, A.R.; Marechal, E.; Falciatore, A. Investigating mixotrophic metabolism in the model diatom Phaeodactylum tricornutum. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160404. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Marchand, J.; Blanckaert, V.; Lukomska, E.; Ulmann, L.; Wielgosz-Collin, G.; Rabesaotra, V.; Moreau, B.; Bougaran, G.; Mimouni, V. Nitrogen and phosphorus limitations induce carbon partitioning and membrane lipid remodelling in the marine diatom Phaeodactylum tricornutum. Eur. J. Phycol. 2019, 54, 342–358. [Google Scholar] [CrossRef]
- Yodsuwan, N.; Sawayama, S.; Sirisansaneeyakul, S. Effect of nitrogen concentration on growth, lipid production and fatty acid profiles of the marine diatom Phaeodactylum tricornutum. Agric. Nat. Resour. 2017, 51, 190–197. [Google Scholar] [CrossRef]
- Nymark, M.; Valle, K.C.; Hancke, K.; Winge, P.; Andresen, K.; Johnsen, G.; Bones, A.M.; Brembu, T. Molecular and photosynthetic responses to prolonged darkness and subsequent acclimation to re-illumination in the diatom Phaeodactylum tricornutum. PLoS ONE 2013, 8, e58722. [Google Scholar] [CrossRef]
- Nymark, M.; Valle, K.C.; Brembu, T.; Hancke, K.; Winge, P.; Andresen, K.; Johnsen, G.; Bones, A.M. An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE 2009, 4, e7743. [Google Scholar] [CrossRef]
- Levitan, O.; Dinamarca, J.; Zelzion, E.; Lun, D.S.; Guerra, L.T.; Kim, M.K.; Kim, J.; Van Mooy, B.A.S.; Bhattacharya, D.; Falkowski, P.G. Remodeling of intermediate metabolism in the diatom Phaeodactylum tricornutum under nitrogen stress. Proc. Natl. Acad. Sci. USA 2015, 112, 412–417. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, M.; Mao, X.; Kou, Y.; Liu, J. Time-resolved carotenoid profiling and transcriptomic analysis reveal mechanism of carotenogenesis for astaxanthin synthesis in the oleaginous green alga Chromochloris zofingiensis. Biotechnol. Biofuels 2019, 12, 1–19. [Google Scholar] [CrossRef]
- Scarsini, M.; Thiriet-Rupert, S.; Veidl, B.; Mondeguer, F.; Hu, H.; Marchand, J.; Schoefs, B. The transition toward nitrogen deprivation in diatoms requires chloroplast stand-by and deep metabolic reshuffling. Front. Plant Sci. 2022, 12, 760516. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Vuong, T.T.; Choi, J.; Lee, T.S.; Um, J.-I.; Koo, S.Y.; Hwang, K.T.; Kim, S.M. Fucoxanthin biosynthesis has a positive correlation with the specific growth rate in the culture of microalga Phaeodactylum tricornutum. J. Appl. Phycol. 2021, 33, 1473–1485. [Google Scholar] [CrossRef]
- Ishika, T.; Moheimani, N.R.; Bahri, P.A.; Laird, D.W.; Blair, S.; Parlevliet, D. Halo-adapted microalgae for fucoxanthin production: Effect of incremental increase in salinity. Algal Res. 2017, 28, 66–73. [Google Scholar] [CrossRef]
- Marella, T.K.; Tiwari, A. Marine diatom Thalassiosira weissflogii based biorefinery for co-production of eicosapentaenoic acid and fucoxanthin. Bioresour. Technol. 2020, 307, 123245. [Google Scholar] [CrossRef] [PubMed]
- Pereira, H.; Sá, M.; Maia, I.; Rodrigues, A.; Teles, I.; Wijffels, R.H.; Navalho, J.; Barbosa, M. Fucoxanthin production from Tisochrysis lutea and Phaeodactylum tricornutum at industrial scale. Algal Res. 2021, 56, 102322. [Google Scholar] [CrossRef]
- Xia, S.; Wang, K.; Wan, L.; Li, A.; Hu, Q.; Zhang, C. Production, characterization, and antioxidant activity of fucoxanthin from the marine diatom Odontella aurita. Mar. Drugs 2013, 11, 2667–2681. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Kyu, S.Y.; Pe, P.P.W.; Park, K.I.; Lee, J.M.; Lim, K.B.; Kim, C.K. Silencing of the phytoene desaturase (PDS) gene affects the expression of fruit-ripening genes in tomatoes. Plant Methods 2019, 15, 110. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Babu, S.; Gothandam, K.M. Accumulation of phytoene, a colorless carotenoid by inhibition of phytoene desaturase (PDS) gene in Dunaliella salina V-101. Bioresour. Technol. 2017, 242, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Cordero, B.F.; Couso, I.; León, R.; Rodríguez, H.; Vargas, M.Á. Enhancement of carotenoids biosynthesis in Chlamydomonas reinhardtii by nuclear transformation using a phytoene synthase gene isolated from Chlorella zofingiensis. Appl. Microbiol. Biotechnol. 2011, 91, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Longworth, J.; Wu, D.; Huete-Ortega, M.; Wright, P.C.; Vaidyanathan, S. Proteome response of Phaeodactylum tricornutum, during lipid accumulation induced by nitrogen depletion. Algal Res. 2016, 18, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Büchel, C. How diatoms harvest light. Science 2019, 365, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Pajot, A.; Lavaud, J.; Carrier, G.; Garnier, M.; Saint-Jean, B.; Rabilloud, N.; Baroukh, C.; Bérard, J.-B.; Bernard, O.; Marchal, L. The fucoxanthin chlorophyll a/c-binding protein in Tisochrysis lutea: Influence of nitrogen and light on fucoxanthin and chlorophyll a/c-binding protein gene expression and fucoxanthin synthesis. Front. Plant Sci. 2022, 13, 830069. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-K.; Niu, Y.-F.; Ma, Y.-H.; Xue, J.; Zhang, M.-H.; Yang, W.-D.; Liu, J.-S.; Lu, S.-H.; Guan, Y.; Li, H.-Y. Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol. Biofuels 2013, 6, 67. [Google Scholar] [CrossRef] [PubMed]
- Conceição, D.; Lopes, R.G.; Derner, R.B.; Cella, H.; do Carmo, A.P.B.; Montes D’Oca, M.G.; Petersen, R.; Passos, M.F.; Vargas, J.V.C.; Galli-Terasawa, L.V.; et al. The effect of light intensity on the production and accumulation of pigments and fatty acids in Phaeodactylum tricornutum. J. Appl. Phycol. 2020, 32, 1017–1025. [Google Scholar] [CrossRef]
- Lv, J.; Guo, J.; Feng, J.; Liu, Q.; Xie, S. Effect of sulfate ions on growth and pollutants removal of self-flocculating microalga Chlorococcum sp. GD in synthetic municipal wastewater. Bioresour. Technol. 2017, 234, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, R.J. Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth. Res. 2006, 89, 27–41. [Google Scholar] [CrossRef]
- Levitan, O.; Chen, M.; Kuang, X.; Cheong Kuan, Y.; Jiang, J.; Banal, M.; Nambiar, N.; Gorbunov Maxim, Y.; Ludtke Steven, J.; Falkowski Paul, G.; et al. Structural and functional analyses of photosystem II in the marine diatom Phaeodactylum tricornutum. Proc. Natl. Acad. Sci. USA 2019, 116, 17316–17322. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Prot. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Kang, K.; Song, D.-G.; Lee, E.H.; Lee, K.-M.; Park, Y.G.; Jung, S.H.; Pan, C.-H.; Nho, C.W. Secretome profiling reveals the signaling molecules of apoptotic HCT116 cells induced by the dietary polyacetylene gymnasterkoreayne B. J. Agric. Food Chem. 2014, 62, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
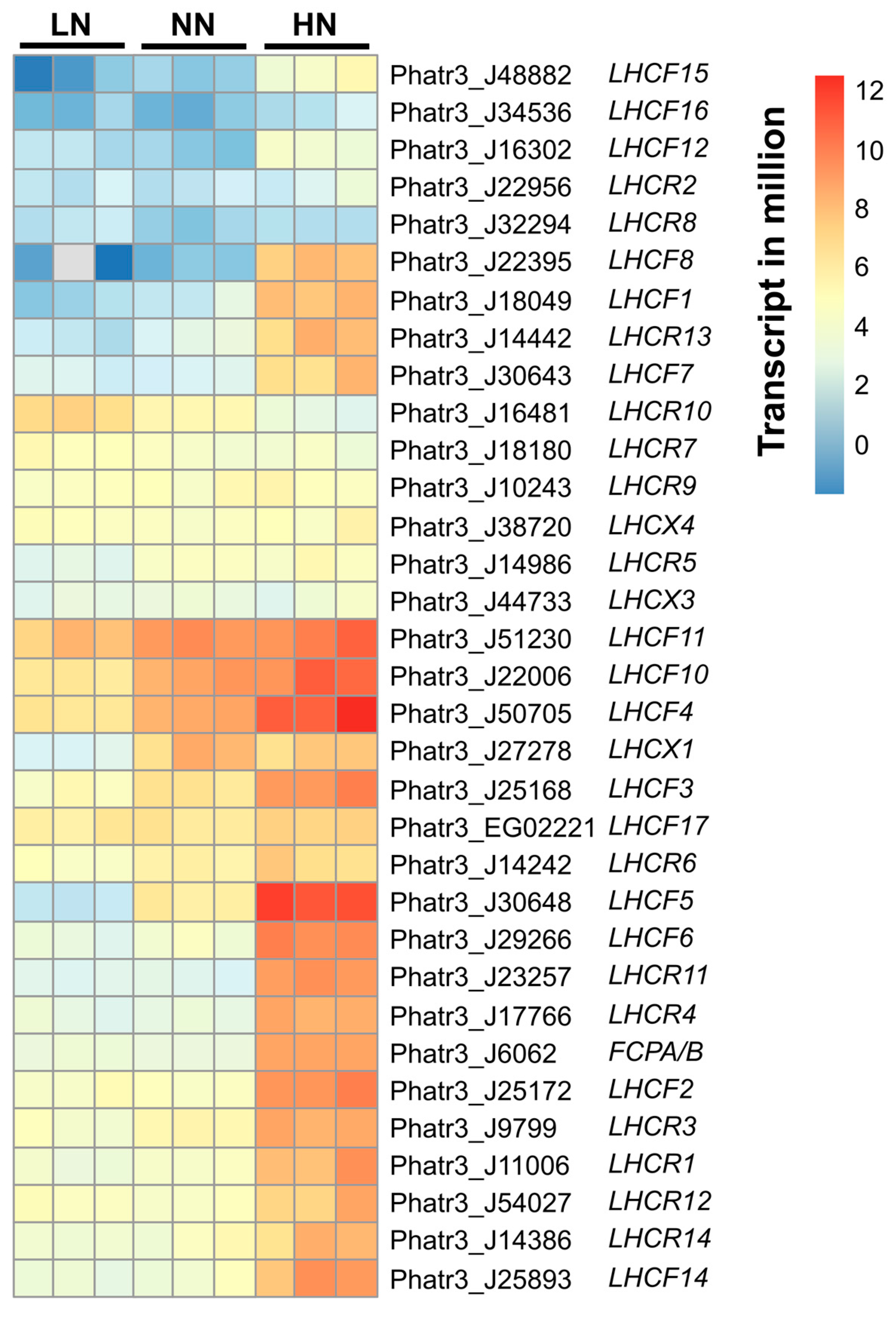
| Accession No. | Alternate ID | LN | NN | HN |
|---|---|---|---|---|
| B7FRW5 | LHCF1 | 2857 ± 273 c | 11,243 ± 1525 b | 21,399 ± 1584 a |
| B7FRW4 | LHCF2 | 2844 ± 328 c | 12,718 ± 1318 b | 22,923 ± 1385 a |
| B7FRW2 | LHCF3/4 | 13,016 ± 1422 c | 39,244 ± 945 b | 67,185 ± 3149 a |
| B7GBK7 | LHCF5 | 3124 ± 433 c | 16,442 ± 2394 b | 31,459 ± 1594 a |
| B7G5S7 | LHCF6 | 2270 ± 267 c | 7624 ± 1040 b | 15,851 ± 1660 a |
| B7G6Y1 | LHCF8 | 5580 ± 1122 c | 18,586 ± 1326 b | 32,556 ± 2839 a |
| B7G955 | LHCF9 | 401 ± 69 c | 1230 ± 399 b | 2804 ± 1421 a |
| B7G5B6 | LHCF10 | 3297 ± 718 c | 14,299 ± 1525 b | 26,277 ± 1850 a |
| B7GBK6 | LHCF11 | 4659 ± 1123 c | 15,459 ± 669 b | 25,301 ± 1245 a |
| B7GCA2 | LHCF12 | 3484 ± 984 c | 11,945 ± 791 b | 19,205 ± 1901 a |
| B7G871 | LHCF13 | 254 ± 46 c | 843 ± 279 b | 1646 ± 317 a |
| B5Y5L4 | LHCF14 | 1415 ± 231 c | 4813 ± 580 b | 9206 ± 460 a |
| B7FV42 | LHCF16 | 80 ± 0 c | 351 ± 161 b | 610 ± 381 a |
| B7GC99 | LHCF17 | 334 ± 61 c | 1124 ± 219 b | 1890 ± 761 a |
| B7FUM6 | LHCR1 | 587 ± 101 c | 1124 ± 122 b | 1585 ± 211 a |
| B7FSP4 | LHCR3 | 935 ± 23 c | 2846 ± 105 b | 4938 ± 317 a |
| B7FQE1 | LHCR4 | 267 ± 23 c | 1230 ± 61 b | 1890 ± 381 a |
| B7FQE0 | LHCR12 | 1695 ± 311 c | 5059 ± 422 b | 7682 ± 1018 a |
| B7G502 | LHCR13 | 854 ± 46 c | 2846 ± 658 b | 4694 ± 1598 a |
| B7G503 | LHCR14 | 1268 ± 228 c | 5973 ± 896 b | 9511 ± 2420 a |
| B7FYL0 | LHCX1 | 240 ± 40 c | 773 ± 265 b | 1280 ± 484 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Truong, T.Q.; Park, Y.J.; Winarto, J.; Huynh, P.K.; Moon, J.; Choi, Y.B.; Song, D.-G.; Koo, S.Y.; Kim, S.M. Understanding the Impact of Nitrogen Availability: A Limiting Factor for Enhancing Fucoxanthin Productivity in Microalgae Cultivation. Mar. Drugs 2024, 22, 93. https://doi.org/10.3390/md22020093
Truong TQ, Park YJ, Winarto J, Huynh PK, Moon J, Choi YB, Song D-G, Koo SY, Kim SM. Understanding the Impact of Nitrogen Availability: A Limiting Factor for Enhancing Fucoxanthin Productivity in Microalgae Cultivation. Marine Drugs. 2024; 22(2):93. https://doi.org/10.3390/md22020093
Chicago/Turabian StyleTruong, To Quyen, Yun Ji Park, Jessica Winarto, Phuong Kim Huynh, Jinyoung Moon, Yeong Bin Choi, Dae-Geun Song, Song Yi Koo, and Sang Min Kim. 2024. "Understanding the Impact of Nitrogen Availability: A Limiting Factor for Enhancing Fucoxanthin Productivity in Microalgae Cultivation" Marine Drugs 22, no. 2: 93. https://doi.org/10.3390/md22020093
APA StyleTruong, T. Q., Park, Y. J., Winarto, J., Huynh, P. K., Moon, J., Choi, Y. B., Song, D.-G., Koo, S. Y., & Kim, S. M. (2024). Understanding the Impact of Nitrogen Availability: A Limiting Factor for Enhancing Fucoxanthin Productivity in Microalgae Cultivation. Marine Drugs, 22(2), 93. https://doi.org/10.3390/md22020093







