Proteomic Analysis and Biochemical Characterization of the Nematocyst Extract of the Hydrozoan Velella velella
Abstract
1. Introduction
2. Results
2.1. Quantification of Total Proteins in the Crude Nematocyst Extract
2.2. Transcriptome Assembly and Analysis
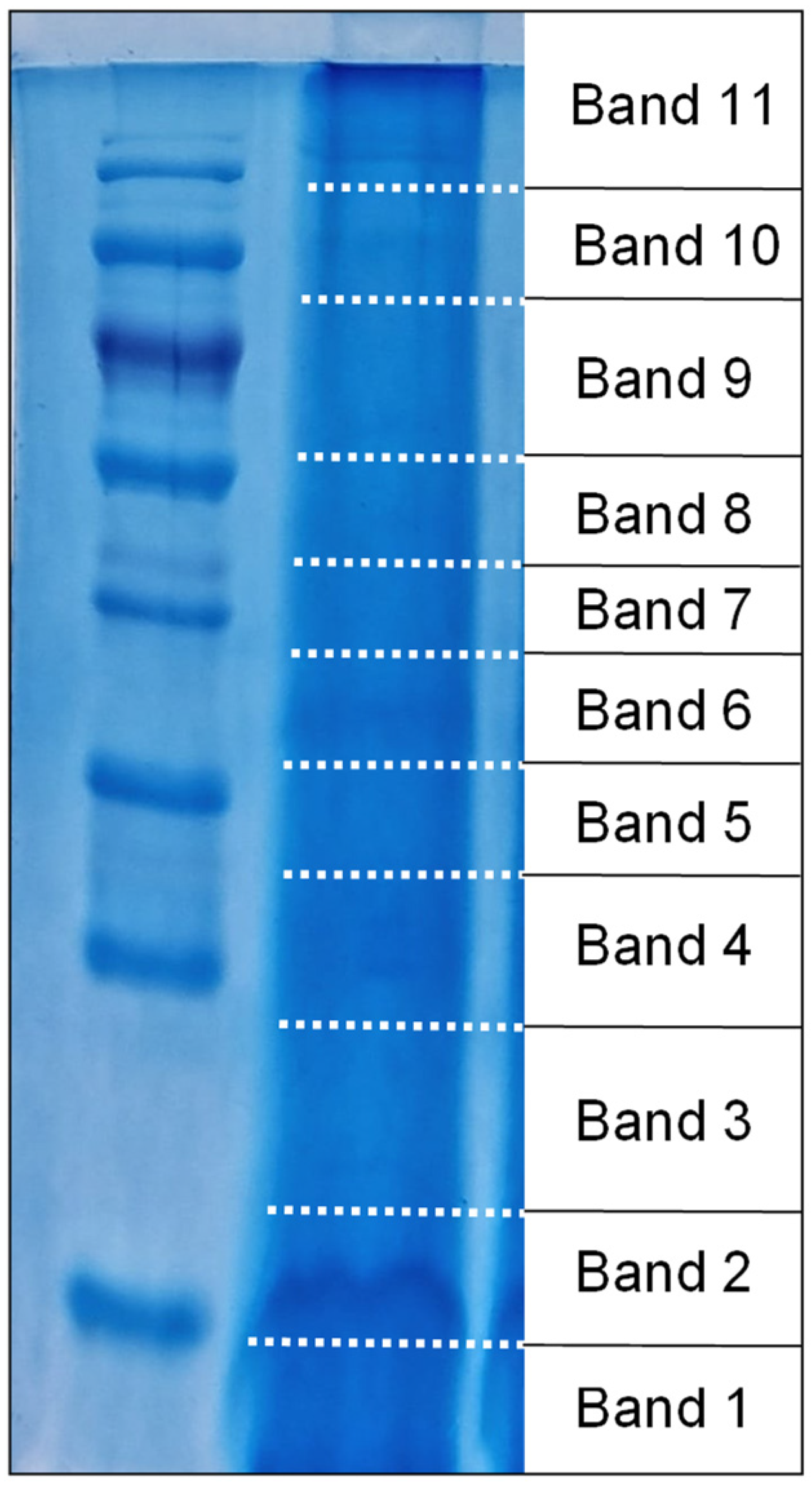
2.3. Sodium Dodecyl Sulphate–PolyAcrylamide Gel Electrophoresis (SDS–PAGE) of the Crude Venom Extract
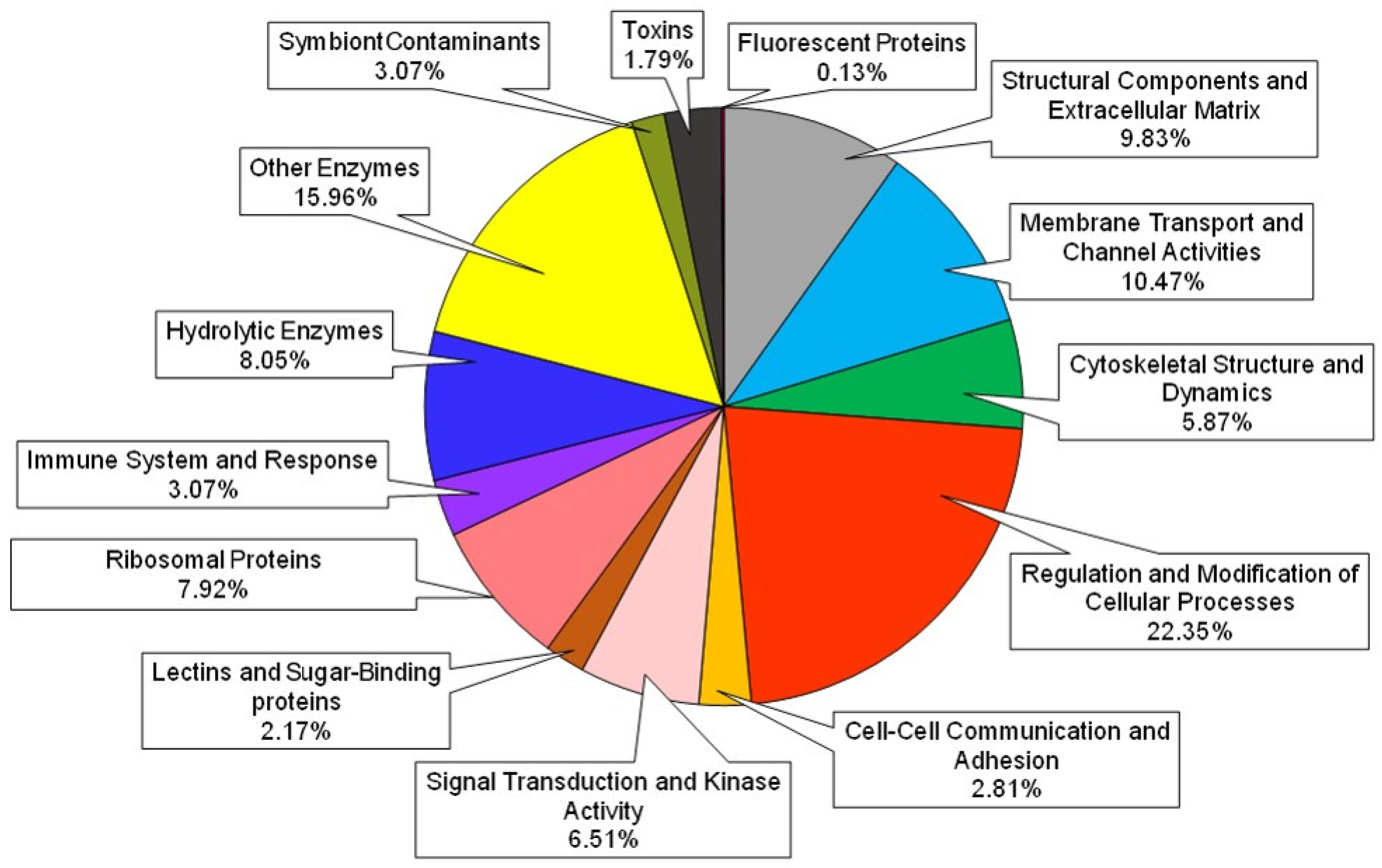
2.4. Characterization of Proteins Obtained from Proteomic Analysis
2.5. Evaluation of the Main Venom Hydrolytic Enzymes Activities in the Crude Nematocyst Extract
2.5.1. Proteases Activity Evaluation
Generic Protease Activity Evaluation
Collagenase Activity Evaluation
Zymographic Analysis of Active Proteases
2.5.2. Evaluation of Other Enzymatic Activities
Hyaluronidase Activity Evaluation
Phospholipase A2 Activity Evaluation
DNase Activity Evaluation
Chitinase Activity Evaluation
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Velella velella Sampling
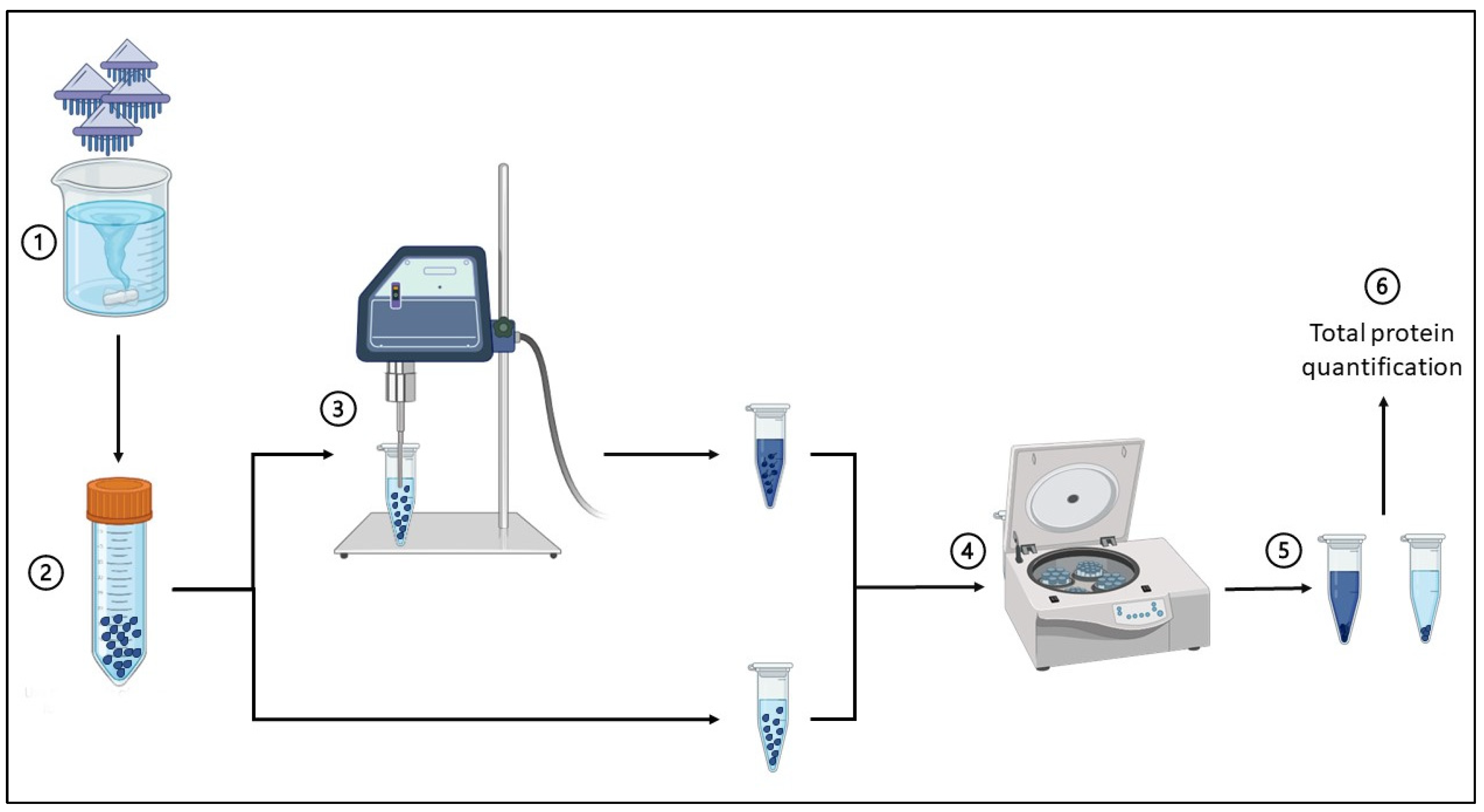
4.3. Crude Venom Extraction
4.4. Quantification of Total Proteins in the Crude Nematocyst Extract
4.5. Transcriptome Assembly and Putative Protein Database Construction
4.6. Proteomic Analysis
4.6.1. Sample Preparation
4.6.2. Mass Spectrometry Analysis
4.6.3. Protein Characterization
4.7. In Vitro Evaluation of the Main Crude Venom Hydrolytic Enzymes Activities
4.7.1. Generic Protease Activity
4.7.2. Collagenase Activity
4.7.3. Zymographic Analysis of Active Proteases
4.7.4. Hyaluronidase Activity
4.7.5. Phospholipase A2 Activity
4.7.6. DNase Activity
4.7.7. Chitinase Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slobodkin, L.B.; Bossert, P.E. Chapter 5—Cnidaria. In Ecology and Classification of North American Freshwater Invertebrates, 3rd ed.; Thorp, J.H., Covich, A.P., Eds.; Academic Press: San Diego, CA, USA, 2010; pp. 125–142. ISBN 978-0-12-374855-3. [Google Scholar]
- Technau, U.; Genikhovich, G.; Kraus, J.E.M. Cnidaria. In Evolutionary Developmental Biology of Invertebrates 1: Introduction, Non-Bilateria, Acoelomorpha, Xenoturbellida, Chaetognatha; Wanninger, A., Ed.; Springer: Vienna, Austria, 2015; pp. 115–163. ISBN 978-3-7091-1862-7. [Google Scholar]
- Atkinson, S.D.; Bartholomew, J.L.; Lotan, T. Myxozoans: Ancient Metazoan Parasites Find a Home in Phylum Cnidaria. Zoology 2018, 129, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Berzins, I.K.; Yanong, R.P.E.; LaDouceur, E.E.B.; Peters, E.C. Cnidaria. In Invertebrate Histology; LaDouceur, E.E.B., Ed.; Wiley: Hoboken, NJ, USA, 2021; pp. 55–86. ISBN 978-1-119-50765-9. [Google Scholar]
- Beckmann, A.; Özbek, S. The Nematocyst: A Molecular Map of the Cnidarian Stinging Organelle. Int. J. Dev. Biol. 2012, 56, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, R. Venomology of Marine Cnidarians. In Biology and Ecology of Venomous Marine Cnidarians; Santhanam, R., Ed.; Springer: Singapore, 2020; pp. 287–320. ISBN 9789811516030. [Google Scholar]
- Jouiaei, M.; Yanagihara, A.A.; Madio, B.; Nevalainen, T.J.; Alewood, P.F.; Fry, B.G. Ancient Venom Systems: A Review on Cnidaria Toxins. Toxins 2015, 7, 2251–2271. [Google Scholar] [CrossRef] [PubMed]
- Trim, C.M.; Byrne, L.J.; Trim, S.A. Chapter One—Utilisation of Compounds from Venoms in Drug Discovery. In Progress in Medicinal Chemistry; Witty, D.R., Cox, B., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; Volume 60, pp. 1–66. [Google Scholar]
- Frazão, B.; Vasconcelos, V.; Antunes, A. Sea Anemone (Cnidaria, Anthozoa, Actiniaria) Toxins: An Overview. Mar. Drugs 2012, 10, 1812–1851. [Google Scholar] [CrossRef]
- Podobnik, M.; Anderluh, G. Pore-Forming Toxins in Cnidaria. Semin. Cell Dev. Biol. 2017, 72, 133–141. [Google Scholar] [CrossRef]
- Cooper, R.A.; De Freitas, J.; Porreca, F.; Eisenhour, C.M.; Lukas, R.; Huxtable, R.J. The Sea Anemone Purine, Caissarone: Adenosine Receptor Antagonism. Toxicon 1995, 33, 1025–1031. [Google Scholar] [CrossRef]
- Tibballs, J.; Yanagihara, A.A.; Turner, H.C.; Winkel, K. Immunological and Toxinological Responses to Jellyfish Stings. Inflamm. Allergy-Drug Targets (Former. Curr. Drug Targets-Inflamm. Allergy) 2011, 10, 438–446. [Google Scholar] [CrossRef]
- Lee, H.; Jung, E.; Kang, C.; Yoon, W.D.; Kim, J.-S.; Kim, E. Scyphozoan Jellyfish Venom Metalloproteinases and Their Role in the Cytotoxicity. Toxicon 2011, 58, 277–284. [Google Scholar] [CrossRef]
- Mariottini, G.L.; Grice, I.D. Natural Compounds and Drug Discovery: Can Cnidarian Venom Play a Role? Cent. Nerv. Syst. Agents Med. Chem.-Cent. Nerv. Syst. Agents) 2019, 19, 114–118. [Google Scholar] [CrossRef]
- D’Ambra, I.; Lauritano, C. A Review of Toxins from Cnidaria. Mar. Drugs 2020, 18, 507. [Google Scholar] [CrossRef]
- Cegolon, L.; Heymann, W.C.; Lange, J.H.; Mastrangelo, G. Jellyfish Stings and Their Management: A Review. Mar. Drugs 2013, 11, 523–550. [Google Scholar] [CrossRef] [PubMed]
- Killi, N.; Bonello, G.; Mariottini, G.L.; Pardini, P.; Pozzolini, M.; Cengiz, S. Nematocyst Types and Venom Effects of Aurelia aurita and Velella velella from the Mediterranean Sea. Toxicon 2020, 175, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Amreen Nisa, S.; Vinu, D.; Krupakar, P.; Govindaraju, K.; Sharma, D.; Vivek, R. Jellyfish Venom Proteins and Their Pharmacological Potentials: A Review. Int. J. Biol. Macromol. 2021, 176, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Maduraiveeran, H.; Raja, K.; Chinnasamy, A. Antiproliferative and Antioxidant Properties of Nematocysts Crude Venom from Jellyfish Acromitus flagellatus against Human Cancer Cell Lines. Saudi J. Biol. Sci. 2021, 28, 1954–1961. [Google Scholar] [CrossRef]
- D’Ambra, I.; Merquiol, L. Jellyfish from Fisheries By-Catches as a Sustainable Source of High-Value Compounds with Biotechnological Applications. Mar. Drugs 2022, 20, 266. [Google Scholar] [CrossRef]
- de Oliveira, S.A.M.; Magalhães, M.R.; de Oliveira, L.P.; da Cunha, L.C. Identification of Antinociceptive Fraction of Snake Venom from Crotalus durissus collilineatus Crotamine-Negative and Its Acute Toxicity Evaluation. Toxicon 2016, 122, 145–151. [Google Scholar] [CrossRef]
- Cardoso, F.C.; Dekan, Z.; Rosengren, K.J.; Erickson, A.; Vetter, I.; Deuis, J.R.; Herzig, V.; Alewood, P.F.; King, G.F.; Lewis, R.J. Identification and Characterization of ProTx-III [μ-TRTX-Tp1a], a New Voltage-Gated Sodium Channel Inhibitor from Venom of the Tarantula Thrixopelma pruriens. Mol. Pharmacol. 2015, 88, 291–303. [Google Scholar] [CrossRef]
- Lima, P.C.; Bordon, K.C.F.; Pucca, M.B.; Cerni, F.A.; Zoccal, K.F.; Faccioli, L.H.; Arantes, E.C. Partial Purification and Functional Characterization of Ts19 Frag-I, a Novel Toxin from Tityus serrulatus Scorpion Venom. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 49. [Google Scholar] [CrossRef]
- Nguyen, B.; Le Caer, J.-P.; Aráoz, R.; Thai, R.; Lamthanh, H.; Benoit, E.; Molgó, J. Isolation, Purification and Functional Characterization of Alpha-BnIA from Conus bandanus Venom. Toxicon 2014, 91, 155–163. [Google Scholar] [CrossRef]
- Weston, A.J.; Chung, R.; Dunlap, W.C.; Morandini, A.C.; Marques, A.C.; Moura-da-Silva, A.M.; Ward, M.; Padilla, G.; da Silva, L.F.; Andreakis, N.; et al. Proteomic Characterisation of Toxins Isolated from Nematocysts of the South Atlantic Jellyfish Olindias sambaquiensis. Toxicon 2013, 71, 11–17. [Google Scholar] [CrossRef]
- Brinkman, D.L.; Jia, X.; Potriquet, J.; Kumar, D.; Dash, D.; Kvaskoff, D.; Mulvenna, J. Transcriptome and Venom Proteome of the Box Jellyfish Chironex fleckeri. BMC Genom. 2015, 16, 407. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, I.; Hwang, D.H.; Lee, H.; Yoon, W.D.; Chae, J.; Han, C.H.; Yum, S.; Kang, C.; Kim, E. Proteomic Analysis of Novel Components of Nemopilema nomurai Jellyfish Venom: Deciphering the Mode of Action. Toxins 2019, 11, 153. [Google Scholar] [CrossRef] [PubMed]
- Leung, T.C.N.; Qu, Z.; Nong, W.; Hui, J.H.L.; Ngai, S.M. Proteomic Analysis of the Venom of Jellyfishes Rhopilema esculentum and Sanderia malayensis. Mar. Drugs 2020, 18, 655. [Google Scholar] [CrossRef] [PubMed]
- Modahl, C.M.; Durban, J.; Mackessy, S.P. Exploring Toxin Evolution: Venom Protein Transcript Sequencing and Transcriptome-Guided High-Throughput Proteomics. In Snake and Spider Toxins: Methods and Protocols; Priel, A., Ed.; Springer: New York, NY, USA, 2020; pp. 97–127. ISBN 978-1-4939-9845-6. [Google Scholar]
- Modahl, C.M.; Saviola, A.J.; Mackessy, S.P. Integration of Transcriptomic and Proteomic Approaches for Snake Venom Profiling. Expert Rev. Proteom. 2021, 18, 827–834. [Google Scholar] [CrossRef]
- Betti, F.; Bo, M.; Enrichetti, F.; Manuele, M.; Cattaneo-Vietti, R.; Bavestrello, G. Massive Strandings of Velella velella (Hydrozoa: Anthoathecata: Porpitidae) in the Ligurian Sea (North-Western Mediterranean Sea). Eur. Zool. J. 2019, 86, 343–353. [Google Scholar] [CrossRef]
- Kirkpatrick, P.A.; Pugh, P.R. Siphonophores and Velellids: Keys and Notes for the Identification of the Species; Brill Archive: Boston, MA, USA, 1984; ISBN 978-90-04-07470-5. [Google Scholar]
- Jaimes-Becerra, A.; Chung, R.; Morandini, A.C.; Weston, A.J.; Padilla, G.; Gacesa, R.; Ward, M.; Long, P.F.; Marques, A.C. Comparative Proteomics Reveals Recruitment Patterns of Some Protein Families in the Venoms of Cnidaria. Toxicon 2017, 137, 19–26. [Google Scholar] [CrossRef]
- Doonan, L.B.; Lynham, S.; Quinlan, C.; Ibiji, S.C.; Winter, C.E.; Padilla, G.; Jaimes-Becerra, A.; Morandini, A.C.; Marques, A.C.; Long, P.F. Venom Composition Does Not Vary Greatly Between Different Nematocyst Types Isolated from the Primary Tentacles of Olindias sambaquiensis (Cnidaria: Hydrozoa). Biol. Bull. 2019, 237, 26–35. [Google Scholar] [CrossRef]
- Jaimes-Becerra, A.; Gacesa, R.; Doonan, L.B.; Hartigan, A.; Marques, A.C.; Okamura, B.; Long, P.F. “Beyond Primary Sequence”—Proteomic Data Reveal Complex Toxins in Cnidarian Venoms. Integr. Comp. Biol. 2019, 59, 777–785. [Google Scholar] [CrossRef]
- Holstein, T.W.; Benoit, M.; Herder, G.V.; David, C.N.; Wanner, G.; Gaub, H.E. Fibrous Mini-Collagens in Hydra Nematocysts. Science 1994, 265, 402–404. [Google Scholar] [CrossRef]
- David, C.N.; Özbek, S.; Adamczyk, P.; Meier, S.; Pauly, B.; Chapman, J.; Hwang, J.S.; Gojobori, T.; Holstein, T.W. Evolution of Complex Structures: Minicollagens Shape the Cnidarian Nematocyst. Trends Genet. 2008, 24, 431–438. [Google Scholar] [CrossRef]
- Watson, G.M.; Mire-Thibodeaux, P. The Cell Biology of Nematocysts. In International Review of Cytology; Jeon, K.W., Jarvik, J., Eds.; Academic Press: Cambridge, MA, USA, 1994; Volume 156, pp. 275–300. [Google Scholar]
- Kass-Simon, G.; Scappaticci, A.A., Jr. The Behavioral and Developmental Physiology of Nematocysts. Can. J. Zool. 2002, 80, 1772–1794. [Google Scholar] [CrossRef]
- Berking, S.; Herrmann, K. Formation and Discharge of Nematocysts Is Controlled by a Proton Gradient across the Cyst Membrane. Helgol. Mar. Res. 2006, 60, 180–188. [Google Scholar] [CrossRef]
- Sabatier, J.-M.; Kovacic, H.; Shahbazzadeh, D.; Wang, J.-L.; Cao, Z.; McNutt, P.M.; Wulff, H.; Utkin, Y.N. Venoms, Animal and Microbial Toxins, Volume II; Frontiers Media SA: Lausanne, Switzerland, 2022; ISBN 978-2-8325-0114-6. [Google Scholar]
- Fox, J.W.; Serrano, S.M.T. Structural Considerations of the Snake Venom Metalloproteinases, Key Members of the M12 Reprolysin Family of Metalloproteinases. Toxicon 2005, 45, 969–985. [Google Scholar] [CrossRef]
- Li, A.; Yu, H.; Li, R.; Yue, Y.; Yu, C.; Geng, H.; Liu, S.; Xing, R.; Li, P. Jellyfish Nemopilema nomurai Causes Myotoxicity through the Metalloprotease Component of Venom. Biomed. Pharmacother. 2022, 151, 113192. [Google Scholar] [CrossRef]
- García-Arredondo, A.; Rojas-Molina, A.; Ibarra-Alvarado, C.; Lazcano-Pérez, F.; Arreguín-Espinosa, R.; Sánchez-Rodríguez, J. Composition and Biological Activities of the Aqueous Extracts of Three Scleractinian Corals from the Mexican Caribbean: Pseudodiploria strigosa, Porites astreoides and Siderastrea siderea. J. Venom. Anim. Toxins Incl. Trop. Dis. 2016, 22, 32. [Google Scholar] [CrossRef]
- Liu, G.; Zhou, Y.; Liu, D.; Wang, Q.; Ruan, Z.; He, Q.; Zhang, L. Global Transcriptome Analysis of the Tentacle of the Jellyfish Cyanea capillata Using Deep Sequencing and Expressed Sequence Tags: Insight into the Toxin- and Degenerative Disease-Related Transcripts. PLoS ONE 2015, 10, e0142680. [Google Scholar] [CrossRef]
- Ponce, D.; Brinkman, D.L.; Potriquet, J.; Mulvenna, J. Tentacle Transcriptome and Venom Proteome of the Pacific Sea Nettle, Chrysaora fuscescens (Cnidaria: Scyphozoa). Toxins 2016, 8, 102. [Google Scholar] [CrossRef]
- Koh, C.Y.; Kini, R.M. From Snake Venom Toxins to Therapeutics—Cardiovascular Examples. Toxicon 2012, 59, 497–506. [Google Scholar] [CrossRef]
- Ra, H.-J.; Parks, W.C. Control of Matrix Metalloproteinase Catalytic Activity. Matrix Biol. 2007, 26, 587–596. [Google Scholar] [CrossRef]
- Lal, D.M.; Calton, G.J.; Neeman, I.; Burnett, J.W. Characterization of Physalia physalis (Portugese Man-o’war) Nematocyst Venom Collagenase. Comp. Biochem. Physiol. Part B Comp. Biochem. 1981, 70, 635–638. [Google Scholar] [CrossRef]
- Nevalainen, T.J.; Peuravuori, H.J.; Quinn, R.J.; Llewellyn, L.E.; Benzie, J.A.H.; Fenner, P.J.; Winkel, K.D. Phospholipase A2 in Cnidaria. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 139, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Rachamim, T.; Morgenstern, D.; Aharonovich, D.; Brekhman, V.; Lotan, T.; Sher, D. The Dynamically Evolving Nematocyst Content of an Anthozoan, a Scyphozoan, and a Hydrozoan. Mol. Biol. Evol. 2015, 32, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Kamikado, N.; Fujimoto, R.; Hamaguchi, K.; Nakamura, H.; Chijiwa, T.; Ohno, M.; Oda-ueda, N. A [Lys49]Phospholipase A2 from Protobothrops flavoviridis Venom Induces Caspase-Independent Apoptotic Cell Death Accompanied by Rapid Plasma-Membrane Rupture in Human Leukemia Cells. Biosci. Biotechnol. Biochem. 2011, 75, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Neeman, I.; Calton, G.J.; Burnett, J.W. Purification and Characterization of the Endonuclease Present in Physalia physalis Venom. Comp. Biochem. Physiol. Part B Comp. Biochem. 1980, 67, 155–158. [Google Scholar] [CrossRef]
- Neeman, I.; Calton, G.J.; Burnett, J.W. Purification of an Endonuclease Present in Chrysaora quinquecirrha Venom. Proc. Soc. Exp. Biol. Med. 1981, 166, 374–382. [Google Scholar] [CrossRef]
- Purcell, J.E.; Clarkin, E.; Doyle, T.K. Foods of Velella velella (Cnidaria: Hydrozoa) in Algal Rafts and Its Distribution in Irish Seas. Hydrobiologia 2012, 690, 47–55. [Google Scholar] [CrossRef]
- Patil, R.S.; Ghormade, V.; Deshpande, M.V. Chitinolytic Enzymes: An Exploration. Enzym. Microb. Technol. 2000, 26, 473–483. [Google Scholar] [CrossRef]
- Matsui, T.; Fujimura, Y.; Titani, K. Snake Venom Proteases Affecting Hemostasis and Thrombosis. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 2000, 1477, 146–156. [Google Scholar] [CrossRef]
- Sulca, M.A.; Remuzgo, C.; Cárdenas, J.; Kiyota, S.; Cheng, E.; Bemquerer, M.P.; Machini, M.T. Venom of the Peruvian Snake Bothriopsis oligolepis: Detection of Antibacterial Activity and Involvement of Proteolytic Enzymes and C-Type Lectins in Growth Inhibition of Staphylococcus aureus. Toxicon 2017, 134, 30–40. [Google Scholar] [CrossRef]
- Fiorotti, H.B.; Soares, T.G.; Borges, M.H.; Matavel, A.; Campos, F.V.; Figueiredo, S.G.D. Preliminary Report on the Hemagglutinating Activity of the Scorpaena plumieri Fish Venom. An. Acad. Bras. Ciênc. 2022, 94, e20200976. [Google Scholar] [CrossRef]
- East, L.; Isacke, C.M. The Mannose Receptor Family. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2002, 1572, 364–386. [Google Scholar] [CrossRef]
- von Reumont, B.M.; Anderluh, G.; Antunes, A.; Ayvazyan, N.; Beis, D.; Caliskan, F.; Crnković, A.; Damm, M.; Dutertre, S.; Ellgaard, L.; et al. Modern Venomics—Current Insights, Novel Methods, and Future Perspectives in Biological and Applied Animal Venom Research. GigaScience 2022, 11, giac048. [Google Scholar] [PubMed]
- Ribeiro, C.; Vignes, M.; Brehélin, M. Xenorhabdus nematophila (Enterobacteriacea) Secretes a Cation-Selective Calcium-Independent Porin Which Causes Vacuolation of the Rough Endoplasmic Reticulum and Cell Lysis. J. Biol. Chem. 2003, 278, 3030–3039. [Google Scholar] [CrossRef] [PubMed]
- Anderluh, G.; Maček, P. Cytolytic Peptide and Protein Toxins from Sea Anemones (Anthozoa: Actiniaria). Toxicon 2002, 40, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Kuhn-Nentwig, L.; Langenegger, N.; Heller, M.; Koua, D.; Nentwig, W. The Dual Prey-Inactivation Strategy of Spiders—In-Depth Venomic Analysis of Cupiennius salei. Toxins 2019, 11, 167. [Google Scholar] [CrossRef]
- Campos, F.V.; Fiorotti, H.B.; Coitinho, J.B.; Figueiredo, S.G. Fish Cytolysins in All Their Complexity. Toxins 2021, 13, 877. [Google Scholar] [CrossRef]
- Castañeda, O.; Sotolongo, V.; Amor, A.M.; Stöcklin, R.; Anderson, A.J.; Harvey, A.L.; Engström, Å.; Wernstedt, C.; Karlsson, E. Characterization of a Potassium Channel Toxin from the Caribbean Sea Anemone Stichodactyla helianthus. Toxicon 1995, 33, 603–613. [Google Scholar] [CrossRef]
- Escoubas, P.; Rash, L. Tarantulas: Eight-Legged Pharmacists and Combinatorial Chemists. Toxicon 2004, 43, 555–574. [Google Scholar] [CrossRef]
- Rangaraju, S.; Khoo, K.; Feng, Z.; Crossley, G.; Nugent, D.; Khaytin, I.; Pennington, M.; Norton, R.; Chandy, K.G. Potassium Channel Modulation by A Toxin Domain in Matrix Metalloprotease 23. Biophys. J. 2010, 98, 212a. [Google Scholar] [CrossRef][Green Version]
- Terstappen, G.C.; Schlüpen, C.; Raggiaschi, R.; Gaviraghi, G. Target Deconvolution Strategies in Drug Discovery. Nat. Rev. Drug Discov. 2007, 6, 891–903. [Google Scholar] [CrossRef]
- Botelho, D.; Wall, M.J.; Vieira, D.B.; Fitzsimmons, S.; Liu, F.; Doucette, A. Top-Down and Bottom-Up Proteomics of SDS-Containing Solutions Following Mass-Based Separation. J. Proteome Res. 2010, 9, 2863–2870. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-Gel Digestion for Mass Spectrometric Characterization of Proteins and Proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot Program: A Central Hub of Integrated Venom Protein Data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef]
- Junior, N.; Barros, R.D.A.; Vital, C.E.; Barbosa, S.L.; de Oliveira, J.V.A.; da Rocha, G.C.; Oliveira, C.N.; Assis, J.V.M.G. Enzymatic Assay of Protease Using Azocasein as Substrate; Springer Nature: Berlin, Germany, 1970. [Google Scholar]
- Fu, X.; Xue, C.; Miao, B.; Li, Z.; Gao, X.; Yang, W. Characterization of Proteases from the Digestive Tract of Sea Cucumber (Stichopus japonicus): High Alkaline Protease Activity. Aquaculture 2005, 246, 321–329. [Google Scholar] [CrossRef]
- Van Wart, H.E.; Steinbrink, D.R. A Continuous Spectrophotometric Assay for Clostridium histolyticum Collagenase. Anal. Biochem. 1981, 113, 356–365. [Google Scholar] [CrossRef]
- Wilkesman, J.; Kurz, L. Advances in Zymography Techniques and Patents Regarding Protease Analysis. Recent Pat. Biotechnol. 2012, 6, 106–114. [Google Scholar] [CrossRef]
- Pukrittayakamee, S.; Warrell, D.A.; Desakorn, V.; McMichael, A.J.; White, N.J.; Bunnag, D. The Hyaluronidase Activities of Some Southeast Asian Snake Venoms. Toxicon 1988, 26, 629–637. [Google Scholar] [CrossRef]
- Laxme, R.R.S.; Khochare, S.; de Souza, H.F.; Ahuja, B.; Suranse, V.; Martin, G.; Whitaker, R.; Sunagar, K. Beyond the ‘Big Four’: Venom Profiling of the Medically Important yet Neglected Indian Snakes Reveals Disturbing Antivenom Deficiencies. PLoS Negl. Trop. Dis. 2019, 13, e0007899. [Google Scholar] [CrossRef]
- Shen, C.-R.; Chen, Y.-S.; Yang, C.-J.; Chen, J.-K.; Liu, C.-L. Colloid Chitin Azure Is a Dispersible, Low-Cost Substrate for Chitinase Measurements in a Sensitive, Fast, Reproducible Assay. J. Biomol. Screen. 2010, 15, 213–217. [Google Scholar] [CrossRef]
- Hussain, M.H.M.; Ngaini, Z.; Chong, N.F.M. Modified Bicinchoninic Acid Assay for Accurate Determination of Variable Length Reducing Sugars in Carbohydrates. Int. Food Res. J. 2018, 25, 2614. [Google Scholar]
- Holding, M.L.; Margres, M.J.; Mason, A.J.; Parkinson, C.L.; Rokyta, D.R. Evaluating the Performance of De Novo Assembly Methods for Venom-Gland Transcriptomics. Toxins 2018, 10, 249. [Google Scholar] [CrossRef]

| Category | GO Analysis | N° of Proteins |
|---|---|---|
| Uncharacterized | - | 116 |
| Structural Components and Extracellular Matrix | GO:0005198—Structural molecule activity GO:0005201—Extracellular matrix structural constituent | 65 |
| Membrane Transport and Channel Activities | GO:0016020—Membrane GO:0015297—Antiporter activity GO:0022803—Passive transmembrane transporter activity GO:0005216—Ion channel activity | 76 |
| Cytoskeletal Structure and Dynamics | GO:0005856—Cytoskeleton GO:0003779—Actin binding | 43 |
| Regulation and Modification of Cellular Processes | GO:0006464—Cellular protein modification process GO:0003677—DNA binding transcription factor activity GO:0004842—Ubiquitin-protein transferase activity GO:0050789—Regulation of biological process GO:0065007—Biological regulation GO:0019222—Regulation of metabolic process | 170 |
| Cell-Cell Communication and Adhesion | GO:0007155—Cell adhesion GO:0005921—Gap junction | 19 |
| Signal Transduction and Kinase Activity | GO:0007165—Signal transduction GO:0004672—Protein kinase activity | 44 |
| Lectins and Sugar-Binding proteins | GO:0030246—Carbohydrate binding | 11 |
| Ribosomal proteins | GO:0005840—Ribosome GO:0003735—Structural constituent of ribosome | 58 |
| Immune System and Response | GO:0006955—Immune response GO:0002376—Immune system process | 19 |
| Hydrolytic Enzymes | GO:0016787—Hydrolase activity | 47 |
| Other Enzymes | GO:0003824—Catalytic activity | 115 |
| Category | N° of Proteins |
|---|---|
| Structural Components and Extracellular Matrix | 12 |
| Membrane Transport and Channel Activities | 6 |
| Cytoskeletal Structure and Dynamics | 3 |
| Regulation and Modification of Cellular Processes | 5 |
| Cell-Cell Communication and Adhesion | 3 |
| Signal Transduction and Kinase Activity | 7 |
| Lectins and Sugar-Binding Proteins | 6 |
| Ribosomal Proteins | 4 |
| Immune System and Response | 5 |
| Hydrolytic Enzymes | 16 |
| Other Enzymes | 10 |
| Toxins | 14 |
| Symbiont Contaminants | 24 |
| Fluorescent Proteins | 1 |
| Category of Potential Venom-Related Element | Proteins (Best Hit in Terms of Homology and Structure) |
|---|---|
| Proteases | A disintegrin and metalloproteinase Aminopeptidase N Pro-X carboxypeptidase Aminopeptidase 1-like N-acetylated-alpha-linked acidic dipeptidase 2-like Trypsin-1 isoform X3-like Carboxypeptidase D-like Carboxypeptidase B-like Neprilysin-4 isoform X2 Puromycin-sensitive aminopeptidase-like Zinc metalloproteinase nas-13-like CAAX prenyl protease 1 Furin-like protease 1 C5a peptidase Gelatinase |
| Phospholipases | Phospholipase D1-like PI-Phospholipase-C X domain-containing protein 3-like Group XIIA secretory phospholipase A2-like N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D-like |
| Glycosidases | Alpha-mannosidase-like Chitinase Chitooligosaccharidolytic beta-N-acetylglucosaminidase Beta-hexosaminidase Non-lysosomal glucosylceramidase-like Chitotriosidase-1 |
| DNases | TatD hydrolase Endonuclease/exonuclease/phosphatase family protein Diphosphonucleotide phosphatase-1 |
| Other hydrolases | Ectonucleoside triphosphate diphosphohydrolase-2 Angiotensin-converting enzyme |
| Lectins | Maltose-binding periplasmic protein Lectin-2 FBP32 f-lectin Cation-independent mannose-6-phosphate receptor Macrophage mannose receptor 1 SUEL-type lectin D-galactoside-specific lectin-like Hemolectin, putative L-rhamnose-binding lectin CSL3-like |
| Anticoagulant proteins | Urokinase plasminogen activator |
| Proteinase inhibitors | Kunitz-type protease inhibitor-1 Pappalysin1 |
| Neurotoxins | Ly-6/neurotoxin-like protein-1 Alpha-elapitoxin-Dpp2a Long neurotoxin OH-55 Antigen 5 like allergen Cul-n-1 |
| Pore-forming toxins (PFTs) | XaxB pore forming toxin |
| Ion channel blockers | Kappa-stichotoxin-She3a ShKT peptide ShKT-Ts1 |
| Coagulation factors | Coagulation factor FVIII-Fc-XTEN Coagulation factor V Coagulation factor VIII Properdin |
| Inflammatory response-related proteins | Complement C1q and TNF-related protein 9B-like Neutrophil cytosol factor-2-like |
| Agglutination and Haemostasis Proteins | Hemagglutinin/amebocyte aggregation factor-like |
| Other potentially toxic proteins | Merozoite surface protein-1 25 kDa ookinete surface antigen Cry1K-like protein chimera Pesticidal crystal protein Cry11Ba |
| Sample | Corresponding Protease Units (U)/mg |
|---|---|
| Crude nematocyst extract | 306.66 ± 11.00 (SD) |
| Crude nematocyst extract treated with PMSF | 178.90 ± 10.90 (SD) |
| Crude nematocyst extract treated with EDTA | 189.93 ± 7.03 (SD) |
| Enzymatic Test | Enzymatic Activity |
|---|---|
| Collagenase | undetected |
| Hyaluronidase | 2.00 ± 0.03 (SD) µg/mg |
| Phospholipase A2 | 12.11 ± 2.20 (SD) U/mg |
| DNase | detectable |
| Chitinase | 0.070 ± 0.001 (SD) U/mg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tassara, E.; Mikšík, I.; Pompach, P.; Mariottini, G.L.; Xiao, L.; Giovine, M.; Pozzolini, M. Proteomic Analysis and Biochemical Characterization of the Nematocyst Extract of the Hydrozoan Velella velella. Mar. Drugs 2024, 22, 468. https://doi.org/10.3390/md22100468
Tassara E, Mikšík I, Pompach P, Mariottini GL, Xiao L, Giovine M, Pozzolini M. Proteomic Analysis and Biochemical Characterization of the Nematocyst Extract of the Hydrozoan Velella velella. Marine Drugs. 2024; 22(10):468. https://doi.org/10.3390/md22100468
Chicago/Turabian StyleTassara, Eleonora, Ivan Mikšík, Petr Pompach, Gian Luigi Mariottini, Liang Xiao, Marco Giovine, and Marina Pozzolini. 2024. "Proteomic Analysis and Biochemical Characterization of the Nematocyst Extract of the Hydrozoan Velella velella" Marine Drugs 22, no. 10: 468. https://doi.org/10.3390/md22100468
APA StyleTassara, E., Mikšík, I., Pompach, P., Mariottini, G. L., Xiao, L., Giovine, M., & Pozzolini, M. (2024). Proteomic Analysis and Biochemical Characterization of the Nematocyst Extract of the Hydrozoan Velella velella. Marine Drugs, 22(10), 468. https://doi.org/10.3390/md22100468










