Abstract
In light of the escalating global energy crisis, microalgae have emerged as highly promising producers of biofuel and high-value products. Among these microalgae, Nannochloropsis has received significant attention due to its capacity to generate not only triacylglycerol (TAG) but also eicosapentaenoic acid (EPA) and valuable carotenoids. Recent advancements in genetic tools and the field of synthetic biology have revolutionized Nannochloropsis into a powerful biofactory. This comprehensive review provides an initial overview of the current state of cultivation and utilization of the Nannochloropsis genus. Subsequently, our review examines the metabolic pathways governing lipids and carotenoids, emphasizing strategies to enhance oil production and optimize carbon flux redirection toward target products. Additionally, we summarize the utilization of advanced genetic manipulation techniques in Nannochloropsis. Together, the insights presented in this review highlight the immense potential of Nannochloropsis as a valuable model for biofuels and synthetic biology. By effectively integrating genetic tools and metabolic engineering, the realization of this potential becomes increasingly feasible.
1. Introduction
Up until now, fossil-derived fuels have served as the main global energy sources. The ever-increasing energy demand, depleted reserves of fossil fuels, and environmental concerns associated with fossil fuel burning, however, have led to the exploration of alternative energy that is green, renewable, and sustainable. Biomass-derived oils have a similar structure to fossil fuels, are energy rich, and represent important sources for making biodiesel. Currently, plant oils contribute to the major feedstocks for biodiesel production. Biodiesel from plants, however, is a long way from meeting the existing demand and will cause the conflict of food versus fuel. Algae has gradually come to people’s attention because of its advantages of fast growth, no competition with grain, and higher oil production potential than plants [1,2]. However, it remains economically infeasible for algae to serve as a separate biofuel producer. Combining oil production with high-value co-products using synthetic biology is a solution.
Nannochloropsis, a genus of Eustigmatophyceae microalgae, holds immense promise due to its unique attributes, including rapid growth, a high lipid content, and the ability to thrive in diverse environmental conditions; these characteristics make it an ideal candidate for producing oils, omega-3 fatty acids, and a wide array of high-value compounds [3]. Nannochloropsis, with its lipid content exceeding 30% of its dry weight, emerges as a highly promising feedstock for biodiesel production. In a study, the type 1 diacylglycerol acyltransferase (DGAT) gene derived from Arabidopsis thaliana (AtDGAT) was heterologously expressed in Nannochloropsis oceanica using electroporation. In comparison to the wild-type N. oceanica, the overexpression of AtDGAT resulted in a significant increase in C16 (C16:0 + C16:1) levels and a slight decrease in C18 (C18:0 + C18:1), providing novel insights into the genetic engineering of oleaginous microalgae [4]. In addition, there have been many studies exploring Nannochloropsis as feed for Pacific white shrimp, Atlantic salmon, kuruma shrimp, and European sea bass [5,6]. Adissin et al. [7] used algal powder to take the place of fish oil and finally improved shrimp growth. More interestingly, this strategy changed the fatty acid profile of kuruma shrimp, with a big increase in polyunsaturated fatty acids (PUFAs). Nannochloropsis can be used in food as well. Lafarga et al. [8] tried to add 2.0% Nannochloropsis to functional breads and performed an evaluation of their physicochemical properties, organoleptic properties, and nutritional properties. The new breads with a high amount of bioaccessible polyphenols showed a highly improved antioxidant capacity. Additionally, the non-living cells of N. oceanica can be effective adsorbents for treating textile wastewater containing azo dyes [9]. The bioactive extracts of Nannochloropsis are interesting targets in the medical and cosmetic industries. Khattib et al. [10] isolated an active compound of lyso-diacylglyceryl-N,N,N-trimethyl-homoserine (lyso-DGTS) from Nannochloropsis, which can interact with high-density lipoprotein to improve its anti-atherosclerosis abilities. Moreover, violaxanthin has significant skin-protective activity against induced oxidative stress, such as UVB-mediated cellular damage [11]. Due to the high abundance of violaxanthin (32% of total carotenoids), Nannochloropsis can be used as a functional ingredient in cosmetic formulations [11]. It has been found that Nannochloropsis produces high levels of vitamin D3 naturally, which is not common in other algae [12].
Synthetic biology, with its genetic and metabolic manipulative prowess, allows for the fine tuning of Nannochloropsis to optimize the production of lipids, proteins, carotenoids, and other high-value compounds [13,14]. Synthetic biology provides the essential toolbox for reprogramming Nannochloropsis at the genetic level. It involves a spectrum of tools, such as the revolutionary CRISPR-Cas9 gene-editing system, custom-designed promoters, and metabolic pathway engineering [15,16]. These tools empower researchers to tailor Nannochloropsis strains for trait improvements, including but not restricted to growth, oils, omega-3 PUFAs, and high-value carotenoids.
This review aims to provide a comprehensive overview of the current state of Nannochloropsis-based biorefining driven by synthetic biology [13]. It will delve into the taxonomy and growth characteristics of Nannochloropsis, the fundamental principles of synthetic biology, the tools and techniques employed in genetic and metabolic engineering, and the various biorefining strategies being pursued. Furthermore, it will explore the challenges faced in scaling up Nannochloropsis-based biorefining processes, address environmental considerations and sustainability, and touch upon the regulatory and ethical aspects governing this rapidly advancing field.
2. Growth Physiology of Nannochloropsis
Nannochloropsis is a genus of unicellular and non-motile marine microalgae classified within the phylum Heterokontophyta, the class Eustigmatophyceae, and the family Eustigmataceae. This genus was initially defined by Hibberd and comprises six well-documented species: Nannochloropsis gaditana, Nannochloropsis granulata, N. oceanica, Nannochloropsis oculata, Nannochloropsis salina, and Nannochloropsis australis [17,18]. Nannochloropsis is characterized by small, unicellular, non-motile, and spherical or ovoid cells. It is a single-celled organism, typically ranging from 2 to 8 μm in size, constituting an entire living entity within a solitary cell. This distinction sets it apart from multicellular organisms, which are formed through collaboration between multiple cells [3]. The cells are encased within a unique cell-wall structure composed of complex polysaccharides, offering protection and stability. The inner layer of the cell wall in Nannochloropsis displays a porous structure with a delicate fibrous substructure and support structures linking this layer to the plasma membrane [19]. In certain Nannochloropsis species, small amounts of additional sugars (such as rhamnose, mannose, ribose, xylose, fucose, and galactose) may be present [20]. The ultrastructure of Nannochloropsis cells, including the organization of chloroplasts and other organelles, has been studied in great detail using electron microscopy techniques, shedding light on the intricacies of its cellular architecture [3].
Nannochloropsis exhibits rapid growth rates under optimal conditions, making it an attractive candidate for biotechnological purposes. Its growth physiology encompasses several key factors, including environmental requirements, nutrient uptake, photosynthesis, and metabolic pathways. The efficiency of photosynthesis in Nannochloropsis is influenced by the light intensity, quality, and photoperiod.
2.1. Environmental Factors
The cultivation of Nannochloropsis is highly dependent on various environmental factors. These factors play a crucial role in determining the growth rate, lipid content, and overall health of Nannochloropsis cultures. Understanding and managing these environmental conditions are essential for successful and efficient cultivation. Adequate light is essential for their growth.
As the light intensity increased from low to high levels, the growth rates of N. oceanica decreased in both autotrophic and mixotrophic cultures [21]. Comparatively, under light intensities of 40, 80, 120, and 160 µmol m−2 s−1, the biomass concentration and productivity of N. gaditana were highest at the light intensity of 120 µmol m−2 s−1 and a photoperiod comprising a 16 h light/8 h dark cycle [22]. Similarly, Nannochloropsis sp. demonstrated optimal growth with the maximum cell concentration under the conditions of a light intensity of 100 µmol m−2 s−1 and a photoperiod comprising a 18 h light/6 h dark cycle [23]. The light exposure significantly influences the production of compounds, particularly lipid synthesis, in Nannochloropsis. When subjected to light at an intensity of 500 µmol m−2 s−1, N. oceanica cells exhibited a 45% reduction in total protein content and a 38% decrease in sugar content, accompanied by a substantial 44% increase in total lipid content and more than 2-fold increases in β-carotene, zeaxanthin, astaxanthin, and canthaxanthin. The analysis using isobaric tags for relative and absolute quantification (iTRAQ) revealed that in carbon metabolism, glycolysis, the Calvin cycle, and the tricarboxylic acid (TCA) cycle were activated to channel more acetyl-CoA towards lipid and carotenoid biosynthesis [24]. The lower light intensity (30 and 50 µmol m−2 s−1), on the other hand, could promote the eicosapentaenoic acid (EPA) content in N. oculata to rise by 126% [25]. A similar trend is observed in Nitzschia laevis. Under the influence of elongases, C16:0-acyl carrier protein (ACP) can convert into C18:0, followed by the formation of C18:1 under the action of Δ9 desaturase. Additionally, C18:0 has the potential to generate C18:2 through stearoyl-ACP desaturase and Δ12-fatty acid desaturase (FAD). The up-regulation of these pathways suggests that low light conditions promote the biosynthesis of C18:1 and C18:2, which is beneficial for EPA biosynthesis [26]. Light wavelengths can influence the growth physiology of microalgal cells as well. It has been reported that low-light red light-emitting diodes (LEDs) could lead to a higher cell density and more fatty acids [27]. Analyzing different light wavelengths from red, green, blue, and white LEDs, it was observed that under blue light, Nannochloropsis sp. in both phototrophic and mixotrophic cultures exhibited the highest growth rates, with the fatty acid content reaching 20.45% and 15.11%, respectively [28]. The establishment of light–dark cycles contributes to the enhancement of cell photosynthetic efficiency; continuous light illumination, one the other hand, can result in energy loss and a decrease in cell density [28]. A significant correlation between metabolic status and light absorption rate was observed by Matsui et al. [29].
Temperature is another significant element of Nannochloropsis growth. Study has shown that the optimal growth temperature for N. oceanica falls between 25 and 29 °C, while growth ceases completely at temperatures above 31 °C and below 9 °C [30]. The suboptimal temperatures (<20 °C) were investigated, and the lowest growth rate but highest quantity of EPA occurred at 5 °C for N. salina [31]. Similarly, low-temperature stress at 10 °C favored the accumulation of EPA in N. oculata, enabling the EPA content to reach 12.8 mg L−1, which was 158% more than that obtained under 25 °C conditions [25]. By contrast, high temperature promoted lipid accumulation, with N. gaditana reaching the highest lipid content at 30 °C within the examined temperature ranges [32]. A sequential treatment approach was employed for N. oceanica, involving the initial addition of bicarbonate as the carbon source, followed by the application of low temperature to facilitate the redirection of carbon flux towards PUFA synthesis [33]. A study utilized a dual strategy involving random mutagenesis and adaptive laboratory evolution to acquire thermotolerant strains of N. oculata. Metabolomics and lipidomics unveiled a reconfiguration of the central carbon metabolism and membrane lipid synthesis in these thermotolerant strains [34].
Furthermore, it is imperative to consider the impact of symbiotic bacteria and fungi on the Nannochloropsis culture process. Fulbright et al. [35], for instance, isolated Bacillus pumilus from a underperforming N. salina culture in a 200 L system, and demonstrated its inhibitory effect on cell growth. On the other hand, there are symbiotic organisms that can be beneficial to Nannochloropsis growth. Du et al. [36], for example, combined the marine alga N. oceanica with the oleaginous fungus Mortierella elongata, resulting in increased biomass and production of triacylglycerols (TAGs).
2.2. Nutritional Factors
To harness their substantial potential in enhancing biomass and lipid production, carbon sources are frequently employed as supplements during cultivation. These carbon sources can be categorized into inorganic carbon, such as carbon dioxide [37] and bicarbonate [38], as well as organic carbon, including acetate [39], glucose [40], and glycerol [41]. Notably, a synergistic effect can be achieved by combining organic carbon and inorganic carbon. For instance, the addition of glycerol and carbon dioxide in the culture medium of N. oculata resulted in a significant increase in biomass and lipid production [42]. Furthermore, the interaction of carbon and nitrogen is of paramount importance for algae growth. The presence of acetate can greatly reduce the toxicity of ammonium, allowing it to be used as a nitrogen source in N. oculata. With 1 mM of ammonium and 32 mM of acetate in the culture, biomass and lipid production could be increased 1.5-fold and 9.5-fold, respectively [43]. It is worth noting that the presence of carbon promotes nitrogen utilization, and vice versa. When nitrate was introduced, glycerol consumption in N. salina was observed to shift from 60% to 79% [44].
Nitrogen and phosphorus comprise two of the most limiting factors in medium composition, and their deprivation often leads to lipid overproduction. Nitrogen availability is crucial for cell metabolism and the physiological and biochemical state in algae [45]. Phosphorus, on the other hand, is a component of several essential molecules that play a vital role in lipid turnover and remodeling [46]. The luxury uptake of phosphorus can act as a regulator of gene expression and an energy source [47]. The simultaneous occurrence of phosphorus stress and nitrogen stress has garnered interest. When the N:P supply ratio is raised to at least 32:1, phosphorus usage can be halved without harming the biomass productivity [48].
The presence of certain metal ions at relatively low concentrations is crucial for regulating cellular activities and plays important roles in growth and oil metabolism; these ions include Fe2+. Increasing the concentration of Fe2+ to 0.5 mmol L−1 results in a significant increase in PUFAs as a proportion of the total fatty acids in Nannochloropsis [49]. The optimal salinity range for Nannochloropsis growth and EPA accumulation falls between 27% and 29% [50]. Within a certain range, increasing the carbon dioxide concentration significantly boosts the EPA content in Nannochloropsis [51].
Moreover, combined factors have the potential to perform better than individual ones in lipid production. For example, the combination of salinity and light intensity could promote increased dry weight and lipid production in Nannochloropsis [52]. With combined nitrogen deprivation and high light exposure, N. oculata showed enhanced TAG accumulation compared to either nitrogen deprivation or high light exposure alone [53].
3. Biosynthesis of Lipids and High-Value Products by Nannochloropsis
3.1. TAG Biosynthesis
TAGs are the main storage lipids in eukaryotic cells and have been widely used for foods, light industrial products, and biofuels. In the biofuel sector, TAGs are harnessed as a source of fatty acids for the transesterification process, which converts them into biodiesel. This chemical transformation involves reacting TAGs with an alcohol, typically methanol or ethanol, in the presence of a catalyst, resulting in the production of biodiesel (fatty acid methyl or ethyl esters) and glycerol as a byproduct. The renewable and sustainable nature of TAGs, especially those derived from sources like microalgae, makes them highly desirable for biofuels [54]. It is crucial to understand the pathways and key enzymes involved in TAG biosynthesis, as outlined in Figure 1.

Figure 1.
Reactions known or thought to be involved in lipid metabolism in Nannocloropsis. Enzymes are in bold. Characterized enzymes are highlighted in red, while those that have not yet been characterized are shown in blue. Abbreviations: ABC, ATP-binding cassette; ACCase, acetyl-CoA carboxylase; ATG8, ATG autophagy-related protein 8; BTA, betaine lipid synthase; CCT1, CTP:phosphocholine cytidylyltransferase 1; CDS, mitochondrial half-size ABC transporter; DGAT, diacylglycerol acyltransferase; DGD1, digalactosyldiacylglycerol synthase; FAD, fatty acid desaturase; FAS, fatty acid synthase; FAE, fatty acid elongase; GPAT, glycerol 3-phosphate acyltransferase; G6PDH, glucose-6-phosphate dehydrogenase; LACS, long-chain acyl-CoA synthetase; LPAT, lysophosphatidic acid acyltransferase; MCMT, malonyl-CoA:acyl carrier protein malonyltransferase; MGD1, monogalactosyldiacylglycerol synthase; PAP, phosphatidic acid phosphatase; PDAT, phospholipid:diacylglycerol acyltransferase; PDHC, pyruvate dehydrogenase complex; PDK, pyruvate dehydrogenase kinase; PECT1, CTP:phosphoethanolamine cytidylyltransferase 1; PGPS, phosphatidylglycerolphosphate synthase; PIS1, phosphatidylinositol synthase 1; SQD1, UDP-sulfoquinovose synthase 1; ACP, acyl carrier protein; CoA, coenzyme A; CDP, cytidine 50-diphosphate; DAG, diacylglycerol; DGDG, digalactosyldiacylglycerol; DGTS, diacylglycerol-N,N,N-trimethylhomoserine; ER, endoplasmic reticulum; FAT, fatty acyl-ACP thioesterase; FFA, free fatty acid; G3P, glycerol 3-phosphate; LPA, lysophosphatidic acid; MGDG, monogalactosyldiacylglycerol; NADPH, nicotinamide adenine dinucleotide phosphate; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; SQDG, sulfoquinovosyldiacylglycerol; TAG, triacylglycerol.
Acetyl-CoA serves as the crucial carbon precursor for de novo fatty acid biosynthesis and can be derived from the conversion of pyruvate [55]. This conversion is negatively regulated by the enzyme pyruvate dehydrogenase kinase (PDK). Attenuating PDK has the potential to enhance TAG accumulation without impairing cell growth for N. salina [56]. Acetyl-CoA carboxylase (ACCase) is involved in the first step of de novo fatty acid biosynthesis, converting acetyl-CoA to malonyl CoA-ACP; N. oceanica is predicted to include multiple isoforms of ACCase [55]. As a key enzyme in the type II fatty acid synthesis (FAS II) pathway, malonyl CoA-ACP transacylase (MCAT) has been found to contribute to the increased lipid production and cell growth in N. oceanica [57]. The synthesized acyl-ACPs are either utilized in the chloroplast or released as free fatty acids, which are transported to the endoplasmic reticulum (ER) via transport proteins and long-chain acyl-CoA synthetase (LACS) [58]. In Arabidopsis, the half-size ATP-binding cassette (ABC) transporter subfamily A (ABCA) located in the ER is responsible for transporting acyl-CoA to the ER [59]. In Nannochloropsis, LACSs are believed to play a critical role in activating and facilitating the formation of acyl-CoA, which is essential for TAG synthesis, lipid elongation, and β-oxidation [60]. Transporter proteins are also predicted to exist in Nannochloropsis but have yet to be functionally identified [55].
There are multiple pathways towards TAG assembly. The Kennedy pathway serves as the primary TAG assembly pathway, comprising four enzymes of glycerol 3-phosphate acyltransferase (GPAT), lysophosphatidic acid acyltransferase (LPAT), phosphatidic acid phosphatase (PAP), and DGAT [61]. GPAT, the initial enzyme in the Kennedy pathway, catalyzes the transacylation of glycerol-3-phosphate (G3P) at the sn-1 position to lysophosphatidic acid (LPA). There are two GPAT isoforms in N. oceanica; the efforts to generate a disruption mutant of each GPAT have failed, indicating that both are essential for the alga [62]. Moreover, the overexpression of the ER-located GPAT in N. oceanica can lead to an increase in non-polar lipids [63]. In N. oceanica, four LPATs (LPAT1-LPAT4) have been identified, and they catalyze the transacylation of LPA at the sn-2 position to form phosphatidic acid (PA), with a preference for C16:0-CoA over C18:0-CoA as the acyl donor [62], similar to LPATs from the green alga Chlamydomonas reinhardtii [58]. The dephosphorylation of PA at the sn-3 position yields diacylglycerol (DAG), the precursor of TAG, which is catalyzed by PAP. It has been reported in N. oceanica that four putative cytoplasmic PAP2 genes are stimulated by TAG-induction conditions, indicative of their roles in TAG biosynthesis [55].
The final step in TAG assembly involves the transacylation of DAG to TAG, catalyzed by DGAT. In N. oceanica, 13 putative homologs of DGAT have been identified, including 2 DGAT1 genes (DGAT1A-B) and 11 DGAT2 genes (DGAT2A-K, also known as DGTT1-11) [17]. Among these, seven DGAT genes are upregulated under nitrogen-depleted conditions, including DGAT1A and six DGAT2 genes [55]. DGAT1A prefers C16 and C18 saturated/monounsaturated fatty acids as substrates, and its overexpression can result in a ~39% increase in the TAG content of N. oceanica under nitrogen deprivation [64]. As for DGAT2 genes, they have been reported to show different preferences on the substrates of acyl-CoA and DAG, play roles in TAG synthesis, and have great potential in the manipulation of N. oceanica for enhancing TAG levels and/or modifying fatty acid compositions [65,66,67,68,69].
Another well-characterized pathway for TAG synthesis in microalgae is acyl-independent and is mediated by phospholipid:diacylglycerol acyltransferase (PDAT). This enzyme transfers an acyl group from the sn-2 position of phospholipids (PLs) to form TAG [70]. Similar to in C. reinhardtii, PDAT in N. oceanica functions parallel to the DGAT-mediated pathway for TAG biosynthesis and shows preferences for the acyl donors [71,72]. It is worth mentioning that N. oceanica PDAT contributes more to the stress-associated TAG synthesis than to the basal TAG synthesis, seemingly distinct from C. reinhardtii PDAT [71].
3.2. EPA Synthesis
EPA, a ω3 long-chain polyunsaturated fatty acid (LC-PUFA), has long been recognized for its potential in preventing and treating human diseases [73]. Nannochloropsis is capable of producing EPA, and its biosynthesis pathway, starting from stearic acid (C18:0), involves a series of fatty acid desaturates (Δ9-FAD, Δ12-FAD, Δ6-FAD, Δ5-FAD, ω3-FAD) and a fatty acid elongase (Δ6-FAE) (Figure 1). These enzymes from Nannochloropsis have been functionally characterized to varying extents, via heterologous expression in the yeast Saccharomyces cerevisiae, overexpression, and/or knockdown [74,75,76,77,78,79]. It is believed that N. oceanica utilizes the ω6 pathway towards EPA biosynthesis, namely, from C18:0 via the intermediates C18:1Δ9, C18:2Δ9,12, C18:3Δ6,9,12, C20:3Δ8,11,14, and C20:4Δ5,8,11,14 [74,75]. By contrast, the diatom alga Phaeodactylum tricornutum employs both the ω6 and ω3 pathways for EPA biosynthesis [80], while Thalassiosira pseudonana likely uses only the ω3 pathway to produce EPA [81]. Additionally, an elongase (Δ0-ELO) that elongates C16:0 to C18:0 is involved in EPA biosynthesis, as its knockout disruption leads to a decreased EPA level in N. gaditana [82]. The overexpression of this elongase in N. oceanica, on the other hand, benefits EPA accumulation in TAG [69]. Understanding the key enzymes involved in the EPA synthesis pathway and their regulation is crucial for the engineering of EPA content and/or distribution in Nannochloropsis and also provides insights into manipulating other organisms for EPA biosynthesis.
It is generally accepted that EPA biosynthesis in Nannochloropsis occurs in the ER, where the key desaturases and elongases are predicted to be located. This is further confirmed via the ER subcellular localization experiment for Δ6-FAE, a key enzyme in EPA biosynthesis in N. oceanica [75]. It should be noted that in Nannochloropsis, similar to in diatoms, EPA is mainly distributed in the chloroplast membrane lipids [69], raising the question that how EPA is channeled from the ER into chloroplasts. While there have been several studies dealing with this, it remains largely unknown [83,84]. Additionally, EPA in chloroplast membrane lipids can be transferred to neutral lipids (NLs) like TAG during nitrogen deprivation, as demonstrated through 13C isotopic labeling in the microalgae N. gaditana [85]. The abundance of EPA in TAG, nevertheless, is relatively low for Nannochloropsis [69]. Nannochloropsis also produces long-chain hydroxy fatty acids (LCHFAs), long-chain alkenols (LCAs), and long-chain alkyl diols (LCDs); some enzymes involved in the biosynthesis of these compounds have been identified, including polyketide synthases (PKSs) and 3-hydroxyacyl dehydratases (HADs) [86].
3.3. Carotenoid Biosynthesis
The Nannochloropsis chloroplast evolves from the secondary endosymbiosis and is surrounded by four membranes. Unlike green algae with chlorophylls a and b or diatoms with chlorophylls a and c, Nannochloropsis contains only chlorophyll a in its light-harvesting pigment–protein complexes (LHCs) [87]. The proteomic analysis identified 17 LHC-type proteins among the 21 known in the genus Nannochloropsis, including LHCR-type proteins (related to red algae LHCI), LHCV proteins, and others [88]. LHCV also refers to violaxanthin/vaucheriaxanthin chlorophyll protein (VCP), where the two central carotenoids are surrounded by five chlorophyll a molecules, preserving the LHC superfamily structure [89]. The energy transfer efficiency of the VCP complex, from carotenoids to chlorophyll a, can reach up to 90% [90]. The two carotenoids serve as the accessory pigments in light harvesting for Nannochloropsis [91]. β-carotene is another prominent pigment in Nannochloropsis. Other carotenoids, such as neoxanthin, antheraxanthin, zeaxanthin, canthaxanthin, and astaxanthin, are also present in Nannochloropsis but at a low level [24,92]. Compared to green algae, Nannochloropsis lacks α-carotene and lutein [92].
The carotenoid composition of Nannochloropsis can be substantially impacted by nutritional and environmental factors, with N. oceanica being the most well studied. Under high light illumination, N. oceanica shows a reduction in chlorophyll a and most carotenoids (e.g., violaxanthin, vaucheriaxanthin, β-carotene, and neoxanthin), accompanied by an increase in zeaxanthin [24,92,93]. Based on the carotenoid profiles and in silico analysis of carotenogenic genes, carotenoid biosynthetic pathways for N. oceanica have been proposed [92], including the production of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) via the 2-C-methylerythritol 4-phosphate (MEP) pathway, β-carotene biosynthesis from IPP/DMAPP, and β-carotene-derived xanthophyll biosynthesis (Figure 2). Briefly, IPP/DMAPP molecules produced in the MEP pathway are condensed to the 20-carbon geranylgeranyl diphosphate (GGPP) and then to the 40-carbon phytoene through the action of GGPP synthase (GGPPS) and phytoene synthase (PYS), respectively. Phytoene is then desaturated, isomerized, and cyclized to β-carotene, mediated by phytoene desaturase (PDS), ζ-carotene desaturase (ZDS), carotenoid isomerase (CRTISO), and lycopene β-cyclase (LCYB). β-carotene, catalyzed by the heme-containing cytochrome P450 enzymes, is hydroxylated to zeaxanthin, which can be epoxidated to violaxanthin and de-epoxidated back to zeaxanthin, by the enzyme pair zeaxanthin epoxidase (ZEP) and violaxanthin de-epoxidase (VDE). Violaxanthin can be further converted to neoxanthin by a violaxanthin-de-epoxidase-like enzyme (VDL) and then to vaucheriaxanthin via yet-to-be-identified enzymes.
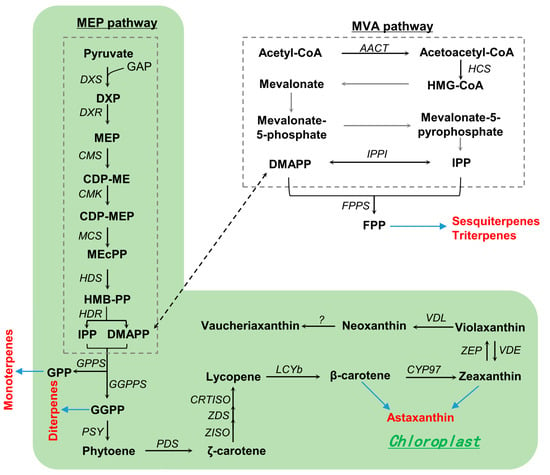
Figure 2.
Proposed carotenoid biosynthetic pathways in N. ocenica. Abbreviations: AACT, acetoacetyl-CoA thiolase; CDP-ME, 4-diphosphocytidyl-2-C-methylerythritol; CDP-MEP, 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate; CMK, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; CMS, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CRTISO, carotenoid isomerase; CYP97, cytochrome P450 beta-hydroxylase; DMAPP, dimethylallyl pyrophosphate; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; DXP, 1-deoxy-D-xylulose 5-phosphate; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; GAP, glyceraldehyde 3-phosphate; GGPP, geranylgeranyl diphosphate; GGPPS, geranylgeranyl diphosphate synthase; HCS, hydroxymethylglutaryl-CoA synthase; HDR, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase; HDS, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; HMB-PP, (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate; IPP, isopentenyl pyrophosphate; IPPI, isopentenyl-diphosphate Delta-isomerase; LCYB, lycopene beta cyclase; MCS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; mEcPP, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate; MEP, 2-C-methylerythritol 4-phosphate; PDS, phytoene desaturase; PSY, phytoene synthase; VDE, violaxanthin de-epoxidase; VDL, violaxanthin-de-epoxidase-like enzyme; ZDS, zeta-carotene desaturase; ZEP, zeaxanthin epoxidase; ZISO, zeta-carotene isomerase; ?, unknown enzyme.
So far, the functional characterization of carotenogenic enzymes for N. oceanica remains limited. It has been reported that PDS overexpression has little effect on carotenoid profiles, while LCYB overexpression benefits the accumulation of carotenoids, particularly β-carotene; in this context, LCYB, rather than PDS, mediates a rate-limiting step for carotenoid biosynthesis in N. oceanica [92]. There are two phylogenetically distant ZEP isoforms in N. oceanica, namely, ZEP1 and ZEP2. The functional roles of the two ZEPs have been revealed in a recent study, which supports the hypothesis that both ZEP1 and ZEP2, localized in the chloroplast, overlap in epoxidating zeaxanthin to violaxanthin for the light-dependent growth of N. oceanica [93]. It is worth noting that ZEP1 appears to be more functional than NoZEP2 [93]. N. oceanica harbors one VDE; deactivating this enzyme via insertional disruption can attenuate zeaxanthin accumulation substantially in response to high light and thus leads to growth impairment [94]. Likely, zeaxanthin contributes to the induction of the non-photochemical quenching and scavenging of reactive oxygen species, thus playing an essential role in protecting the alga from excess light. While violaxanthin is essential for N. oceanica, neoxanthin and vaucheriaxanthin are not, as a disruption to VDL that leads to the abolishment of the two carotenoids is viable [95]. As expected, the VDL-deficient mutant accumulates more violaxanthin [95].
Violaxanthin exhibits significant antioxidative capacity and anti-inflammatory activity against cancer cells [96]. A previous study has demonstrated that extracts from Nannochloropsis, primarily composed of violaxanthin, can protect against UV-B-induced damage [11]. In this context, the pursuit of violaxanthin enhancement in Nannochloropsis is an intriguing avenue of exploration. The overexpression of ZEPs can enable N. oceanica to synthesize more violaxanthin, yet the increase remains to be enlarged [93]. Although N. oceanica synthesizes astaxanthin, a valuable ketocarotenoid with broad applications [97], the level is very low and generally detectable under stress conditions [24]. The enzymes responsible for astaxanthin biosynthesis in the alga have not been identified. Given the relative abundance of β-carotene, the direct precursor of astaxanthin, introducing a strong β-carotenoid ketolase has the potential to allow N. oceanica for synthesizing astaxanthin in addition to EPA [98,99]. Considering the function of ZEP1/2 [93], suppressing its expression may reallocate carotenoid flux towards astaxanthin synthesis in the engineered N. oceanica. The accumulated astaxanthin may, in turn, endow the alga with an improved production performance under high light, as is the case for C. reinhardtii [100]. Moreover, allowing astaxanthin storage in the cytosolic lipid droplets, like in the astaxanthin-rich Haematococcus pluvialis [101], will be a promising strategy for a further increase in astaxanthin in the engineered N. oceanica.
3.4. Other High-Value Products
IPP/DMAPP has multiple destinations: (1) to farnesyl diphosphate (FPP) by FPP synthase (FPPS)—this can be used for the synthesis of sesquiterpenes (e.g., humulene, artemisinic acid, artemisinin) and triterpenes (e.g., dogfish, olefin, oleanolic acid, aromatic acid); (2) to form geranyl diphosphate (GPP) by GPP synthase (GPPS), which can serve as the precursor to monoterpenes (e.g., limonene, menthol); and (3) to GGPP, which can be used for synthesizing diterpenes (e.g., sclareol, taxadiene, paclitaxel) beyond carotenoids. Nannochloropsis has the potential to synthesize these valuable terpenoids via metabolic engineering as long as the corresponding enzymes can be functionally expressed in the alga (Figure 2). As a proof of concept, the biosynthesis of certain terpenoids has been achieved in the green alga C. reinhardtii via metabolic engineering [102,103,104].
Nannochloropsis can be employed as a potential host for the production of recombinant proteins. For example, N. oceanica has been genetically engineered to produce the surface protein VP2 of the infectious pancreatic necrosis virus, which causes high mortality rates in young salmonids [105]. Nannochloropsis is also a promising candidate for the production of marine antimicrobial peptides. These peptides have garnered attention due to their natural ability to combat various pathogens and microbes in the marine environment. The oral administration of antimicrobial peptide-producing N. oculata can enable substantial improvements in the survival rate of medaka after infection with Vibrio parahaemolyticus [106]. Moreover, the biosynthesis of a fish growth hormone has been achieved in N. oculata; this hormone can promote the growth of red tilapia larvae [107]. Overall, genetic engineering and synthetic biology hold the key to utilizing Nannochloropsis to produce targeted products.
4. Cultivation Strategy for Nannochloropsis Production
To optimize Nannochloropsis production, a well-thought-out cultivation strategy is essential. Nitrogen deprivation is a well-established strategy for inducing lipid accumulation in Nannochloropsis. When nutrients are limited, the algal cells tend to divert energy and carbon toward lipid biosynthesis. Nitrogen deprivation can trigger TAG accumulation in N. gaditana, accompanied by a reduction in polar lipids and the partial translocation of EPA from polar lipids to TAG [108]. A reduction in EPA content is also observed for N. gaditana responding to nitrogen deprivation, which is independent of light regimes and intensities [109]. There is a considerable decrease in chlorophyll content upon nitrogen deprivation, accompanied by changes in carotenoids [54].
Multi-stage cultivation represents an effective strategy for maximizing lipid production from Nannochloropsis. In the initial stage, Nannochloropsis is cultivated under nutrient-replete conditions to grow fast and achieve high cell densities. Following the growth phase, the culture is subjected to stress-inducing conditions. Common stressors include nutrient limitations, such as nitrogen and phosphorus deprivation. Stress conditions divert the energy and metabolic resources of the algal cells away from growth to the synthesis of lipids. Such a two-stage cultivation approach can lead to more than a 2-fold increase in lipids for Nannochloropsis [110]. Another effort using a two-stage continuous cultivation approach for N. gaditana, which involved four photobioreactors connected in series, resulted in a remarkable 7.7-fold increase in biomass and a 46% increase in lipid productivity [111].
A fed-batch culture is a sophisticated strategy for production improvements. In this approach, nutrients are supplied incrementally during the growth phase, allowing for higher cell densities and enhanced productivity. This strategy has been employed for the cultivation of various Nannochloropsis species, allowing a considerably higher EPA abundance (based on total fatty acids) and yield in N. oculata [112] and greater biomass production and EPA yield in N. oceanca [69]. Implementing these cultivation strategies, alongside ongoing research and innovation, will play a pivotal role in realizing the full potential of Nannochloropsis for sustainable and economically viable production.
5. Genetic Tools for Nannochloropsis Manipulation
5.1. Genetic Resources
Several species of Nannochloropsis have been sequenced, with a nuclear genome size ~29 Mb nuclear genome that encodes a total of over 10,000 genes [17,113,114,115,116,117]. There are increasing number of transcriptomes regarding Nannochloropsis under different conditions [45,46,55,118,119,120,121,122,123,124]. These genome and transcriptome data help us to understand the biology of Nannochloropsis and its metabolic adaptability to respond to dynamic environmental changes, and, on the other hand, provide genetic resources for the genetic engineering of this organism.
5.2. Transformation Methods, Selectable Markers, and Reporters
Nannochloropsis species contain a rigid cell wall, which serves a barrier for the penetration of transgenes. The first successful nuclear transformation of Nannochloropsis was conducted in 2011 via electroporation with a very high voltage [125]. Since then, the electroporation-mediated transformation method has been established for various Nannochloropsis species [115]. Compared to linearized plasmids, the use of a PCR-amplified cassette can improve Nannochloropsis’ transformation efficiency [126]. The co-transformation of Nannochloropsis with two genes has also been achieved [126]. With the assistance of secondary metabolites from myxobacteria that help weaken the algal cell wall, a 2.7-fold increase in transformation efficiency can be reached for N. salina [127]. Other nuclear transformation methods besides electroporation have been developed as well, such as agrobacterium-mediated transformation for Nannochloropsis sp. [128] and particle bombardment for N. oceanica [129]. Moreover, an electroporation-mediated chloroplast transformation method has been developed, opening up possibilities for plastid genome editing in Nannochloropsis [130]. These diverse transformation techniques offer researchers tools to engineer Nannochloropsis for target applications.
Screening transformants of Nannochloropsis, regardless of the transformation methods, requires the assistance of proper selectable markers. The most widely used selectable marker is the bleomycin-resistant gene, which confers Nannochloropsis with resistance against zeocin [125]. Additional feasible markers for Nannochloropsis transformation include hygromycin-B phosphotransferase, blasticidin-S deaminase, aminoglycoside 3′-phosphotransferase, and nourseothricin acetyltransferase, which are resistant to hygromycin B, blasticidin, G418, and nourseothricin, respectively [15]. These antibiotic resistance genes are not desirable, as they have their limitations and face regulatory hurdles, particularly for food uses, driving the development of endogenous genes as alternative selectable markers. One promising candidate is the mutated PDS gene, which has been employed in several algae to confer resistance to herbicides [131,132,133,134]. While selectable markers facilitate the screening of transgene integration into the nuclear genome, reporters play a pivotal role in monitoring transgene expression at the protein level or subcellular localization when fused with transgenes. The workable reporters in Nannochloropsis include green fluorescent protein [135], sfCherry fluorescent protein [136], luciferases (Nlux, Flux) [74], and purple chromoprotein [137], among others.
To enable the efficient expression of transgenes in Nannochloropsis, regulatory elements are needed, for example, promoters, terminators, enhancers, and transit peptides. Generally, strong endogenous promoters are used for transgene expression in Nannochloropsis, including those from ubiquitin extension protein (UEP) [115,136], β-tubulin (β-tub) [56,138], lipid droplet surface protein (LDSP) [17,74], elongation factor (EF) [66,74], violaxanthin/chlorophyll a-binding protein 2 (VCP2) [64], and ribosomal subunits (Ribi) [74]. The frequently used terminators are those from LDSP, VCP1, and heat shock protein (HS) [15]. To facilitate the transcriptional expression of transgenes, a leader-enhancing sequence may be placed upstream of the transgene [65]. To guide the compartmentation of expressed proteins, transit peptides are needed, for example, the chloroplast transit peptides from glycine cleavage system protein L (GCSL) and VCP1 [135,139]. There is an increasing need for the characterization of more regulatory elements (e.g., inducible promoters, dose-dependent promoters) to meet the needs of diverse uses in the genetic engineering of Nannochloropsis.
5.3. Genetic Tools
The implementation of Nannochloropsis engineering for trait manipulation requires sophisticated genetic tools. Overexpression represents a frequently employed approach for the engineering of Nannochloropsis, which typically involves a single gene of interest. To achieve the expression of multiple genes, these are assembled in separate vectors for Nannochloropsis transformation [69,74]. This, however, requires the use of multiple antibiotic resistance markers. To reduce the use of antibiotic resistance markers, a high-capacity gene-stacking toolkit has recently been developed that takes advantage of the Gateway cloning technique and allows the easy assembly of multiple gene expression cassettes (with individual promoters and terminators) into a single vector for N. oceanica transformation [15]. The transgenes can also be assembled in a single vector, fused by self-cleaving 2A peptides, and have proved workable for multigene expression in N. oceanica [74] and N. salina [140]. This technique eliminates the use of multiple promoters and terminators and thus can reduce the size of vectors. Nevertheless, even with the optimization of 2A types, the cleavage rate of 2A-fused proteins is not high, reaching no more than 50% in N. oceanica [74]. To enable the synthetic biology of Nannochloropsis, a more sophisticated and standardized toolkit is needed. A modular cloning toolkit has been developed for the green alga Chlamydomonas; the toolkit is based on the Golden Gate cloning method with standard syntax and comprises over 100 openly distributed genetic parts [141]. This provides insights into the development of such a toolkit for Nannochloropsis.
Besides overexpression, suppression is a common employed strategy to understand gene function and can be achieved via knockdown or knockout. In Nannochloropsis, RNA interference (RNAi)-mediated gene knockdown has been proven feasible, with the knockdown efficiency reaching up to 80% [56,64,71,93,142,143]. As for gene knockout, the homologous recombination (HR) approach has long been applied to Nannochloropsis, with a starting effort in inactivating the nitrate reductase gene [125]. However, due to the low HR efficiency in Nannochloropsis, HR-mediated gene knockout has only been achieved by a limited number of studies [62,144]. The CRISPR/Cas system represents an emerging technology in gene editing and has been widely applied to various organisms. Using the nitrate reductase gene as the target, the CRISPR/Cas9-mediated gene editing of Nannochloropsis was first developed in N. oceanica but with a very low efficiency of less than 1% [138]. With the assistance of homologous DNA templates, the CRISPR/Cas9-mediated gene knockout efficiency in Nannochloropsis can be improved and varied depending on the target genes [145]. To eliminate the restriction of selectable markers, the combination of CRISPR/Cas9 with an inducible Cre recombinase has been introduced into N. gaditana; this can enable the removal of marker genes and thus the potentially unlimited stacking of gene knockouts in the alga [146]. As well as being integrated into the nuclear genome, the CRISPR/Cas9 in the plasmid with the presence of the CEN/ARS6 region from S. cerevisiae can be maintained as circular extrachromosal DNA in N. oceanica; once targeted mutations are generated, the episomal DNA can be removed in the absence of selection pressure, resulting in marker-free non-transgenic engineered strains [147]. Furthermore, Cas ribonucleoprotein can be delivered into N. oceanica via electroporation for gene editing, thus eliminating the involvement of the plasmid; the editing efficiency using the Cas12a from Francisella novicida can reach up to 90% [148]. With this Cas12a, an episomal plasmid-based CRISPR/Cas12a system has been developed, capable of performing multiplexed gene editing in N. oceanica [149]. Moreover, CRISPR/Cas9-based transcription interference and activation systems have also been established for N. oceanica [149,150].
6. Conclusions
Nannochloropsis harbors advantageous features and has attracted increasing interest from academic and commercial stakeholders. It has emerged as a focal point in synthetic biology due to its versatile genetic characteristics and potential applications in biofuel production, bioremediation, and the synthesis of valuable compounds. Yet there are many things to do to increase the production potential of Nannochloropsis. Firstly, the cultivation strategies for Nannochloropsis need to be upgraded, with an emphasis on large-scale production for various applications. The development of next-generation photobioreactors and corresponding cultivation strategies will be instrumental in achieving commercial-scale biorefining. Secondly, the application of advanced omics technologies will further our understanding of Nannochloropsis’ biology and metabolism, enabling more precise engineering for trait improvements. Thirdly, synthetic biology tools will continue to play a pivotal role in tailoring Nannochloropsis. This will depend on the development of versatile and robust toolkits. Learning from other algae such as C. reinhardtii and P. tricornutum would provide valuable insights and solutions for advancing the synthetic biology of Nannochloropsis. Nannochloropsis-based biorefineries hold great potential for the sustainable production of biofuels, high-value chemicals, and nutraceuticals. A key focus will be to optimize production processes to ensure economic viability and to meet the growing demand for renewable and environmentally friendly products. Navigating the potential applications of Nannochloropsis in synthetic biology is not without its set of challenges. One of the primary hurdles lies in achieving a meticulous level of control over the genetic modifications made to this microalgae genus. Ensuring the stability and predictability of the outcomes is essential for the successful integration of Nannochloropsis-based technologies into industrial processes. The precision required in genetic manipulation becomes increasingly critical as efforts are made to scale up production and consistently meet the demands of various industries.
In conclusion, Nannochloropsis represents a promising light-driven cell factory in the realm of synthetic biology. The inherent ability to synthesize abundant lipids and valuable pigments makes Nannochloropsis an ideal candidate for the sustainable production of biofuels and high-value compounds. Through the concerted efforts of researchers and the continued development of synthetic biology tools, we anticipate substantial advancements in strain engineering, large-scale production, and biorefining. With ongoing research and innovation, Nannochloropsis is set to contribute to shaping a more sustainable and greener future.
Author Contributions
Y.Y.: conceptualization, writing—original draft, resources; M.L.: writing—original draft; L.Y.: resources, supervision; H.S.: writing—review and editing; J.L.: conceptualization, funding acquisitions, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work is partially supported by a grant from National Key R&D Program of China (2018YFA0902500), National Natural Science Foundation of China (32072183), Guangdong Province Zhujiang Talent Program (2019ZT08H476); and Science and Technology Innovation Commission of Shenzhen (KQTD20180412181334790).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yang, S.; Fan, Y.; Cao, Y.; Wang, Y.; Mou, H.; Sun, H. Technological readiness of commercial microalgae species for foods. Crit. Rev. Food Sci. Nutr. 2023, 1–25. [Google Scholar] [CrossRef]
- Yang, S.; Wang, Y.; Wang, J.; Cheng, K.; Liu, J.; He, Y.; Zhang, Y.; Mou, H.; Sun, H. Microalgal protein for sustainable and nutritious foods: A joint analysis of environmental impacts, health benefits and consumer’s acceptance. Trends Food Sci. Technol. 2024, 143, 104278. [Google Scholar] [CrossRef]
- Ma, X.; Chen, T.; Yang, B.; Liu, J.; Chen, F. Lipid Production from Nannochloropsis. Mar. Drugs 2016, 14, 61. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, S.; Han, J.-C.; Yang, G.-P.; Pan, K.-H.; Xu, J.-L. Manipulation of triacylglycerol biosynthesis in Nannochloropsis oceanica by overexpressing an Arabidopsis thaliana diacylglycerol acyltransferase gene. Algal Res.-Biomass Biofuels Bioprod. 2022, 61, 102590. [Google Scholar] [CrossRef]
- Zahran, E.; Elbahnaswy, S.; Ahmed, F.; Ibrahim, I.; Khaled, A.A.; Eldessouki, E.A. Nutritional and immunological evaluation of Nannochloropsis oculata as a potential Nile tilapia-aquafeed supplement. BMC Vet. Res. 2023, 19, 65. [Google Scholar] [CrossRef] [PubMed]
- Soto-Sanchez, O.; Hidalgo, P.; Gonzalez, A.; Oliveira, P.E.; Arias, A.J.H.; Dantagnan, P. Microalgae as Raw Materials for Aquafeeds: Growth Kinetics and Improvement Strategies of Polyunsaturated Fatty Acids Production. Aquac. Nutr. 2023, 2023, 5110281. [Google Scholar] [CrossRef]
- Adissin, T.O.O.; Manabu, I.; Shunsuke, K.; Saichiro, Y.; Moss, A.S.; Dossou, S. Effects of dietary Nannochloropsis sp. powder and lipids on the growth performance and fatty acid composition of larval and postlarval kuruma shrimp, Marsupenaeus japonicus. Aquac. Nutr. 2020, 26, 186–200. [Google Scholar] [CrossRef]
- Lafarga, T.; Mayre, E.; Echeverria, G.; Vinas, I.; Villaro, S.; Gabriel Acien-Fernandez, F.; Castellari, M.; Aguilo-Aguayo, I. Potential of the microalgae Nannochloropsis and Tetraselmis for being used as innovative ingredients in baked goods. Lwt-Food Sci. Technol. 2019, 115, 108439. [Google Scholar] [CrossRef]
- Zuorro, A.; Maffei, G.; Lavecchia, R. Kinetic modeling of azo dye adsorption on non-living cells of Nannochloropsis oceanica. J. Environ. Chem. Eng. 2017, 5, 4121–4127. [Google Scholar] [CrossRef]
- Khattib, A.; Atrahimovich, D.; Dahli, L.; Vaya, J.; Khatib, S. Lyso-diacylglyceryltrimethylhomoserine (lyso-DGTS) isolated from Nannochloropsis microalgae improves high-density lipoprotein (HDL) functions. Biofactors 2020, 46, 146–157. [Google Scholar] [CrossRef]
- Kim, H.-M.; Jung, J.H.; Kim, J.Y.; Heo, J.; Cho, D.-H.; Kim, H.-S.; An, S.; An, I.-S.; Bae, S. The Protective Effect of Violaxanthin from Nannochloropsis oceanica against Ultraviolet B-Induced Damage in Normal Human Dermal Fibroblasts. Photochem. Photobiol. 2019, 95, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Ljubic, A.; Jacobsen, C.; Holdt, S.L.; Jakobsen, J. Microalgae Nannochloropsis oceanica as a future new natural source of vitamin D-3. Food Chem. 2020, 320, 126627. [Google Scholar] [CrossRef] [PubMed]
- Poliner, E.; Farre, E.M.; Benning, C. Advanced genetic tools enable synthetic biology in the oleaginous microalgae Nannochloropsis sp. Plant Cell Rep. 2018, 37, 1383–1399. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wu, T.; Chen, S.H.Y.; Ren, Y.; Yang, S.; Huang, J.; Mou, H.; Chen, F. Powerful tools for productivity improvements in microalgal production. Renew. Sustain. Energy Rev. 2021, 152, 111609. [Google Scholar] [CrossRef]
- Poliner, E.; Clark, E.; Cummings, C.; Benning, C.; Farre, E.M. A high-capacity gene stacking toolkit for the oleaginous microalga, Nannochloropsis oceanica CCMP1779. Algal Res.-Biomass Biofuels Bioprod. 2020, 45, 101664. [Google Scholar] [CrossRef]
- Belshaw, N.; Grouneva, I.; Aram, L.; Gal, A.; Hopes, A.; Mock, T. Efficient gene replacement by CRISPR/Cas-mediated homologous recombination in the model diatom Thalassiosira pseudonana. New Phytol. 2023, 238, 438–452. [Google Scholar] [CrossRef]
- Vieler, A.; Wu, G.; Tsai, C.H.; Bullard, B.; Cornish, A.J.; Harvey, C.; Reca, I.B.; Thornburg, C.; Achawanantakun, R.; Buehl, C.J.; et al. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 2012, 8, e1003064. [Google Scholar] [CrossRef]
- Hibberd, D.J. Notes on the taxonomy and nomenclature of the algal classes Eustigmatophyceae and Tribophyceae (synonym Xanthophyceae). Bot. J. Linn. Soc. 1981, 82, 93–119. [Google Scholar] [CrossRef]
- Scholz, M.J.; Weiss, T.L.; Jinkerson, R.E.; Jing, J.; Roth, R.; Goodenough, U.; Posewitz, M.C.; Gerken, H.G. Ultrastructure and Composition of the Nannochloropsis gaditana Cell Wall. Eukaryot. Cell 2014, 13, 1450–1464. [Google Scholar] [CrossRef]
- Brown, M.R. The Amino acid and sugar composition of 16 species of microalgae used in mariculture. J. Exp. Mar. Biol. Ecol. 1991, 145, 79–100. [Google Scholar] [CrossRef]
- Li, Y.; Huang, A.; Gu, W.; Wu, S.; Xie, X.; Wang, G. Effects of inorganic carbon and light on acetate assimilation by Nannochloropsis oceanica (Eustigmatophyceae) in mixotrophic cultivation. Eur. J. Phycol. 2020, 55, 64–75. [Google Scholar] [CrossRef]
- Onay, M. Enhancing carbohydrate productivity from Nannochloropsis gaditana for bio-butanol production. Energy Rep. 2020, 6, 63–67. [Google Scholar] [CrossRef]
- Wahidin, S.; Idris, A.; Shaleh, S.R.M. The influence of light intensity and photoperiod on the growth and lipid content of microalgae Nannochloropsis sp. Bioresour. Technol. 2013, 129, 7–11. [Google Scholar] [CrossRef]
- Wang, B.B.; Jia, J. Photoprotection mechanisms of Nannochloropsis oceanica in response to light stress. Algal Res.-Biomass Biofuels Bioprod. 2020, 46, 101784. [Google Scholar] [CrossRef]
- Sousa, S.; Freitas, A.C.; Gomes, A.M.; Carvalho, A.P. Modulated stress to balance Nannochloropsis oculata growth and eicosapentaenoic acid production. Appl. Microbiol. Biotechnol. 2022, 106, 4017–4027. [Google Scholar] [CrossRef]
- Lu, X.; Sun, H.; He, Y.; Yang, S.; Chen, F. System metabolic tools reveal fucoxanthin metabolism in Nitzschia laevis for the improvement of fucoxanthin productivity. Front. Mar. Sci. 2023, 10, 1182777. [Google Scholar] [CrossRef]
- Ma, R.; Thomas-Hall, S.R.; Chua, E.T.; Eltanahy, E.; Netzel, M.E.; Netzel, G.; Lu, Y.; Schenk, P.M. LED power efficiency of biomass, fatty acid, and carotenoid production in Nannochloropsis microalgae. Bioresour. Technol. 2018, 252, 118–126. [Google Scholar] [CrossRef]
- Das, P.; Lei, W.; Aziz, S.S.; Obbard, J.P. Enhanced algae growth in both phototrophic and mixotrophic culture under blue light. Bioresour. Technol. 2011, 102, 3883–3887. [Google Scholar] [CrossRef]
- Matsui, H.; Anraku, K.; Kotani, T. Spectrophotometry can monitor changes in algal metabolism triggered by nutrient deficiency in Nannochloropsis oculata cultured under various light-emitting diode light regimes. Fish. Sci. 2019, 85, 167–176. [Google Scholar] [CrossRef]
- Ferrer-Ledo, N.; Stegemueller, L.; Janssen, M.; Wijffels, R.H.; Barbosa, M.J. Growth and fatty acid distribution over lipid classes in Nannochloropsis oceanica acclimated to different temperatures. Front. Plant Sci. 2023, 14, 1078998. [Google Scholar] [CrossRef]
- Gill, S.S.; Willette, S.; Dungan, B.; Jarvis, J.M.; Schaub, T.; VanLeeuwen, D.M.; St Hilaire, R.; Holguin, F.O. Suboptimal Temperature Acclimation Affects Kennedy Pathway Gene Expression, Lipidome and Metabolite Profile of Nannochloropsis salina during PUFA Enriched TAG Synthesis. Mar. Drugs 2018, 16, 425. [Google Scholar] [CrossRef]
- MarKose, S.; Chellappan, A.; Thangamani, P.; George, S.; Thangaswamy, S.; Thavasimuthu, C.; Mariavincent, M. Optimization of physical parameters for the growth and lipid production in Nannochloropsis gaditana (Lubian, 1982). J. Appl. Biol. Biotechnol. 2020, 8, 6–12. [Google Scholar] [CrossRef]
- Yuan, W.; Ma, Y.; Wei, W.; Liu, W.; Ding, Y.; Balamurugan, S. Sequential treatment with bicarbonate and low-temperature to potentiate both biomass and lipid productivity in Nannochloropsis oceanica. J. Chem. Technol. Biotechnol. 2019, 94, 3413–3419. [Google Scholar] [CrossRef]
- Arora, N.; Lo, E.; Philippidis, G.P. A two-prong mutagenesis and adaptive evolution strategy to enhance the temperature tolerance and productivity of Nannochloropsis oculata. Bioresour. Technol. 2022, 364, 128101. [Google Scholar] [CrossRef]
- Fulbright, S.P.; Chisholm, S.; Reardon, K.F. Growth inhibition of Nannochloropsis species by Bacillus pumilus. Algal Res. 2016, 20, 70–76. [Google Scholar] [CrossRef]
- Du, Z.-Y.; Alvaro, J.; Hyden, B.; Zienkiewicz, K.; Benning, N.; Zienkiewicz, A.; Bonito, G.; Benning, C. Enhancing oil production and harvest by combining the marine alga Nannochloropsis oceanica and the oleaginous fungus Mortierella elongata. Biotechnol. Biofuels 2018, 11, 174. [Google Scholar] [CrossRef]
- Pedersen, T.C.; Gardner, R.D.; Gerlach, R.; Peyton, B.M. Assessment of Nannochloropsis gaditana growth and lipid accumulation with increased inorganic carbon delivery. J. Appl. Phycol. 2018, 30, 2155–2166. [Google Scholar] [CrossRef]
- Nunez, M.; Quigg, A. Changes in growth and composition of the marine microalgae Phaeodactylum tricornutum and Nannochloropsis salina in response to changing sodium bicarbonate concentrations. J. Appl. Phycol. 2016, 28, 2123–2138. [Google Scholar] [CrossRef]
- Hu, H.H.; Gao, K.S. Optimization of growth and fatty acid composition of a unicellular marine picoplankton, Nannochloropsis sp., with enriched carbon sources. Biotechnol. Lett. 2003, 25, 421–425. [Google Scholar] [CrossRef]
- Sun, H.; Ren, Y.; Fan, Y.; Lu, X.; Zhao, W.; Chen, F. Systematic metabolic tools reveal underlying mechanism of product biosynthesis in Chromochloris zofingiensis. Bioresour. Technol. 2021, 337, 125406. [Google Scholar] [CrossRef]
- Matos, A.P.; Feller, R.; Siegel Moecke, E.H.; Sant’Anna, E.S. Biomass, lipid productivities and fatty acids composition of marine Nannochloropsis gaditana cultured in desalination concentrate. Bioresour. Technol. 2015, 197, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Shene, C.; Chisti, Y.; Vergara, D.; Burgos-Diaz, C.; Rubilar, M.; Bustarnante, M. Production of eicosapentaenoic acid by Nannochloropsis oculata: Effects of carbon dioxide and glycerol. J. Biotechnol. 2016, 239, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Li, P.; Liao, Z.; Luo, J. Detoxification of ammonium to Nannochloropsis oculata and enhancement of lipid production by mixotrophic growth with acetate. Bioresour. Technol. 2017, 227, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Poddar, N.; Sen, R.; Martin, G.J.O. Glycerol and nitrate utilisation by marine microalgae Nannochloropsis salina and Chlorella sp and associated bacteria during mixotrophic and heterotrophic growth. Algal Res.-Biomass Biofuels Bioprod. 2018, 33, 298–309. [Google Scholar] [CrossRef]
- Zienkiewicz, A.; Zienkiewicz, K.; Poliner, E.; Pulman, J.A.; Du, Z.Y.; Stefano, G.; Tsai, C.H.; Horn, P.; Feussner, I.; Farre, E.M.; et al. The Microalga Nannochloropsis during Transition from Quiescence to Autotrophy in Response to Nitrogen Availability. Plant Physiol. 2020, 182, 819–839. [Google Scholar] [CrossRef] [PubMed]
- Mühlroth, A.; Winge, P.; El Assimi, A.; Jouhet, J.; Maréchal, E.; Hohmann-Marriott, M.F.; Vadstein, O.; Bones, A.M. Mechanisms of Phosphorus Acquisition and Lipid Class Remodeling under P Limitation in a Marine Microalga. Plant Physiol. 2017, 175, 1543. [Google Scholar] [CrossRef]
- Sforza, E.; Calvaruso, C.; La Rocca, N.; Bertucco, A. Luxury uptake of phosphorus in Nannochloropsis salina: Effect of P concentration and light on P uptake in batch and continuous cultures. Biochem. Eng. J. 2018, 134, 69–79. [Google Scholar] [CrossRef]
- Mayers, J.J.; Flynn, K.J.; Shields, R.J. Influence of the N:P supply ratio on biomass productivity and time-resolved changes in elemental and bulk biochemical composition of Nannochloropsis sp. Bioresour. Technol. 2014, 169, 588–595. [Google Scholar] [CrossRef]
- Ruishan, W.U.; Dong, W.E.I. Effects of iron(II) concentration on growth and fatty acid composition of Nannochloropsis oculata. Mar. Sci. 2008, 32, 93–96. [Google Scholar]
- Gu, N.; Lin, Q.; Li, G.; Qin, G.; Lin, J.; Huang, L. Effect of Salinity Change on Biomass and Biochemical Composition of Nannochloropsis oculata. J. World Aquac. Soc. 2012, 43, 97–106. [Google Scholar] [CrossRef]
- Hoshida, H.; Ohira, T.; Minematsu, A.; Akada, R.; Nishizawa, Y. Accumulation of eicosapentaenoic acid in Nannochloropsis sp. in response to elevated CO2 concentrations. J. Appl. Phycol. 2005, 17, 29–34. [Google Scholar] [CrossRef]
- Pal, D.; Khozin-Goldberg, I.; Cohen, Z.; Boussiba, S. The effect of light, salinity, and nitrogen availability on lipid production by Nannochloropsis sp. Appl. Microbiol. Biot. 2011, 90, 1429–1441. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, J.; Liu, B.; Chen, T.; Yang, B.; Chen, F. Physiological and biochemical changes reveal stress-associated photosynthetic carbon partitioning into triacylglycerol in the oleaginous marine alga Nannochloropsis oculata. Algal Res.-Biomass Biofuels Bioprod. 2016, 16, 28–35. [Google Scholar] [CrossRef]
- Simionato, D.; Block, M.A.; La Rocca, N.; Jouhet, J.; Marechal, E.; Finazzi, G.; Morosinotto, T. The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryot. Cell 2013, 12, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, D.; Wang, D.; Ning, K.; Jia, J.; Wei, L.; Jing, X.; Huang, S.; Chen, J.; Li, Y.; et al. Choreography of Transcriptomes and Lipidomes of Nannochloropsis Reveals the Mechanisms of Oil Synthesis in Microalgae. Plant Cell 2014, 26, 1645–1665. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Yao, L.; Yang, B.; Lee, Y.K.; Chen, F.; Liu, J. RNAi-mediated silencing of a pyruvate dehydrogenase kinase enhances triacylglycerol biosynthesis in the oleaginous marine alga Nannochloropsis salina. Sci. Rep. 2017, 7, 11485. [Google Scholar] [CrossRef]
- Chen, H.; Zheng, Y.; Zhan, J.; He, C.; Wang, Q. Comparative metabolic profiling of the lipid-producing green microalga Chlorella reveals that nitrogen and carbon metabolic pathways contribute to lipid metabolism. Biotechnol. Biofuels 2017, 10, 153. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Beisson, F.; Riekhof, W. Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 2015, 82, 504–522. [Google Scholar] [CrossRef]
- Kim, S.; Yamaoka, Y.; Ono, H.; Kim, H.; Shim, D.; Maeshima, M.; Martinoia, E.; Cahoon, E.B.; Nishida, I.; Lee, Y. AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2013, 110, 773–778. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, X.; Yang, G.; Zhu, B.; Han, J.; Yu, W.; Pan, K. Isolation and characterization of a long-chain acyl-coenzyme A synthetase encoding gene from the marine microalga Nannochloropsis oculata. J. Appl. Phycol. 2012, 24, 873–880. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed]
- Nobusawa, T.; Hori, K.; Mori, H.; Kurokawa, K.; Ohta, H. Differently localized lysophosphatidic acid acyltransferases crucial for triacylglycerol biosynthesis in the oleaginous alga Nannochloropsis. Plant J. 2017, 90, 547–559. [Google Scholar] [CrossRef] [PubMed]
- Südfeld, C.; Kiyani, A.; Wefelmeier, K.; Wijffels, R.H.; Barbosa, M.J.; D’Adamo, S. Expression of glycerol-3-phosphate acyltransferase increases non-polar lipid accumulation in Nannochloropsis oceanica. Microb. Cell Factories 2023, 22, 12. [Google Scholar] [CrossRef]
- Wei, H.; Shi, Y.; Ma, X.; Pan, Y.; Hu, H.; Li, Y.; Luo, M.; Gerken, H.; Liu, J. A type-I diacylglycerol acyltransferase modulates triacylglycerol biosynthesis and fatty acid composition in the oleaginous microalga, Nannochloropsis oceanica. Biotechnol. Biofuels 2017, 10, 174. [Google Scholar] [CrossRef]
- Li, D.W.; Cen, S.Y.; Liu, Y.H.; Balamurugan, S.; Zheng, X.Y.; Alimujiang, A.; Yang, W.D.; Liu, J.S.; Li, H.Y. A type 2 diacylglycerol acyltransferase accelerates the triacylglycerol biosynthesis in heterokont oleaginous microalga Nannochloropsis oceanica. J. Biotechnol. 2016, 229, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Zienkiewicz, K.; Zienkiewicz, A.; Poliner, E.; Du, Z.-Y.; Vollheyde, K.; Herrfurth, C.; Marmon, S.; Farré, E.M.; Feussner, I.; Benning, C. Nannochloropsis, a rich source of diacylglycerol acyltransferases for engineering of triacylglycerol content in different hosts. Biotechnol. Biofuels 2017, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Lu, Y.; Lee, Y.-Y.; Wei, L.; Jia, J.; Wang, Q.; Wang, D.; Bai, F.; Hu, H.; Hu, Q.; et al. Producing designer oils in industrial microalgae by rational modulation of co-evolving type-2 diacylglycerol acyltransferases. Mol. Plant. 2017, 10, 1523–1539. [Google Scholar] [CrossRef]
- Xin, Y.; Shen, C.; She, Y.; Chen, H.; Wang, C.; Wei, L.; Yoon, K.; Han, D.; Hu, Q.; Xu, J. Biosynthesis of triacylglycerol molecules with a tailored pufa profile in industrial microalgae. Mol. Plant 2019, 12, 474–488. [Google Scholar] [CrossRef]
- Liu, J.; Liu, M.; Pan, Y.; Shi, Y.; Hu, H. Metabolic engineering of the oleaginous alga Nannochloropsis for enriching eicosapentaenoic acid in triacylglycerol by combined pulling and pushing strategies. Metab. Eng. 2022, 69, 163–174. [Google Scholar] [CrossRef]
- Boyle, N.R.; Page, M.D.; Liu, B.; Blaby, I.K.; Casero, D.; Kropat, J.; Cokus, S.J.; Hong-Hermesdorf, A.; Shaw, J.; Karpowicz, S.J.; et al. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 2012, 287, 15811–15825. [Google Scholar] [CrossRef]
- Yang, J.; Liu, J.; Pan, Y.; Maréchal, E.; Amato, A.; Liu, M.; Gong, Y.; Li, Y.; Hu, H. PDAT regulates PE as transient carbon sink alternative to triacylglycerol in Nannochloropsis. Plant Physiol. 2022, 189, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.; Han, D.; Li, Y.; Sommerfeld, M.; Hu, Q. Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 2012, 24, 3708–3724. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.D.; Srirama, K.; Carani, B.S. Omega-3 Fatty Acids for Nutrition and Medicine: Considering Microalgae Oil as a Vegetarian Source of EPA and DHA. Curr. Diabetes Rev. 2007, 3, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Poliner, E.; Pulman, J.A.; Zienkiewicz, K.; Childs, K.; Benning, C.; Farré, E.M. A toolkit for Nannochloropsis oceanica CCMP1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production. Plant Biotechnol. J. 2018, 16, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, M.; Pan, Y.; Hu, H.; Liu, J. Δ6 fatty acid elongase is involved in eicosapentaenoic acid biosynthesis via the ω6 pathway in the marine alga Nannochloropsis oceanica. J. Agr. Food Chem. 2021, 69, 9837–9848. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ye, S.-C.; Chen, W.-B.; Han, J.-C.; Tian, J.-J.; Zhang, Y.-B.; Xu, J.-L.; Cao, J.-Y.; Qin, C. Screening the rate-limiting genes in the ω6 polyunsaturated fatty acid biosynthesis pathway in Nannochloropsis oceanica. Algal Res. 2021, 57, 102342. [Google Scholar] [CrossRef]
- Kaye, Y.; Grundman, O.; Leu, S.; Zarka, A.; Zorin, B.; Didi-Cohen, S.; Khozin-Goldberg, I.; Boussiba, S. Metabolic engineering toward enhanced LC-PUFA biosynthesis in Nannochloropsis oceanica: Overexpression of endogenous Δ12 desaturase driven by stress-inducible promoter leads to enhanced deposition of polyunsaturated fatty acids in TAG. Algal Res.-Biomass Biofuels Bioprod. 2015, 11, 387–398. [Google Scholar] [CrossRef]
- Yang, F.; Yuan, W.; Ma, Y.; Balamurugan, S.; Li, H.-Y.; Fu, S.; Wu, L. Harnessing the Lipogenic Potential of Δ6-Desaturase for Simultaneous Hyperaccumulation of Lipids and Polyunsaturated Fatty Acids in Nannochloropsis oceanica. Front. Mar. Sci. 2019, 6, 682. [Google Scholar] [CrossRef]
- Guo, M.; Chen, G.; Chen, J.; Zheng, M. Identification of a Long-Chain Fatty Acid Elongase from Nannochloropsis sp. Involved in the Biosynthesis of Fatty Acids by Heterologous Expression in Saccharomyces cerevisiae. J. Ocean. Univ. China 2019, 18, 1199–1206. [Google Scholar] [CrossRef]
- Arao, T.; Yamada, M. Biosynthesis of polyunsaturated fatty acids in the marine diatom, Phaeodactylum tricornutum. Phytochemistry 1994, 35, 1177–1181. [Google Scholar] [CrossRef]
- Cook, O.; Hildebrand, M. Enhancing LC-PUFA production in Thalassiosira pseudonana by overexpressing the endogenous fatty acid elongase genes. J. Appl. Phycol. 2016, 28, 897–905. [Google Scholar] [CrossRef]
- Dolch, L.-J.; Rak, C.; Perin, G.; Tourcier, G.; Broughton, R.; Leterrier, M.; Morosinotto, T.; Tellier, F.; Faure, J.-D.; Falconet, D.; et al. A palmitic acid elongase affects eicosapentaenoic acid and plastidial monogalactosyldiacylglycerol levels in Nannochloropsis. Plant Physiol. 2017, 173, 742–759. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Jia, J.; Li, J.; Sommerfeld, M.; Xu, J.; Hu, Q. Metabolic remodeling of membrane glycerolipids in the microalga Nannochloropsis oceanica under nitrogen deprivation. Front. Mar. Sci. 2017, 4, 242. [Google Scholar] [CrossRef]
- Meng, Y.; Cao, X.; Yang, M.; Liu, J.; Yao, C.; Xue, S. Glycerolipid remodeling triggered by phosphorous starvation and recovery in Nannochloropsis oceanica. Algal Res. 2019, 39, 101451. [Google Scholar] [CrossRef]
- Janssen, J.H.; Lamers, P.P.; de Vos, R.C.H.; Wijffels, R.H.; Barbosa, M.J. Translocation and de novo synthesis of eicosapentaenoic acid (EPA) during nitrogen starvation in Nannochloropsis gaditana. Algal Res. 2019, 37, 138–144. [Google Scholar] [CrossRef]
- Balzano, S.; Villanueva, L.; de Bar, M.; Sahonero Canavesi, D.X.; Yildiz, C.; Engelmann, J.C.; Marechal, E.; Lupette, J.; Sinninghe Damsti, J.S.; Schouten, S. Biosynthesis of Long Chain Alkyl Diols and Long Chain Alkenols in Nannochloropsis spp. (Eustigmatophyceae). Plant Cell Physiol. 2019, 60, 1666–1682. [Google Scholar] [CrossRef] [PubMed]
- Umetani, I.; Kunugi, M.; Yokono, M.; Takabayashi, A.; Tanaka, A. Evidence of the supercomplex organization of photosystem II and light-harvesting complexes in Nannochloropsis granulata. Photosynth. Res. 2018, 136, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Litvin, R.; Bina, D.; Herbstova, M.; Gardian, Z. Architecture of the light-harvesting apparatus of the eustigmatophyte alga Nannochloropsis oceanica. Photosynth. Res. 2016, 130, 137–150. [Google Scholar] [CrossRef]
- Carbonera, D.; Agostini, A.; Di Valentin, M.; Gerotto, C.; Basso, S.; Giacometti, G.M.; Morosinotto, T. Photoprotective sites in the violaxanthin–chlorophyll a binding Protein (VCP) from Nannochloropsis gaditana. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 1235–1246. [Google Scholar] [CrossRef]
- Keşan, G.; Litvín, R.; Bína, D.; Durchan, M.; Šlouf, V.; Polívka, T. Efficient light-harvesting using non-carbonyl carotenoids: Energy transfer dynamics in the VCP complex from Nannochloropsis oceanica. Biochim. Biophys. Acta (BBA)-Bioenerg. 2016, 1857, 370–379. [Google Scholar] [CrossRef]
- Fu, Y.; Wang, Y.; Yi, L.; Liu, J.; Yang, S.; Liu, B.; Chen, F.; Sun, H. Lutein production from microalgae: A review. Bioresour. Technol. 2023, 376, 128875. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ding, W.; Yu, L.; Shi, Y.; Liu, J. Functional characterization of carotenogenic genes provides implications into carotenoid biosynthesis and engineering in the marine alga Nannochloropsis oceanica. Algal Res. 2022, 67, 102853. [Google Scholar] [CrossRef]
- Liu, M.; Ding, W.; Pan, Y.; Hu, H.; Liu, J. Zeaxanthin epoxidase is involved in the carotenoid biosynthesis and light-dependent growth of the marine alga Nannochloropsis oceanica. Biotechnol. Biofuels Bioprod. 2023, 16, 74. [Google Scholar] [CrossRef] [PubMed]
- Perin, G.; Bellan, A.; Michelberger, T.; Lyska, D.; Wakao, S.; Niyogi, K.K.; Morosinotto, T. Modulation of xanthophyll cycle impacts biomass productivity in the marine microalga Nannochloropsis. Proc. Natl. Acad. Sci. USA 2023, 120, e2214119120. [Google Scholar] [CrossRef]
- Dautermann, O.; Lyska, D.; Andersen-Ranberg, J.; Becker, M.; Fröhlich-Nowoisky, J.; Gartmann, H.; Krämer, L.C.; Mayr, K.; Pieper, D.; Rij, L.M.; et al. An algal enzyme required for biosynthesis of the most abundant marine carotenoids. Sci. Adv. 2020, 6, eaaw9183. [Google Scholar] [CrossRef]
- Pasquet, V.; Morisset, P.; Ihammouine, S.; Chepied, A.; Aumailley, L.; Berard, J.-B.; Serive, B.; Kaas, R.; Lanneluc, I.; Thiery, V.; et al. Antiproliferative activity of violaxanthin isolated from bioguided fractionation of Dunaliella tertiolecta extracts. Mar. Drugs 2011, 9, 819–831. [Google Scholar] [CrossRef]
- Guerin, M.; Huntley, M.E.; Olaizola, M. Haematococcus astaxanthin: Applications for human health and nutrition. Trends Biotechnol. 2003, 21, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ye, Y.; Ding, W.; Mao, X.; Li, Y.; Gerken, H.; Liu, J. Astaxanthin is ketolated from zeaxanthin independent of fatty acid synthesis in Chromochloris zofingiensis. Plant Physiol. 2020, 183, 883–897. [Google Scholar] [CrossRef]
- Perozeni, F.; Cazzaniga, S.; Baier, T.; Zanoni, F.; Zoccatelli, G.; Lauersen, K.J.; Wobbe, L.; Ballottari, M. Turning a green alga red: Engineering astaxanthin biosynthesis by intragenic pseudogene revival in Chlamydomonas reinhardtii. Plant Biotechnol. J. 2020, 18, 2053–2067. [Google Scholar] [CrossRef]
- Cazzaniga, S.; Perozeni, F.; Baier, T.; Ballottari, M. Engineering astaxanthin accumulation reduces photoinhibition and increases biomass productivity under high light in Chlamydomonas reinhardtii. Biotechnol. Biofuels Bioprod. 2022, 15, 77. [Google Scholar] [CrossRef]
- Zhekisheva, M.; Zarka, A.; Khozin-Goldberg, I.; Cohen, Z.; Boussiba, S. Inhibition of astaxanthin synthesis under high irradiance does not abolish triacylglycerol accumulation in the green alga haematococcus pluvialis (chlorophyceae)1. J. Phycol. 2005, 41, 819–826. [Google Scholar] [CrossRef]
- Lauersen, K.J.; Wichmann, J.; Baier, T.; Kampranis, S.C.; Pateraki, I.; Møller, B.L.; Kruse, O. Phototrophic production of heterologous diterpenoids and a hydroxy-functionalized derivative from Chlamydomonas reinhardtii. Metab. Eng. 2018, 49, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Wichmann, J.; Baier, T.; Wentnagel, E.; Lauersen, K.J.; Kruse, O. Tailored carbon partitioning for phototrophic production of (E)-α-bisabolene from the green microalga Chlamydomonas reinhardtii. Metab. Eng. 2018, 45, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Einhaus, A.; Steube, J.; Freudenberg, R.A.; Barczyk, J.; Baier, T.; Kruse, O. Engineering a powerful green cell factory for robust photoautotrophic diterpenoid production. Metab. Eng. 2022, 73, 82–90. [Google Scholar] [CrossRef] [PubMed]
- de Grahl, I.; Reumann, S. Stramenopile microalgae as “green biofactories” for recombinant protein production. World J. Microbiol. Biotechnol. 2021, 37, 163. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-S.; Tsai, H.-J. Transgenic microalgae as a non-antibiotic bactericide producer to defend against bacterial pathogen infection in the fish digestive tract. Fish. Shellfish. Immunol. 2009, 26, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Li, S.S.; Huang, R.; Tsai, H.-J. Conditional production of a functional fish growth hormone in the transgenic line of Nannochloropsis oculata (Eustigmatophyceae). J. Phycol. 2008, 44, 768–776. [Google Scholar] [CrossRef]
- Janssen, J.H.; Spoelder, J.; Koehorst, J.J.; Schaap, P.J.; Wijffels, R.H.; Barbosa, M.J. Time-dependent transcriptome profile of genes involved in triacylglycerol (TAG) and polyunsaturated fatty acid synthesis in Nannochloropsis gaditana during nitrogen starvation. J. Appl. Phycol. 2020, 32, 1153–1164. [Google Scholar] [CrossRef]
- Janssen, J.H.; Wijffels, R.H.; Barbosa, M.J. Lipid Production in Nannochloropsis gaditana during Nitrogen Starvation. Biology 2019, 8, 5. [Google Scholar] [CrossRef]
- Yen Thi Thai, D.; Obbard, J.P. Two-stage cultivation of a Nannochloropsis mutant for biodiesel feedstock. J. Appl. Phycol. 2015, 27, 2203–2208. [Google Scholar] [CrossRef]
- Sung, M.-G.; Lee, B.; Kim, C.W.; Nam, K.; Chang, Y.K. Enhancement of lipid productivity by adopting multi-stage continuous cultivation strategy in Nannochloropsis gaditana. Bioresour. Technol. 2017, 229, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Gharat, K.; Agarwal, A.; Pandit, R.A.; Lali, A.M. Development of fed batch strategies to improve the production of eicosapentaenoic acid from a marine microalga Nannochloropsis oculata. Bioresour. Technol. Rep. 2018, 4, 193–201. [Google Scholar] [CrossRef]
- Wang, D.; Ning, K.; Li, J.; Hu, J.; Han, D.; Wang, H.; Zeng, X.; Jing, X.; Zhou, Q.; Su, X.; et al. Nannochloropsis Genomes Reveal Evolution of Microalgal Oleaginous Traits. PLoS Genet. 2014, 10, e1004094. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, A.S.; Brown, R.; Ajjawi, I.; McCarren, J.; Atilla, S.; Bauman, N.; Richardson, T.H. Complete Genome Sequence of the Model Oleaginous Alga Nannochloropsis gaditana CCMP1894. Genome Announc. 2018, 6, 10–1128. [Google Scholar] [CrossRef]
- Radakovits, R.; Jinkerson, R.E.; Fuerstenberg, S.I.; Tae, H.; Settlage, R.E.; Boore, J.L.; Posewitz, M.C. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 2012, 3, 686. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.Q.; Wang, D.M.; Li, J.; Jing, G.C.; Ning, K.; Xu, J. Genome-wide identification of transcription factors and transcription-factor binding sites in oleaginous microalgae Nannochloropsis. Sci. Rep. 2014, 4, 5454. [Google Scholar] [CrossRef]
- Corteggiani Carpinelli, E.; Telatin, A.; Vitulo, N.; Forcato, C.; D’Angelo, M.; Schiavon, R.; Vezzi, A.; Giacometti, G.M.; Morosinotto, T.; Valle, G. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant 2014, 7, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, M.; Ding, W.; Liu, J. Novel insights into phosphorus deprivation-boosted lipid synthesis in the marine alga Nannochloropsis oceanica without compromising biomass production. J. Agr. Food Chem. 2020, 68, 11488–11502. [Google Scholar] [CrossRef]
- Wei, L.; El Hajjami, M.; Shen, C.; You, W.; Lu, Y.; Li, J.; Jing, X.; Hu, Q.; Zhou, W.; Poetsch, A.; et al. Transcriptomic and proteomic responses to very low CO2 suggest multiple carbon concentrating mechanisms in Nannochloropsis oceanica. Biotechnol. Biofuels 2019, 12, 168. [Google Scholar] [CrossRef]
- Hulatt, C.J.; Smolina, I.; Dowle, A.; Kopp, M.; Vasanth, G.K.; Hoarau, G.G.; Wijffels, R.H.; Kiron, V. Proteomic and transcriptomic patterns during lipid remodeling in Nannochloropsis gaditana. Int. J. Mol. Sci. 2020, 21, 6946. [Google Scholar] [CrossRef]
- Wei, L.; You, W.; Gong, Y.; El Hajjami, M.; Liang, W.; Xu, J.; Poetsch, A. Transcriptomic and proteomic choreography in response to light quality variation reveals key adaption mechanisms in marine Nannochloropsis oceanica. Sci. Total Environ. 2020, 720, 137667. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Wei, L.; Gong, Y.; Hajjami, M.E.; Xu, J.; Poetsch, A. Integration of proteome and transcriptome refines key molecular processes underlying oil production in Nannochloropsis oceanica. Biotechnol. Biofuels 2020, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Poliner, E.; Panchy, N.; Newton, L.; Wu, G.; Lapinsky, A.; Bullard, B.; Zienkiewicz, A.; Benning, C.; Shiu, S.-H.; Farre, E.M. Transcriptional coordination of physiological responses in Nannochloropsis oceanica CCMP1779 under light/dark cycles. Plant J. 2015, 83, 1097–1113. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Iqbal, S.; Wen, F.; Tong, M.; Liu, J. Phosphorus-Induced Lipid Class Alteration Revealed by Lipidomic and Transcriptomic Profiling in Oleaginous Microalga Nannochloropsis sp. PJ12. Mar. Drugs 2019, 17, 519. [Google Scholar] [CrossRef] [PubMed]
- Kilian, O.; Benemann, C.S.; Niyogi, K.K.; Vick, B. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl. Acad. Sci. USA 2011, 108, 21265–21269. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Gao, D.; Hu, H. High-efficiency nuclear transformation of the oleaginous marine Nannochloropsis species using PCR product. Biosci. Biotechnol. Biochem. 2014, 78, 812–817. [Google Scholar] [CrossRef]
- Jeon, S. Optimization of electroporation-based multiple pulses and further improvement of transformation efficiency using bacterial conditioned medium for Nannochloropsis salina. J. Appl. Phycol. 2019, 31, 1153–1161. [Google Scholar] [CrossRef]
- Cha, T.-S.; Chen, C.-F.; Yee, W.; Aziz, A.; Loh, S.-H. Cinnamic acid, coumarin and vanillin: Alternative phenolic compounds for efficient Agrobacterium-mediated transformation of the unicellular green alga, Nannochloropsis sp. J. Microbiol. Methods 2011, 84, 430–434. [Google Scholar] [CrossRef]
- Anley, K.A. Developing Molecular Tools to Genetically Engineer the Microalga Nannochloropsis; NTNU: Trondheim, Norway, 2015. [Google Scholar]
- Gan, Q.; Jiang, J.; Han, X.; Wang, S.; Lu, Y. Engineering the Chloroplast Genome of Oleaginous Marine Microalga Nannochloropsis oceanica. Front. Plant Sci. 2018, 9, 439. [Google Scholar] [CrossRef]
- Liu, J.; Gerken, H.; Huang, J.; Chen, F. Engineering of an endogenous phytoene desaturase gene as a dominant selectable marker for Chlamydomonas reinhardtii transformation and enhanced biosynthesis of carotenoids. Process Biochem. 2013, 48, 788–795. [Google Scholar] [CrossRef]
- Liu, J.; Sun, Z.; Gerken, H.; Huang, J.; Jiang, Y.; Chen, F. Genetic engineering of the green alga Chlorella zofingiensis: A modified norflurazon-resistant phytoene desaturase gene as a dominant selectable marker. Appl. Microbiol. Biotechnol. 2014, 98, 5069–5079. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, J.; Sandmann, G. Transformation of the green alga Haematococcus pluvialis with a phytoene desaturase for accelerated astaxanthin biosynthesis. Appl. Environ. Microbiol. 2006, 72, 7477–7484. [Google Scholar] [CrossRef] [PubMed]
- Taparia, Y.; Zarka, A.; Leu, S.; Zarivach, R.; Boussiba, S.; Khozin-Goldberg, I. A novel endogenous selection marker for the diatom Phaeodactylum tricornutum based on a unique mutation in phytoene desaturase 1. Sci. Rep. 2019, 9, 8217. [Google Scholar] [CrossRef] [PubMed]
- Moog, D.; Stork, S.; Reislohner, S.; Grosche, C.; Maier, U.G. In vivo localization studies in the stramenopile alga Nannochloropsis oceanica. Protist 2015, 166, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.K.; Choi, G.-G.; Kim, E.K.; Shin, S.-E.; Jeon, S.; Park, M.S.; Jeong, K.J.; Jeong, B.-r.; Chang, Y.K.; Yang, J.-W.; et al. Heterologous overexpression of sfCherry fluorescent protein in Nannochloropsis salina. Biotechnol. Rep. 2015, 8, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.-H.; Chen, H.-Y.; Lee, H.-C.; Tsai, H.-J. Purple chromoprotein gene serves as a new selection marker for transgenesis of the microalga Nannochloropsis oculata. PLoS ONE 2015, 10, e0120780. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lu, Y.; Xin, Y.; Wei, L.; Huang, S.; Xu, J. Genome editing of model oleaginous microalgae Nannochloropsis spp. by CRISPR/Cas9. Plant J. 2016, 88, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.G.; Kang, N.K.; Jeon, S.; Shin, S.-E.; Jeong, B.-R.; Chang, Y.K. Heterologous synthesis of chlorophyll b in Nannochloropsis salina enhances growth and lipid production by increasing photosynthetic efficiency. Biotechnol. Biofuels 2019, 12, 122. [Google Scholar] [CrossRef]
- Koh, H.G.; Kang, N.K.; Kim, E.K.; Jeon, S.; Shin, S.-E.; Lee, B.; Chang, Y.K. Advanced multigene expression system for Nannochloropsis salina using 2A self-cleaving peptides. J. Biotechnol. 2018, 278, 39–47. [Google Scholar] [CrossRef]
- Crozet, P.; Navarro, F.J.; Willmund, F.; Mehrshahi, P.; Bakowski, K.; Lauersen, K.J.; Perez-Perez, M.-E.; Auroy, P.; Gorchs Rovira, A.; Sauret-Gueto, S.; et al. Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2074–2086. [Google Scholar] [CrossRef]
- Poliner, E.; Busch, A.W.U.; Newton, L.; Kim, Y.U.; Clark, R.; Gonzalez-Martinez, S.C.; Jeong, B.-r.; Montgomery, B.L.; Farré, E.M. Aureochromes maintain polyunsaturated fatty acid content in Nannochloropsis oceanica. Plant Physiol. 2022, 189, 906–921. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-W.; Balamurugan, S.; Yang, Y.-F.; Zheng, J.-W.; Huang, D.; Zou, L.-G.; Yang, W.-D.; Liu, J.-S.; Guan, Y.; Li, H.-Y. Transcriptional regulation of microalgae for concurrent lipid overproduction and secretion. Sci. Adv. 2019, 5, eaau3795. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Kakutani, N.; Kuroyanagi, Y.; Iwai, M.; Hori, K.; Shimojima, M.; Ohta, H. MYB-like transcription factor NoPSR1 is crucial for membrane lipid remodeling under phosphate starvation in the oleaginous microalga Nannochloropsis oceanica. Febs. Lett. 2020, 594, 3384–3394. [Google Scholar] [CrossRef]
- Ajjawi, I.; Verruto, J.; Aqui, M.; Soriaga, L.B.; Coppersmith, J.; Kwok, K.; Peach, L.; Orchard, E.; Kalb, R.; Xu, W.; et al. Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat. Biotech. 2017, 35, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Verruto, J.; Francis, K.; Wang, Y.; Low, M.C.; Greiner, J.; Tacke, S.; Kuzminov, F.; Lambert, W.; McCarren, J.; Ajjawi, I.; et al. Unrestrained markerless trait stacking in Nannochloropsis gaditana through combined genome editing and marker recycling technologies. Proc. Natl. Acad. Sci. USA 2018, 115, E7015. [Google Scholar] [CrossRef] [PubMed]
- Poliner, E.; Takeuchi, T.; Du, Z.-Y.; Benning, C.; Farré, E.M. Nontransgenic marker-free gene disruption by an episomal CRISPR system in the oleaginous microalga, Nannochloropsis oceanica CCMP1779. ACS Synth. Biol. 2018, 7, 962–968. [Google Scholar] [CrossRef]
- Naduthodi, M.I.S.; Mohanraju, P.; Südfeld, C.; D’Adamo, S.; Barbosa, M.J.; van der Oost, J. CRISPR-Cas ribonucleoprotein mediated homology-directed repair for efficient targeted genome editing in microalgae Nannochloropsis oceanica IMET1. Biotechnol. Biofuels 2019, 12, 66. [Google Scholar] [CrossRef]
- Naduthodi, M.I.S.; Südfeld, C.; Avitzigiannis, E.K.; Trevisan, N.; van Lith, E.; Alcaide Sancho, J.; D’Adamo, S.; Barbosa, M.; van der Oost, J. Comprehensive genome engineering toolbox for microalgae Nannochloropsis oceanica based on CRISPR-Cas systems. ACS Synth. Biol. 2021, 10, 3369–3378. [Google Scholar] [CrossRef]
- Wei, L.; Jiang, Z.; Liu, B. A CRISPR/dCas9-based transcription activation system developed in marine microalga Nannochloropsis oceanica. Aquaculture 2021, 546, 737064. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).