Abstract
Pyrrole–imidazole alkaloids (PIAs) are a class of marine sponge derived natural products which have complex carbon frameworks and broad bioactivities. In this study, four new alkaloids, stylimassalins A–B (1–2), 3, and 5, together with two known compounds (4 and 6), were isolated from Stylissa massa. Compounds 2, 4, and 6 are the C-2 brominated analogues of 1, 3, and 5, respectively. Their structures display three different scaffolds, of which scaffold 1 (compounds 1,2) is new. A new biosynthetic pathway from oroidin, through spongiacidin, to latonduine and scaffold 1 was proposed by our group, in which the C12-N13-cleavaged compounds of spongiacidin (scaffold 2), dubbed seco-spongiacidins (3 and 4), are recognized as a key bridged scaffold, to afford PIA analogues (1,2 and 5,6). An anti-inflammatory evaluation in a zebrafish inflammation model induced by copper sulphate (CuSO4) demonstrated that stylimassalins A and B (1 and 2) could serve as a promising lead scaffold for treating inflammation.
1. Introduction
Pyrrole–imidazole alkaloids (PIAs) are a huge family of marine sponges derived natural products that are marked by structural complexity and biological activity [1,2]. Since the identification of the first PIA, (-)-dibromophakellin, in 1971 [3], hundreds of additional PIAs have been discovered and extensive research on their structural elucidation, biological/chemical syntheses, and bioactivities have been conducted [4,5]. The chemical origin of PIA structures can be traced back to the specific amino acids, proline and lysine, and then transitions to oroidin, a key biogenetic precursor of PIAs. A large majority of documented PIA events could benefit from the chemical transformations of oroidin [6]. Oroidin functions as the basis of structural diversity for the carbon skeleton by undergoing various transformations, such as cyclization and dimerization, to produce complicated PIAs such as palau’amine [7] and stylissadine [8]. Among the multifarious PIA community, monomer ones, including spongiacidin [9], agelastatin [10], phakellin [3], and latonduine [11], have been discovered sequentially in recent decades.
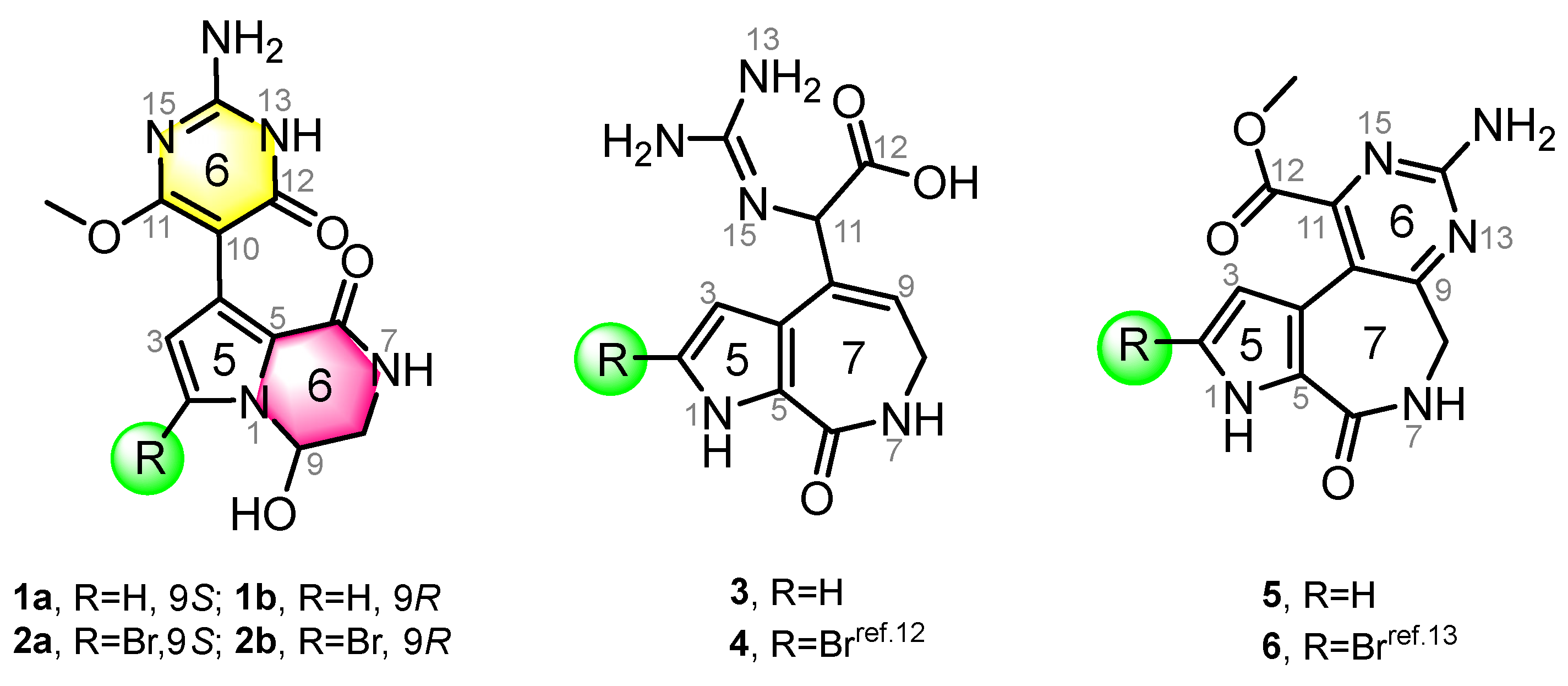
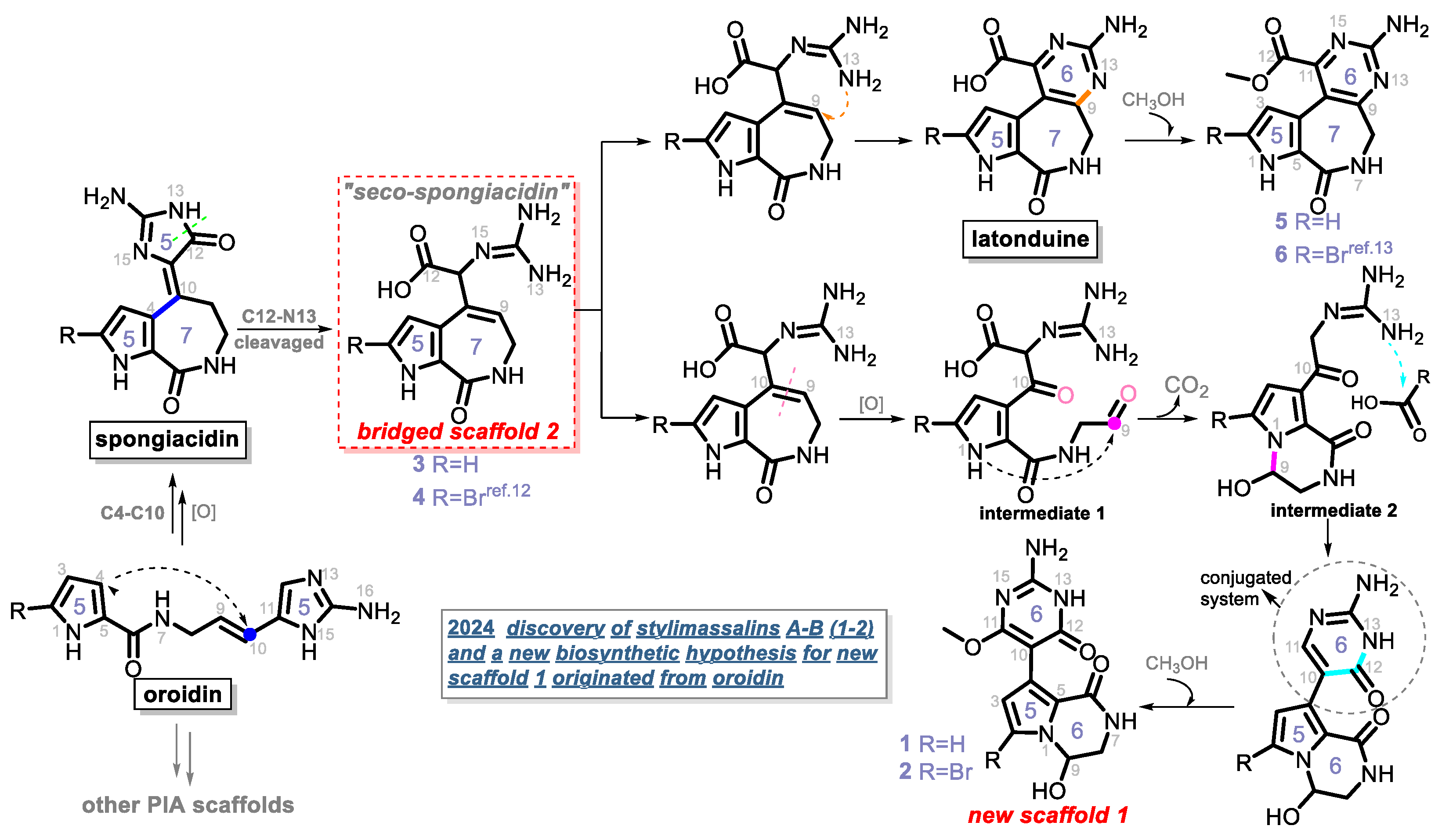
In this study, four new and two known alkaloids (Figure 1) were recovered from the sponge S. massa (Figure S1). Compounds 1 and 2 share the same 5/6/6 tricyclic system, featuring a unique connection between a 3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-one system and a pyrimidine moiety, with the only difference being the presence or absence of bromination at C-2. This is consistent with previous studies noting that PIAs are often accompanied by bromination on their pyrrole ring system. Compounds 1 and 2 (scaffold 1) are not strictly PIAs because they have a rearranged pyrimidine system rather than an imidazole ring. Compounds 3 and 4 [12] are two spongiacidin PIA analogues with a C12-N13 bond cleavage, dubbed secospongiacidin PIAs (scaffold 2). Compounds 5 and 6 [13] belong to the latonduine family of PIA (scaffold 3). A new biosynthetic pathway from oroidin, through spongiacidin, to latonduine and scaffold 1 was proposed in this study, in which seco-spongiacidins (3 and 4, scaffold 2) are recognized as a key bridged scaffold in the formation of latonduine PIA analogues (5 and 6).
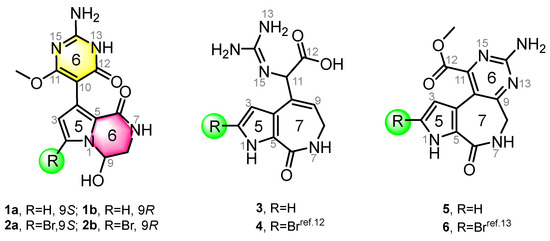
Figure 1.
Chemical structures of compounds 1–6. Stylimassalins A and B (1 and 2) (with a 5/6/6 scaffold) and compounds 3 and 4 are 12,13-secospongiacidin PIAs (with a 5/7 scaffold), and compounds 5 and 6 are latonduine PIAs (with a 5/7/6 scaffold).
2. Results
2.1. Structural Elucidation
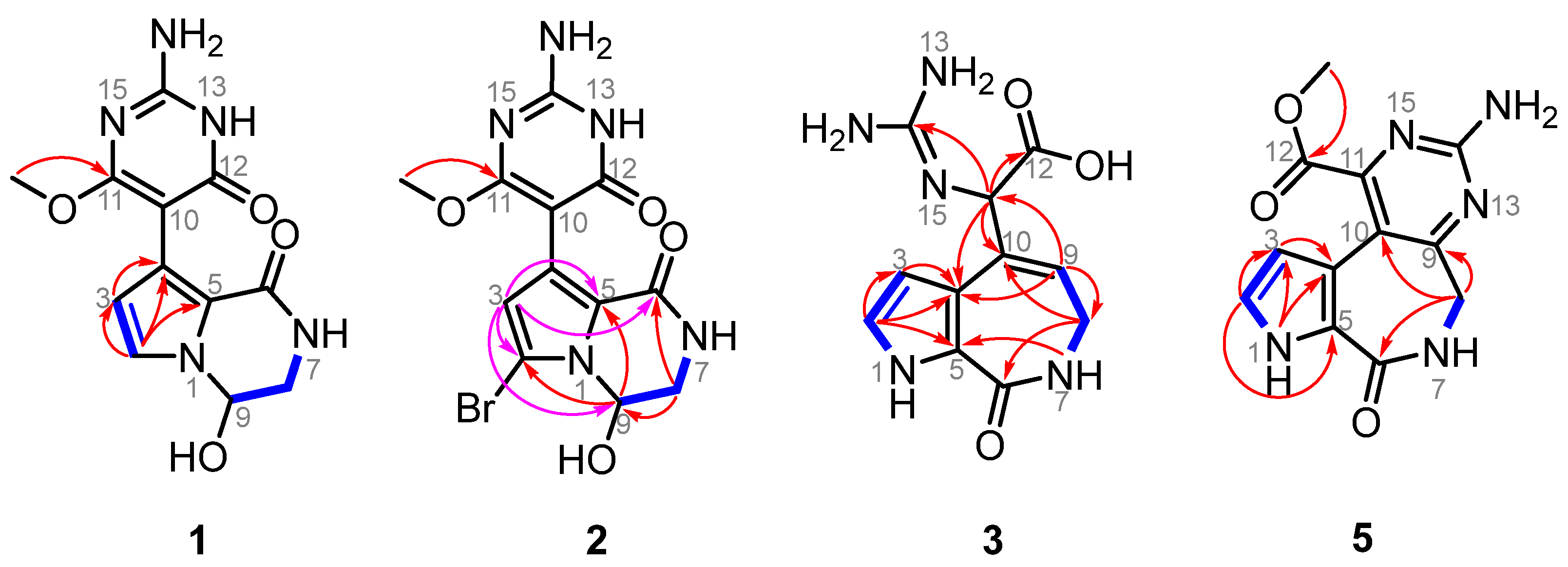
Stylimassalin A (1) was obtained as a yellow oil. In the HRESIMS spectrum of 1, a [M + H]+ ion peak at m/z 292.1036 was observed, which determined its molecular formula to be C12H13N5O4 (calculated for [M + H]+ as 292.1040) with nine degrees of unsaturation. In the 1H NMR spectrum of stylimassalin A (1), acquired in DMSO-d6, there was only one-NH proton signal at δH 7.38, with the regular 1-NH of the pyrrole system at approximately δH 12 not observed. Further analysis of the 1H, 13C NMR (Tables S1 and S2), and HSQC spectra revealed three methines (δH-2 6.93, d, J = 2.6 Hz, δC-2 120.2; δH-3 6.02, d, J = 2.6 Hz, δC-3 112.3; δH-9 5.54, t, J = 3.5 Hz, δC-9 74.4), one methylene group (δH-8a 3.29, ddd, J = 13.2, 7.4, 3.5 Hz, δH-8b 3.54, dd, J = 13.0, 2.1 Hz, δC-8 46.3), one methyl (δ11-OMe 3.65, s, δ11-OMe 53.3), and additional non-protonated carbons (δC-5 119.8; δC-6 119.3; δC-10 89.5; δC-12 159.4; δC-14 153.7; δC-11 167.0). The 1H–1H COSY correlations of H-2 and H-3, together with the HMBC correlations of H-2 with C-3 and C-5 and of H-3 with C-2 and C-5 indicated the presence of a N-substituted pyrrole ring system. The other COSY correlations of 7-NH (δH 7.38), H2-8 (δH 3.29, 3.54), and H-9 (δH 5.54) indicated a NH-CH2-CH molecular fragment. There are no more available 2D NMR correlations in the HMBC spectrum of 1 except for a key correlation between the methyl (δH 3.65) and a quaternary aromatic carbon (δC 167.0). We were confused while determining the structure of stylimassalin A (1), until the discovery of stylimassalin B (2).
Stylimassalin B (2) was obtained as a colourless crystal. Its molecular formula was deduced as C12H12BrN5O4 based on the HRESIMS ions at m/z 370.0135 and 372.0115 (1:1) [M + H]+ (calcd. 370.0145, 372.0125), which indicated the presence of a bromine atom. Detailed analysis of the 1D and 2D NMR spectra (Tables S1 and S2, Figure 2) of both 1 and 2 in DMSO-d6 indicated that their structures were very similar except for the difference in C-2. Obvious high-field movement of the chemical shift of C-2 in compound 2 (2: δC-2 101.9; 1: δC-2 120.2) revealed that the planar structure of 2 is a C-2 brominated analogue of stylimassalin A (1).
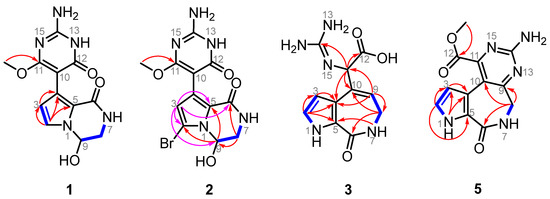
Figure 2.
Structural elucidation of compounds 1, 2, 3, and 5. HMBC correlations are shown using red (acquired via DMSO-d6) and pink (acquired via methanol-d4) arrow lines and COSY correlations are shown using blue bold lines.
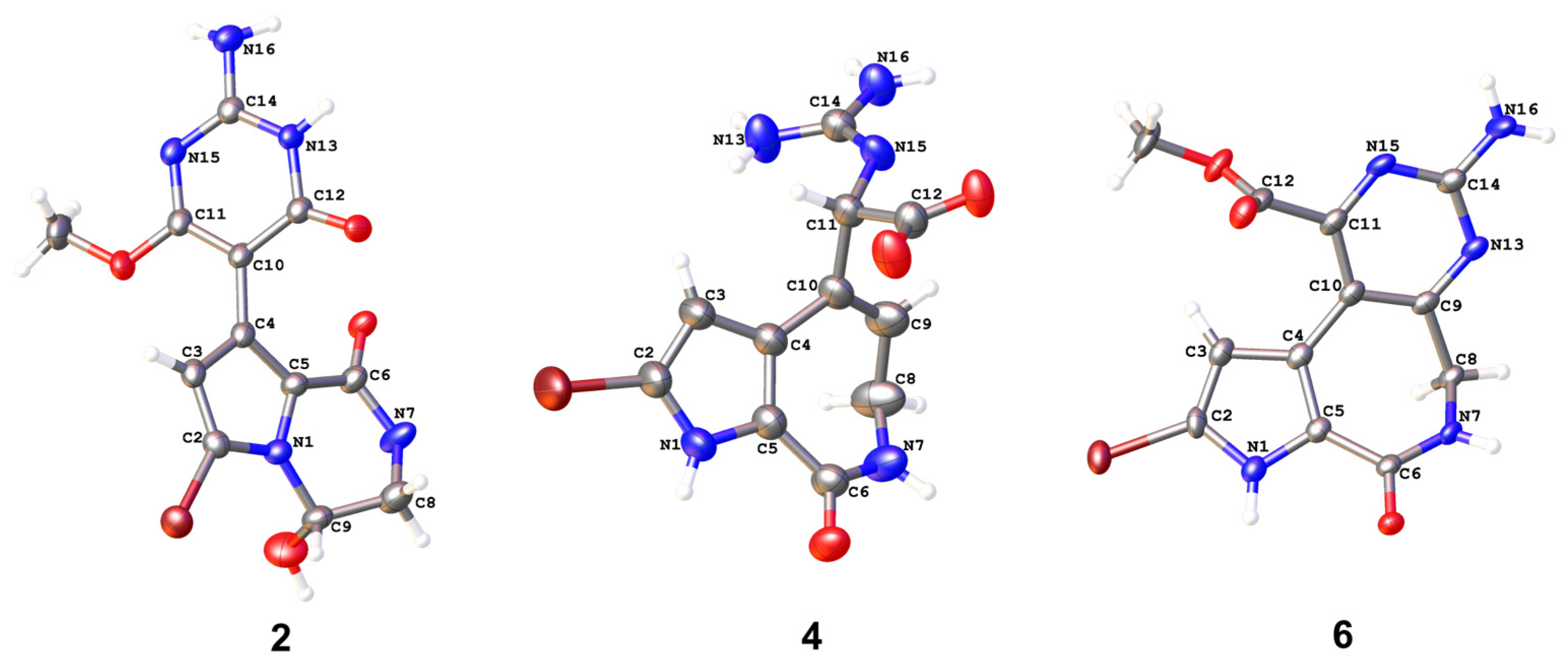
To determine the structures of stylimassalins A-B (1 and 2), single-crystal culturing using a slow solvent evaporation method was carried out over a long time period. Eventually, a qualified colourless crystal of 2 was obtained for crystallographic analyses (90% ethanol aqueous solution, Table S3). The results of its X-ray diffraction experiment gave the structure of 2 (CCDC2249955 for 2, Figure 3). Detailed analysis of the 1D and 2D NMR data of 2 (Figure 2, in methanol-d4 solvent) and the HMBC correlations of H-3 (δH 6.24) with C-2 (δC 105.3), C-4 (δC 124.1), C-5 (δC 123.0), C-6 (δC 161.8), C-9 (δC 74.4), and C-10 (δC 90.4) indicated the presence of a 2twobrominated N-substituted pyrrole ring system. Additionally, the HMBC correlations of H2-8 (δH 3.56, 3.79) with C-6 and C-9 and H-9 (δH 5.75) with C-2 and C-5 disclosed the substructure of 2: a 2-bromo-9-hydroxy-3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-one system. Then, a key HMBC correlation of 11-OMe (δH 3.79) with C-11 (δC 170.3) determined the -OMe moiety substituted at C-11 of 2. Thus, all 1D and 2D NMR data on both 1 and 2 were matched and explainable, which is consistent with the X-ray diffraction results.
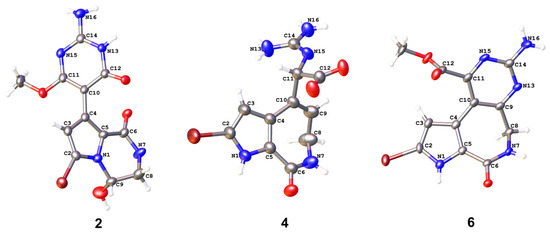
Figure 3.
X-ray structures of compounds 2, 4, and 6, which have three different scaffolds. The data on compound 4 have been published [12].
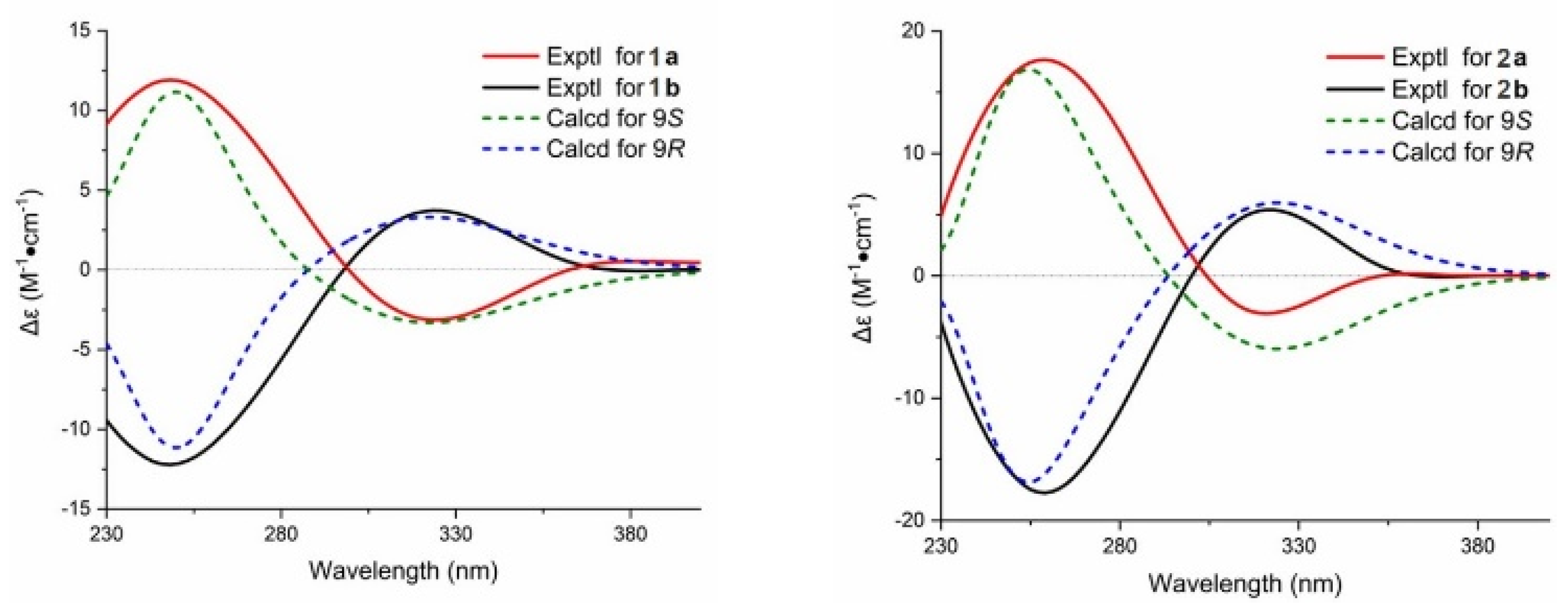
In addition, the crystal of 2 possessed a P1 21/n1 space group, indicating the possible racemic nature of 1 and 2 [14]. To determine the absolute configuration of them, the chiral HPLC isolation of 1 and 2 was carried out, which resulted in an equimolar pair of optically pure enantiomers (Figures S6 and S7). Their absolute configurations were determined by time-dependent density functional theory ECD (TDDFT-ECD) calculations (9S for 1a and 2a, 9R for 1b and 2b; Figure 4 and Figure S8).
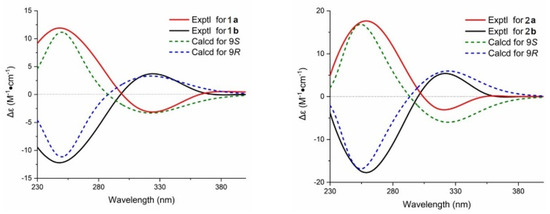
Figure 4.
The absolute configurations of 1 and 2 were determined by time-dependent density functional theory ECD (TDDFT-ECD) calculations (9S for 1a and 2a; 9R for 1b and 2b).
Compound 3 was obtained as a white powder. Detailed analysis of the 1D and 2D NMR spectra of 3 indicated that it is a structure very similar to compound 4, a compound previously isolated by our group (CCDC2172264 for 4; Figure 3 and Table S4) [12]. The molecular formula of 3 was deduced to be C11H13N5O3 based on the HRESIMS ions at m/z 264.1087 [M + H]+ (calcd. 264.1091), which possess one less bromine atom than compound 4 (the HRESIMS ions at m/z 342.0184, 344.0162, 1:1). The only difference between them is that there is either bromination or not at C-2 in 4 (δC-2 104.3) or 3 (δC-2 122.1). Although they are racemates for compound 3, with no ECD curve, the same as with compound 4 [12], the enantiomers of 3 were not separated and then characterized for an absolute configuration assignment due to their limited amount.
In addition, two latonduine-type PIAs (compounds 5 and 6 [13]) were co-isolated from the sponge S. massa. Detailed analysis of their 1D and 2D NMR data revealed compound 6 to be the C-2 brominated analogue of compound 5. The chemical shifts of C-2 in their 13C NMR spectra were identified as δC 122.6 (compound 5) and δC 104.7 (compound 6). The X-ray structure of compound 6 was obtained for the first time in our study (CCDC2331670 for 6, Figure 3, and Table S5).
2.2. Proposed Biosynthetic Pathway
Based on the above description, compounds 1–6, including two different structural skeletons, were isolated from the sponge together. There seems an obvious relationship among them, as they share a common biosynthetic lineage from the same C11N5 biosynthetic precursor, oroidin. Spongiacidin PIAs possess a 5/7/5 tricyclic scaffold, which might be generated by the C4-C10 Michael addition of oroidin. Latonduine is a small PIA family, and it has only contained approximately ten members since the latonduines A and B were first isolated from the sponge Stylissa carteri [11,13]. A previous report [11] suggested that latonduine PIAs cannot be derived from the C11N5 building block of oroidin; however, we have a different opinion (Scheme 1).
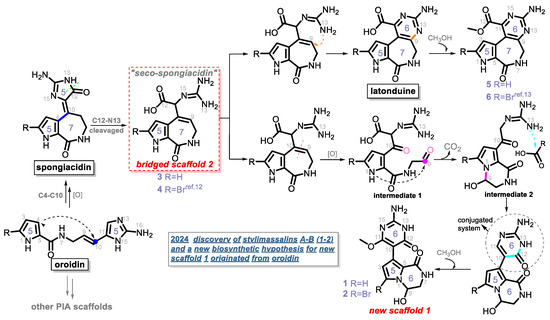
Scheme 1.
A new biosynthetic hypothesis of compounds 1–6.
Compound 4 was previously isolated by our group in 2022 [12]. We ignored its critical role in the biosynthesis of PIAs until the co-isolation of compounds 1–6 in this study. A new biosynthetic pathway from oroidin, through spongiacidin, to latonduine was proposed in Scheme 1, in which the C12-N13-cleavaged compounds of spongiacidin, dubbed seco-spongiacidins (3 and 4), are recognized as a key bridged scaffold that creates latonduine PIA analogues (5 and 6). The biosynthetic pathway from the bridged scaffold 2 to the new scaffold 1 was relatively more complex. Firstly, the cleavage of C9-C10 generated intermediate 1. Then, a mechanism involving intramolecular cyclization and decarboxylation steps was proposed to obtain intermediate 2. Through a new round of nucleophilic addition from 13-NH to a carbonyl moiety, a stable conjugated system of 2-aminopyrimidin-4(3H)-one (indicated by the circular dashed line in Scheme 1) was formed, which obtained the new scaffold 1 (5/6/6 tricyclic system). Then, the compounds 1 and 2 were finally biosynthesised by further methoxylation at C-11.
2.3. Biological Activity
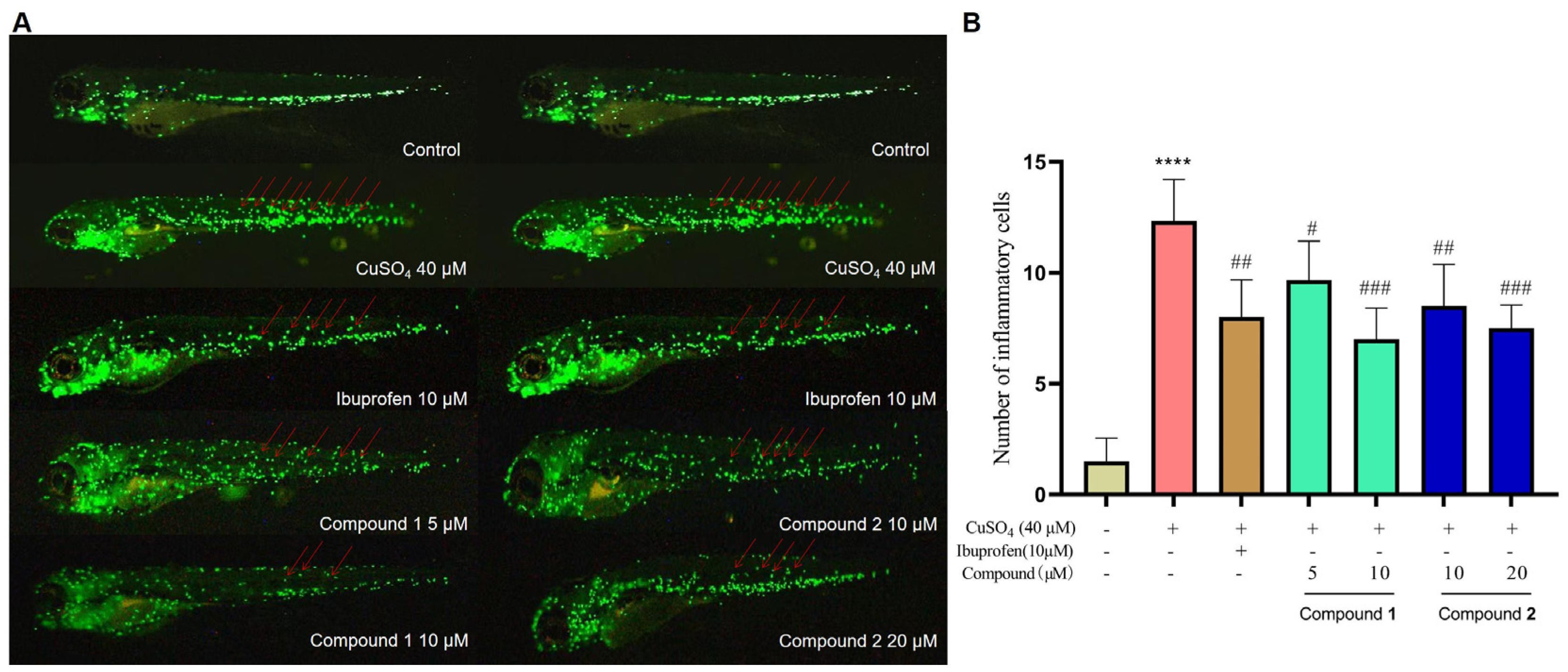
PIAs have been reported to possess a wide range of biological activities. In our earlier study, spongiacidin PIAs were determined to have inhibitory activities against aldose reductase (AKR1B1), a target of complications of diabetes [12,15]. So, compounds 1–6 were subjected to an AKR1B1-targeted inhibitory activity assay in vitro at a concentration of 20 µM (Figure S9). None of them (1–6) shown a significant activity (Figure S10). Then, the anti-inflammatory activities of compounds 1–6 were evaluated in a zebrafish inflammation model induced by copper sulphate (CuSO4). In the CuSO4-induced inflammation model, a significant migration of macrophages towards the lateral line neural crest was observed (Figure 5A). Compared with the CuSO4-induced inflammation model group, treatment with ibuprofen (10 µM) significantly inhibited the migration of macrophages in the positive control group, indicating the establishment of an inflammation model in zebrafish. The results show that stylimassalins A and B (1 and 2) significantly inhibited CuSO4-induced macrophage migration at 5 µM and 10 µM, respectively (Figure 5A,B), while compounds 3–6 had no effect.
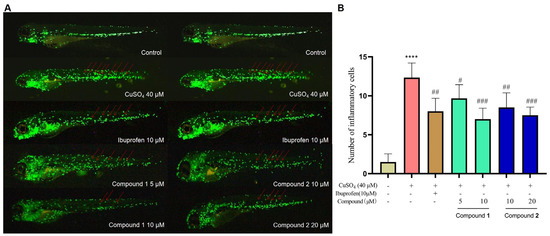
Figure 5.
Compounds 1 and 2 inhibit CuSO4-induced inflammatory responses in zebrafish. (A) Representative fluorometric images of macrophage migration in the CuSO4-induced inflammation model. The red arrows indicate macrophages that migrated to the lateral line neural crest. (B) Statistical analysis of the number of macrophages in the lateral line neural crest. The data are presented as the means ± SEMs (n = 6). # p < 0.05, ## p < 0.01, and ### p < 0.001 compared with the control group; **** p < 0.0001 compared with the model group.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations were obtained on a Jasco P-1020 polarimeter (Tokyo, Japan). UV spectra were recorded using an Agilent 1260 DAD detector (Santa Clara, CA, USA). CD spectra were recorded using an Applied Photophysics Chirascan circular dichroism spectrometer (Bath, UK). NMR spectra were measured on Agilent Pro pulse 500 MHz and Bruker 600 MHz spectrometers (Santa Clara, CA, USA). Chemical shifts are reported with the residuals DMSO-d6 (δH 2.50 ppm) and methanol-d4 (δH 3.31 ppm) as the internal standards for 1H NMR spectroscopy and DMSO-d6 (δC 39.5 ppm) and methanol-d4 (δC 49.0 ppm) for 13C NMR spectroscopy. Electrospray ionization mass spectrometry (ESIMS) and HRESIMS were carried out on a ThermoFisher 480 MS instrument (Waltham, MA, USA). Commercial silica gel (Qingdao Haiyang Chemical Co., Ltd., Qingdao, China, 200–300 mesh, 300–400 mesh) and reversed-phase C18 (Rp-C18) silica gel (12 nm, S-50 μm, YMC Co., Ltd., Tokyo, Japan) were used for column chromatography, and precoated Silica gel GF254 plates (Sinopharm Chemical Reagent Co., Shanghai, China) were used for analytical TLC. Reversed-phase (RP) HPLC was performed on an Agilent 1260 series liquid chromatograph equipped with a DAD G1315D detector at 210, 230, 254, 280, 320, and 360 nm wavelengths (Agilent, Santa Clara, CA, USA). An Agilent ZORBAX Eclipse XDB-C18 column (9.4 × 250 mm, 5 μm) was employed for purification. A chiral HPLC analysis was carried out on a Jasco LC-Net II/ADC HPLC-CD combined instrument with a CHIRALPAK AD-H (5 μm, 4.6 mm I.D. × 250 mm) column. All solvents used for column chromatography and HPLC were of analytical grade (Tianjin Kemio Chemical Reagent Co., Ltd., Tianjin, China) and chromatographic grade (Tianjin Concord Technology Co., Ltd., Tianjin, China), respectively.
3.2. Collection of Sponge Materials
The marine sponge Stylissa massa was collected from the Xisha Islands of the South China Sea in May 2013 and was frozen immediately after collection. The specimen was identified by Dr. Nicole J. de Voogd, National Museum of Natural History, Leiden, The Netherlands. A voucher specimen (No. XS-2013-07) was deposited at lab A1007, School of Pharmacy, Qingdao University, China.
3.3. Extraction and Isolation of Compounds from S. massa
The Stylissa massa sponge (8.0 kg, wet weight) was crushed and then extracted with MeOH four times at room temperature. The combined solutions were concentrated in vacuo and the organic extract was evaporated to produce a brown residue (191.0 g). Then, to remove lipids and other impurities, alkaloid enrichment was carried out by silica gel vacuum liquid chromatography (VLC), eluting with a gradient of petroleum ether/EtOAc (from 10:1 to 0:1, v:v) and, subsequently, CH2Cl2/MeOH (from 10:1 to 0:1, v:v). The alkaloid-containing fractions were combined and evaporated, yielding a crude alkaloid extract (78.9 g).
Guided by the MS and UV-Vis spectra, several runs of purification on the RP-LC [MeOH (0.1%TFA)/H2O (0.1%TFA), from 5:95 to 100:0, v:v, 120 min] afforded the subfraction A (500 mg), which had an undescribed molecular weight and undescribed UV-Vis patterns. Subfraction A (400 mg), which contained compound 2, was purified by semi-preparative HPLC [MeOH (0.1%TFA)/H2O (0.1%TFA), from 5:95 to 100:0, v:v, 1.5 mL/min], yielding compound 2 (2.0 mg, tR = 32.4 min). Subfraction B (200 mg) contained compound 1, which has an identical UV-Vis spectra, and was purified by semi-preparative HPLC [MeOH (0.1%TFA)/H2O (0.1%TFA), from 5:95 to 100:0, v:v, 1.5 mL/min], yielding compound 1 (3.0 mg, tR = 21.8 min). Subfraction C (600 mg) contained compounds 3–4 and was purified by semi-preparative HPLC [MeOH (0.1%TFA)/H2O (0.1%TFA), from 5:95 to 100:0, v:v, 1.5 mL/min], yielding compound 3 (3.0 mg, tR = 16.6 min) and compound 4 (3.0 mg, tR = 21.5 min). Subfraction D (900 mg) contained compounds 5–6 and was purified by semi-preparative HPLC [MeOH (0.1%TFA)/H2O (0.1%TFA), from 5:95 to 100:0, v:v, 1.5 mL/min], yielding compound 5 (5.0 mg, tR = 23.2 min) and compound 6 (11.0 mg, tR = 39.0 min).
3.3.1. Compound 1
3.3.2. Compound 2
3.3.3. Compound 3
3.3.4. Compound 5
3.4. X-Ray Crystallographic Analysis for Compounds 2, 4 and 6
The crystals of 2, 4, and 6 were recrystallized. X-ray analyses were carried out on a Rigaku Xtalab Synergy diffractometer with Cu Kα radiation (λ = 1.54184) at 150 K/293 K. The acquisition parameters for 2, 4, and 6 are provided in the Supporting Information, and their crystallographic data (deposition no. 2: 2249955; 4: 2172264; 6: 2331670) have been deposited at the Cambridge Crystallographic Data Center. Copies of the data can be obtained free of charge via www.ccdc.cam.ac.uk/conts/retrieving.html (accessed on 11 February 2024).
3.5. AKR1B1-Targeted Inhibitory Activity Assay
In the first step of the polyol pathway, glucose reduction is accompanied by the conversion of NADPH to NADP, where NADPH has a spectral absorption around 340 nm and NADP does not. Thus, the decrease in OD340nm can represent the consumption of NADPH. Therefore, we can detect the change in OD340 nm before and after the reaction to screen the effective ARIs or evaluate AR activity; epalrestat was used as the positive control. Briefly, the incubation system contains 10 µL of AKR1B1 enzyme (20 µg/mL), 80 of µL DL-glyceraldehyde (0.16 mmol/L) as the substrate, 1.0 mmol/L of NADPH·4Na as a coenzyme, and 0.1 mol/L of PBS (pH = 6.2). The incubation mixture was minimized to a total volume of 100 µL and transferred into a 96-well ultraviolet plate. Then, the test wells were treated with the tested compounds (at a concentration of 20 µM) for 10 min at 25 °C. Then, the absorbance was measured using FlexStation 3 Reader (Molecular Devices, San Francisco, CA, USA) at 340 nm. The absorbance of wells treated with PBS and NADPH was considered to be 100% (OD1), and the absorbance of wells treated with PBS and DL-glyceraldehyde was considered to be 0% (OD2); the inhibitory rate was calculated by the formula OD compounds − OD2/OD1 − OD2. IC50 represents the concentration that inhibits the AKR1B1 enzyme by 50%.
3.6. The Anti-Inflammatory Effect of the Compounds Was Assessed Utilizing a CuSO4-Induced Inflammation Model
Healthy zebrafish larvae (3 dpf) were transferred to 24-well culture plates (10 per well). The zebrafish were grouped into a control group maintained in zebrafish culture water, a model group, a positive control group treated with a 10 μM ibuprofen solution, and experimental groups treated with compounds at different doses (5, 10, and 20 μM). The plates were maintained at 28 °C in an incubator, and the solution was replaced every 24 h. At 24 h post exposure (hpe), the zebrafish larvae in all groups, except for those in the control group, were treated with a 40 μM CuSO4 solution for 1 h. Finally, the zebrafish larvae in each group were anesthetized with tricaine (0.08%), immobilized with 3% methylcellulose, and positioned laterally on a glass slide. The macrophages were observed under a fluorescence microscope (AXIO Zoom V16, 118 ZEISS, Oberkochen, Germany), and the number of macrophages that migrated to the lateral line of the neural crest (from the cloaca to the tail) was recorded. The above experiments were performed in triplicate.
4. Conclusions
In this paper, we describe the discovery of six marine PIAs, including three different scaffolds. Scaffold 1 (5/6/6), which is new, displays the extraordinary rearrangement diversity of scaffolds. Moreover, discovery of the bridged scaffold, seco-spongicidin (5/7), lead us to propose a new biosynthetic pathway for latonduine PIA metabolites. Compounds 1 and 2 showed considerable anti-inflammatory effects primarily by reducing the macrophage migration induced by CuSO4 damage in zebrafish.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/md22100477/s1. Figure S1: The voucher specimen of Stylissa massa in the lab; Tables S1–S5, and Figures S2–S8: Structural elucidation of compounds 1–6; Figures S9 and S10: Bioactivity experimental procedures for compounds 1–6; Figures S11–S54: NMR and MS spectra of compounds 1–6; Tables S6 and S7: ECD calculations for compounds 1 and 2.
Author Contributions
Conceptualization, H.Z. and Q.W.; methodology, X.L.; software, H.Z. and Q.W.; anti-inflammatory test, Y.Z.; investigation, X.L.; resources, Q.W.; data curation, X.L.; writing—original draft preparation, Q.W.; writing—review and editing, H.Z.; visualization, X.L.; supervision, X.L.; project administration, H.Z.; funding acquisition, Q.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 82273848), the National Key R&D Program for Young Scientists of China (Grant No. 2022YFC2804500), and the key technology tackling and industrialization demonstration projects in Qingdao (Grant No. 22-3-3-hygg-25-hy).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Materials; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jin, Z. Muscarine, Imidazole, oxazole and thiazole alkaloids. Nat. Prod. Rep. 2016, 33, 1268–1317. [Google Scholar] [CrossRef] [PubMed]
- Forte, B.; Malgesini, B.; Piutti, C.; Quartieri, F.; Scolaro, A.; Papeo, G. A submarine journey: The pyrrole imidazole alkaloids. Mar. Drugs 2009, 7, 705–753. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.M.; Burkholder, P.R. Structure of dibromophakellin, a new bromine-containing alkaloid from the marine sponge Phakellia flabellata. Chem. Commun. 1971, 3, 151–152. [Google Scholar] [CrossRef]
- Al-Mourabit, A.; Zancanella, M.A.; Tilvi, S.; Romo, D. Biosynthesis, asymmetric synthesis, and pharmacology, including cellular targets, of the pyrrole-2-aminoimidazole marine alkaloids. Nat. Prod. Rep. 2011, 28, 1229–1260. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, M.S.; Carroll, A.R.; Addepalli, R.; Avery, V.M.; Hooper, J.N.A.; Quinn, R.J. Natural products, stylissadines A and B, specific antagonists of the P2X7 Receptor, an important inflammatory target. J. Org. Chem. 2007, 72, 2309–2317. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, D.P.; Li, K.; Maue, M.; Zografos, A.L.; Baran, P.S. Total synthesis of dimeric pyrrole−imidazole alkaloids: sceptrin, ageliferin, nagelamide E, oxysceptrin, nakamuric acid, and the axinellamine carbon skeleton. J. Am. Chem. Soc. 2007, 129, 4762–4775. [Google Scholar] [CrossRef] [PubMed]
- Kinnel, R.B.; Gehrken, H.P.; Scheuer, P.J. Palau’amine: A cytotoxic and immunosuppressive hexacyclic bisguanidine antibiotic from the sponge Stylotella agminata. J. Am. Chem. Soc. 1993, 115, 3376–3377. [Google Scholar] [CrossRef]
- Grube, A.; Köck, M. Stylissadines A and B: The first tetrameric pyrrole−imidazole alkaloids. Org. Lett. 2006, 8, 4675–4678. [Google Scholar] [CrossRef] [PubMed]
- Inaba, K.; Sato, H.; Tsuda, M.; Kobayashi, J. Spongiacidins A-D, new bromopyrrole alkaloids from Hymeniacidon sponge. J. Nat. Prod. 1998, 61, 693–695. [Google Scholar] [CrossRef] [PubMed]
- Stout, E.P.; Choi, M.Y.; Castro, J.E.; Molinski, T.F. Potent fluorinated agelastatin analogues for chronic lymphocytic leukemia: Design, synthesis, and pharmacokinetic studies. J. Med. Chem. 2014, 57, 5085–5093. [Google Scholar] [CrossRef] [PubMed]
- Linington, R.G.; Williams, D.E.; Tahir, A.; Soest, R.V.; Andersen, R.J. Latonduines A and B, new alkaloids isolated from the marine sponge Stylissa carteri: Structure elucidation, synthesis, and biogenetic implications. Org. Lett. 2003, 5, 2735–2738. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, C.; Wei, Z.; Tang, X.; Ji, L.; Luo, X.; Peng, X.; Li, G.; Lou, H. A series of new pyrrole alkaloids with ALR2 inhibitory activities from the Sponge Stylissa massa. Mar. Drugs 2022, 20, 454. [Google Scholar] [CrossRef] [PubMed]
- Fouad, M.A.; Debbab, A.; Wray, V.; Müller, W.E.G.; Proksch, P. New bioactive alkaloids from the marine sponge Stylissa sp. Tetrahedron 2012, 68, 10176–10179. [Google Scholar] [CrossRef]
- Wang, Q.; Tang, X.L.; Luo, X.C.; Voogd, N.J.D.; Li, P.L.; Li, G.Q. (+)- and (−)-Spiroreticulatine, a pair of unusual spiro bisheterocyclic quinoline-imidazole alkaloids from the South China Sea sponge Fascaplysinopsis reticulata. Org. Lett. 2015, 17, 3458–3461. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Luo, X.C.; Zhu, R.W.; Tong, W.L.; Yang, Y.N.; Li, G.Q.; Wang, Q. Tagpyrrollins A and B and Tagpyrrollidone A: Three Pyrrole Steroid Analogues with AKR1B1-Targeting Inhibitory Activity from the Sponges Stylissa massa and Pseudospongosorites suberitoides. Org. Lett. 2024, 26, 5794–5798. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).