Abstract
With the increase in antimicrobial resistance and the subsequent demand for novel therapeutics, the deep-sea fish microbiome can be a relatively untapped source of antimicrobials, including bacteriocins. Previously, bacterial isolates were recovered from the gut of deep-sea fish sampled from the Atlantic Ocean.In this study, we used in vitro methods to screen a subset of these isolates for antimicrobial activity, and subsequently mined genomic DNA from isolates of interest for bacteriocin and other antimicrobial metabolite genes. We observed antimicrobial activity against foodborne pathogens, including Staphylococcus aureus, Listeria monocytogenes, Enterococcus faecalis and Micrococcus luteus. In total, 147 candidate biosynthetic gene clusters were identified in the genomic sequences, including 35 bacteriocin/RiPP-like clusters. Other bioactive metabolite genes detected included non-ribosomal peptide synthases (NRPS), polyketide synthases (PKS; Types 1 and 3), beta-lactones and terpenes. Moreover, four unique bacteriocin gene clusters were annotated and shown to encode novel peptides: a class IIc bacteriocin, two class IId bacteriocins and a class I lanthipeptide (LanM subgroup). Our dual in vitro and in silico approach allowed for a more comprehensive understanding of the bacteriocinogenic potential of these deep-sea isolates and an insight into the antimicrobial molecules that they may produce.
1. Introduction
The deep sea is one of the largest biomes on Earth, encompassing 95% of the Earth’s oceanic volume and reaching depths of over 10,000 m [1,2]. It is a unique environment that is characterised by low temperatures, high hydrostatic pressure—which increases by one atmosphere with every 10 m increase in water depth—low oxygen, and no sunlight beyond 1000 m [3]. These conditions are relatively constant, with little impact from ocean currents and seasonality [4]. However, it is estimated that less than 0.001% of the deep sea has been explored, and relatively little is known about its microbial inhabitants compared to those of the terrestrial world [2]. The deep sea is, perhaps, one of the last frontiers of biodiscovery and, as such, has been gaining attention as a source of potential novel microbial compounds.
Deep-sea microorganisms are uniquely adapted to the extreme conditions of their environmental niche. Notably, deep-sea microbial proteins are often optimally active at low temperatures [5] and can have unusual amino acid structures [6], leading to an increased tolerance to high pressure [7,8].
Some recently described antimicrobials are produced by novel deep-sea bacteria isolated from deep-sea sediment. These include marthiapeptide A, a cyclic peptide produced by Marinactinospora thermotolerans [9]; lobophorins E, F and K, antibiotics from Streptomyces spp. [10,11]; and anthracimycin, an anti-anthrax (Bacillus anthracis) antibiotic produced by Streptomyces sp. CNH365 [12]. Other such antimicrobials have also been found in deep seawater, for example, brainimycins B and C (macrolides) produced by Pseudonocardia carboxydivorans [13]. Deep-sea bacteria found in association with higher organisms are also now gaining attention as producers of novel antimicrobials, particularly those from deep-sea coral [11] and sponges [14].
Bacteriocins are small (<10 kDa), ribosomally synthesised antimicrobial peptides produced by bacteria that target other bacteria, and to which the producer has immunity. Bacteriocins can be divided into two main classes: class I (lantibiotics or lanthipeptides) and class II (non-lanthionine-containing bacteriocins) [15]. Lanthipeptides can be further subdivided into five subclasses, I–V, based on the biosynthetic enzyme(s) involved in their modification [16]. The class II bacteriocins are subdivided into four groups: the anti-listerial pediocin-like peptides (class IIa), two-component peptides (class IIb), cyclic/circular bacteriocins (class IIc) and the linear and non-pediocin-like peptides (class IId) [17]. Bacteriocins are emerging as alternatives to antibiotics due to their spectrum of activity—which can be broad or narrow—heat stability and their capacity for bioengineering to generate derivatives with value-added properties [18]. This is of particular importance, given the significant and growing threat that antibiotic-resistant organisms pose in the medical and food industries. According to a 2019 report by the Centres for Disease Control and Prevention (CDC), almost three million infections, including foodborne illness, caused by antibiotic-resistant organisms occur each year in the United States of America alone [19].
To date, very few studies have explored the production of bacteriocins from deep-sea fish microbial isolates. One example is BaCf3, a bacteriocin produced by Bacillus amyloliquefaciens BTSS3, isolated from the intestine of a deep-sea shark. This highly thermostable bacteriocin was initially identified through in vitro screening methods and showed inhibitory activity against pathogenic bacteria, including Bacillus cereus, Clostridium perfringens, Staphylococcus aureus and Salmonella enterica Typhimurium [20,21].
Previous work carried out in our laboratory generated metagenome-assembled genomes (MAGs) from shotgun metagenomic sequencing of intestinal microbiomes samples from deep-sea fish from the Atlantic Ocean [22]. The fish were collected as part of a groundfish survey. Analysis of the MAGS revealed information on the taxonomic and functional diversity of these samples, such as a predominance of Pseudomonadota (Proteobacteria), as well as a large abundance of genes involved in DNA repair, protein folding and motility. Bacterial isolates were recovered from the intestines and skin of a subset of these deep-sea fish and were preliminary screened for bioactivity against the Gram-positive target organism, Lactobacillus delbrueckii subsp. bulgaricus. L. bulgaricus is commonly used in the assessment of antimicrobial activity due to its low-pH resistance and sensitivity to a wide range of bacteriocins [23,24,25]. In this study, a cohort of these bacterial isolates were reassessed for bioactivity against the target strain, Lactobacillus delbrueckii subsp. bulgaricus, and were subsequently screened against a range of various target strains.
The aim of this study was to determine the bacteriocinogenic (ability to produce bacteriocins) potential of deep-sea fish microbial isolates (n = 65) through in vitro screening against various target strains, including pathogenic bacteria, and through in silico mining of the genomes of a selection of these isolates (n = 36) for putative bacteriocin biosynthetic gene clusters. We also looked for the presence of other secondary metabolite gene clusters and antimicrobial resistance (AMR) genes.
2. Results
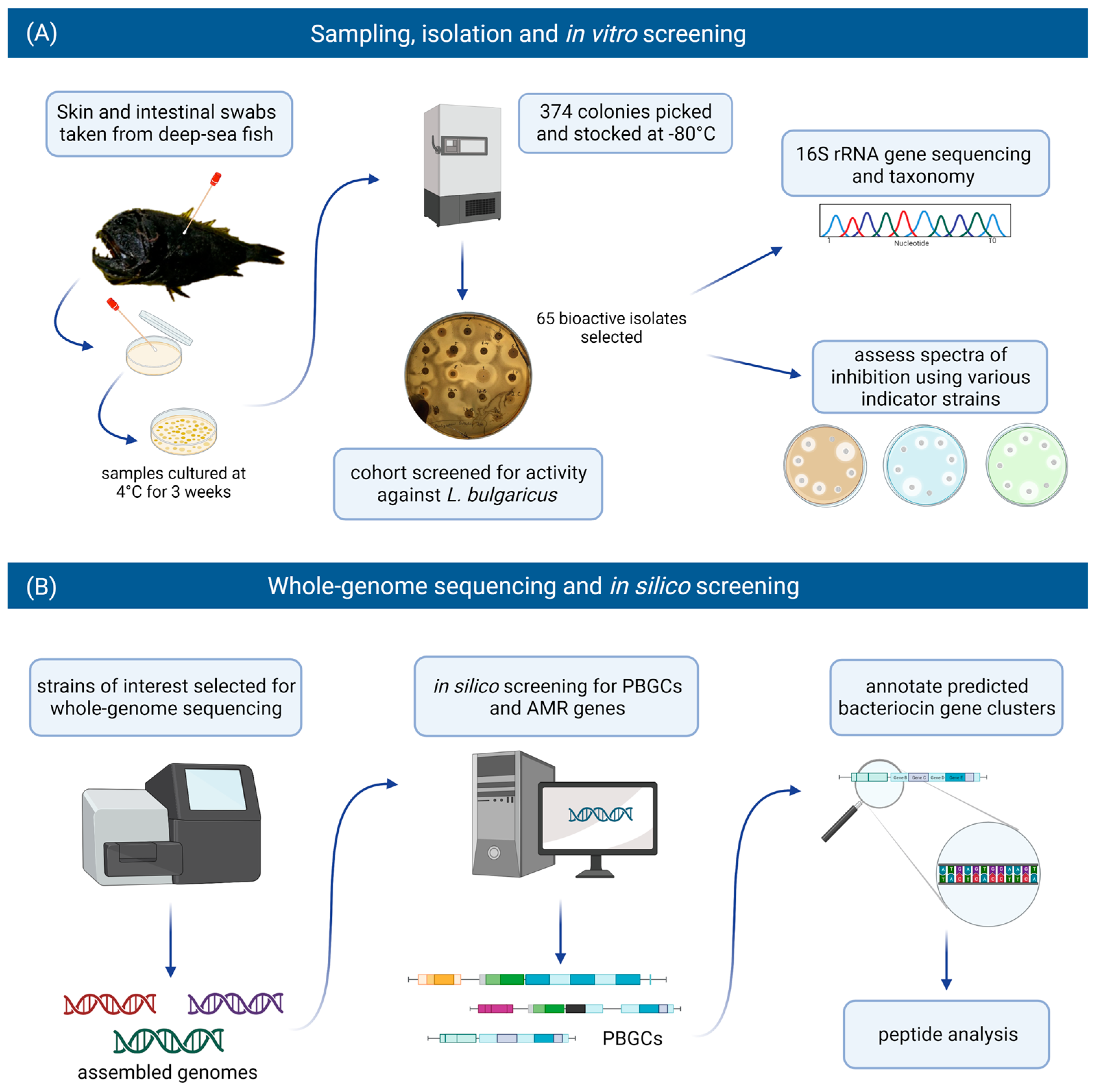
For this study, we included bacteria that had been isolated from the skin and intestines of deep-sea fish and assessed them for antimicrobial activity against various target strains, including foodborne pathogenic bacteria. Those producing antimicrobial activity were identified by 16S rRNA gene analysis. Isolates of interest were selected for whole genome sequencing and subsequent in silico screening for putative bacteriocin gene clusters and putative biosynthetic gene clusters (PBGCs) encoding secondary metabolites. Strains were also assessed for the presence of antimicrobial resistance (AMR) genes. The workflow implemented in this study is illustrated in Figure 1.
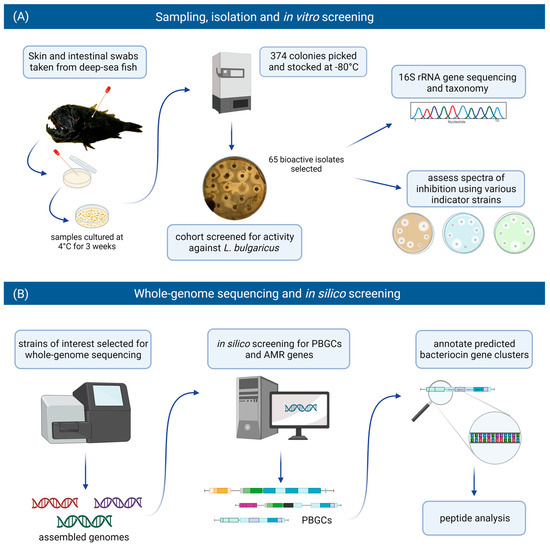
Figure 1.
The in vitro and in silico-based approaches used in this study to assess the bacteriocinogenic potential in the deep-sea fish microbiome. (A) Swabs were taken from the skin and intestines of deep-sea fish. Samples were cultured at 4 °C for three weeks, and 374 isolates were recovered. A cohort of these isolates were screened for activity against Lactobacillus delbrueckii subsp. bulgaricus. Sixty-five bioactive isolates were identified and selected for further in vitro screening and identification through 16S rRNA gene analysis. (B) Strains of interest were selected for whole-genome sequencing and subsequent in silico screening for bacteriocin/RiPP genes, putative biosynthetic gene clusters (PBGCs) and antimicrobial resistance (AMR) genes. Predicted bacteriocin gene clusters were annotated, and the bacteriocin peptide precursors were analysed (figure created with BioRender.com).
2.1. Recovery and Selection of Bioactive Isolates
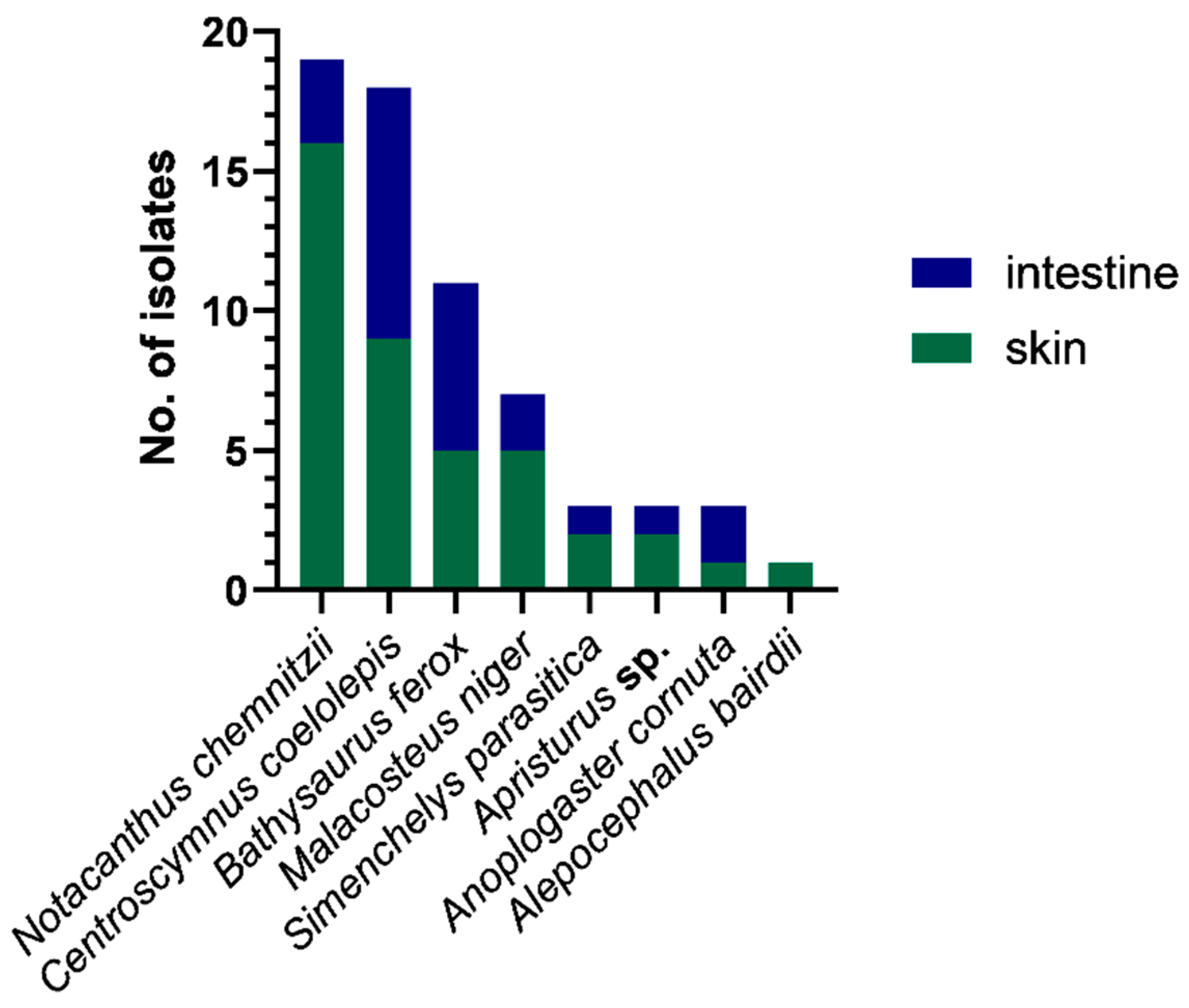
The 65 bacterial isolates came from eight different deep-sea fish, namely: Alepocephalus bairdii, Anoplogaster cornuta, Apristurus sp., Bathysaurus ferox, Centroscymnus coelolepis, Malacosteus niger, Notacanthus chemnitzii, and Simenchelys parasitica. The majority (63%) of the isolates came from skin swabs, with most isolates recovered from the deep-sea fish N. chemnitzii and C. coelolepis (Figure 2).
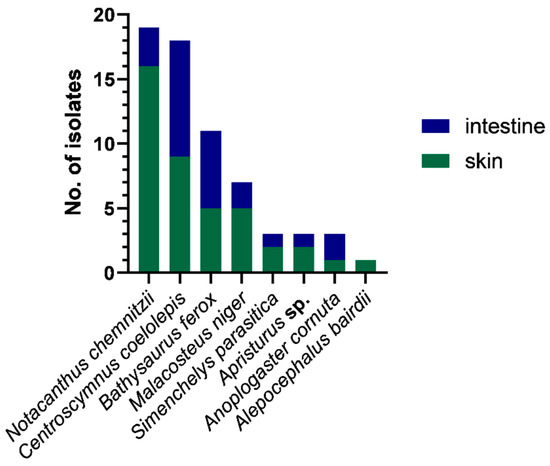
Figure 2.
Total counts of isolates recovered per sample type (skin or intestine swab) and per fish host species. The majority of isolates were from skin swabs of Notacanthus chemnitzii and Centroscymnus coelolepis.
We confirmed that 44 of the 65 isolates were active against L. bulgaricus (Table 1). All the isolates grew on marine agar (MA) with minimal colony swarming, though modified tryptic soy agar (mTSA) resulted in slightly greater inhibitory activity. Twenty-one and eighteen of the isolates did not grow on brain heart infusion (BHI) or mTSA, respectively, under these conditions. In instances where inhibition could not be measured because of colony swarming or poor growth of the indicator, the result was recorded as “no data” (nd; Table 1).

Table 1.
Results of inhibitory activity observed against the target organism Lactobacillus delbrueckii subsp. bulgaricus (L. bulgaricus) on selected media. The isolate sample source and fish host are also given and numbered as follows: 1, Anoplogaster cornuta; 2, Apristurus sp.; 3, Bathysaurus ferox; 4, Centroscymnus coelolepis; 5, Malacosteus niger; 6, Notacanthus chemnitzii; and 7, Alepocephlus bairdii. Inhibition is indicated by + (0.5–2.5 mm), ++ (2.6–5 mm), +++ (5.1–10 mm), or − (no inhibition).
2.2. Taxonomic Diversity
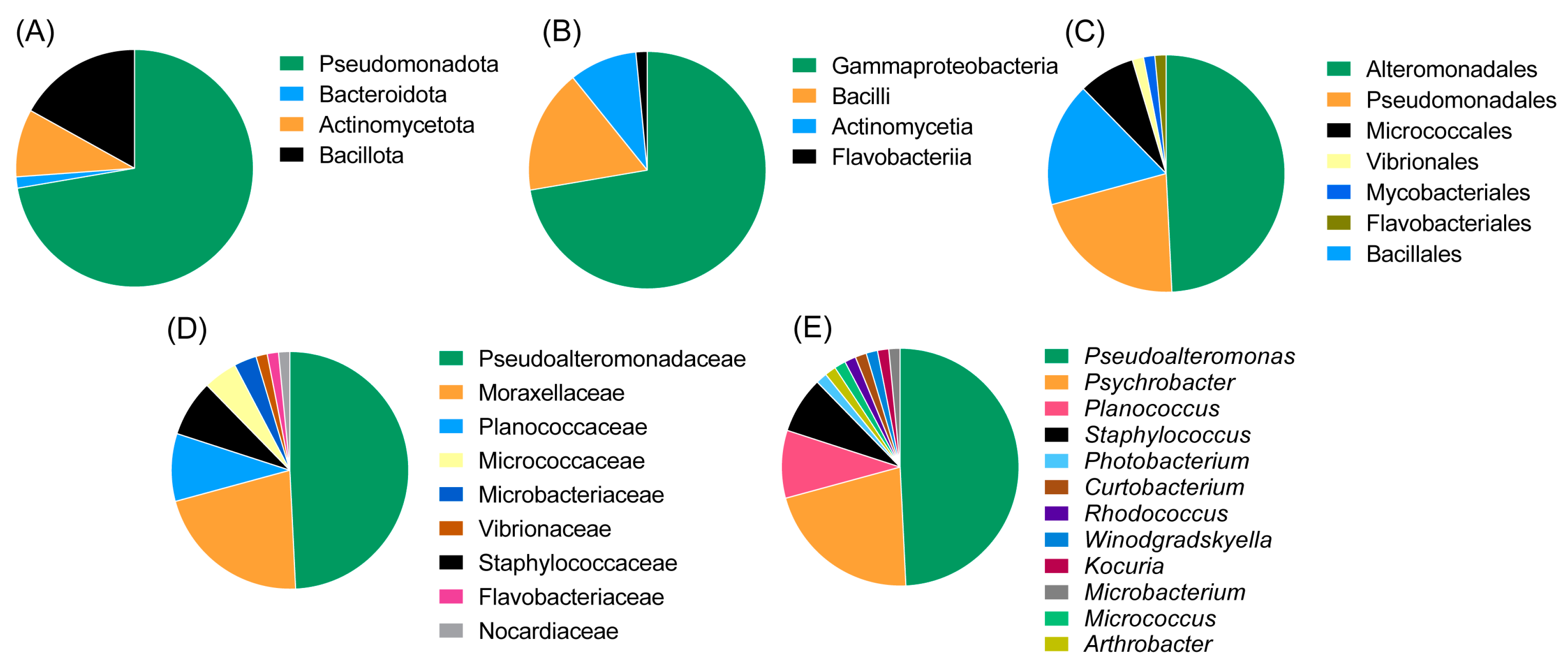
The majority of the strains belonged to the class Gammaproteobacteria (of the phylum Pseudomonadota, at 72.3%) followed by Bacilli (phylum Bacillota 16.9%), Actinomycetia (phylum Actinomycetota, 9.2%) and Flavobacteriia (phylum Bacteroidota, 1.5%). The most abundant orders were Alteromonadales (49.2%), Pseudomonadales (20%) and Bacillales (16.9%). Other represented orders were Micrococcales, Mycobacteriales, Flavobacteriales and Vibrionales. The most abundant genus was Pseudoalteromonas (family Pseudoalteromonadaceae, 51.9%), followed by Psychrobacter (family Moraxellaceae, 21.5%), Planococcus (family Planococcaceae, 9.2%) and Staphylococcus (family Staphylococcaceae, 7.7%). The remaining genera were represented only by a single isolate; these included Winogradskyella, Rhodococcus, Arthrobacter, Kocuria, Curtobacterium, Photobacterium and Micrococcus. An overview of the taxonomic diversity of the isolates is shown in Figure 3.
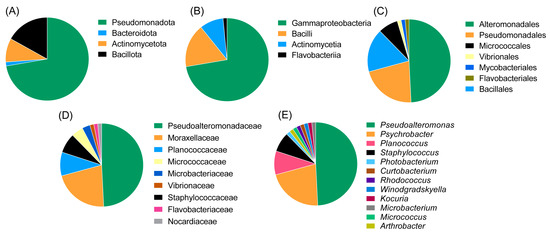
Figure 3.
Diversity of the deep-sea fish microbiome isolates at the phylum level (A), class level (B), order level (C), family level (D) and genus level (E). The most abundant isolates in this study were Pseudoalteromonas and Psychrobacter species of the phylum Pseudomonadota (Proteobacteria).
2.3. Spectrum of Activity
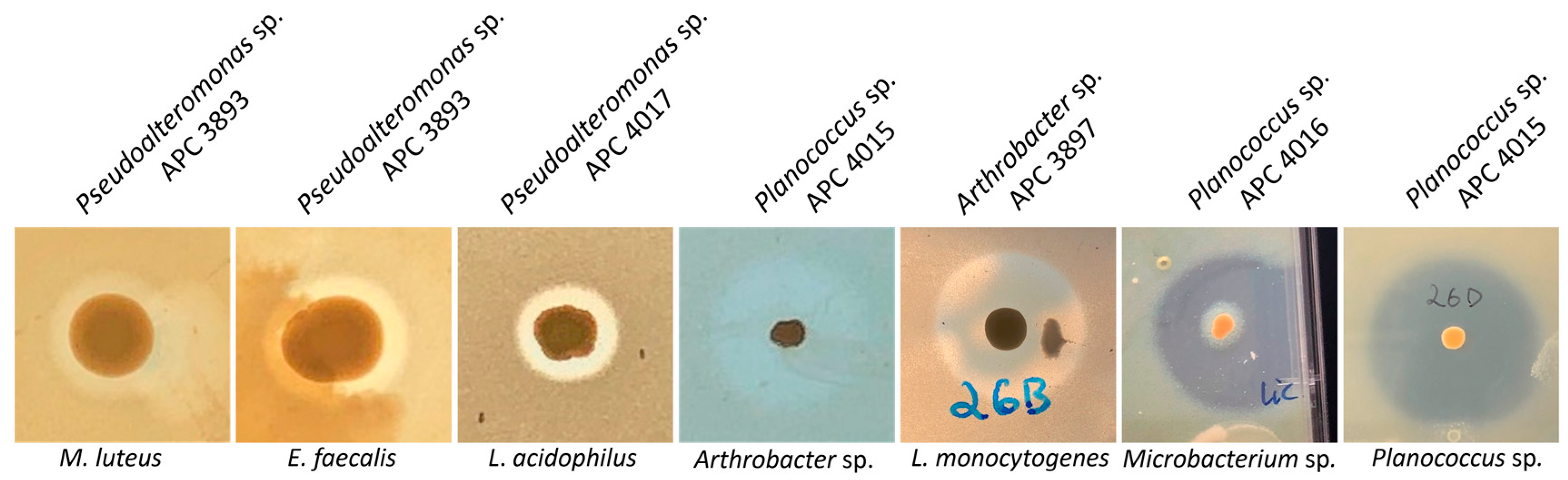
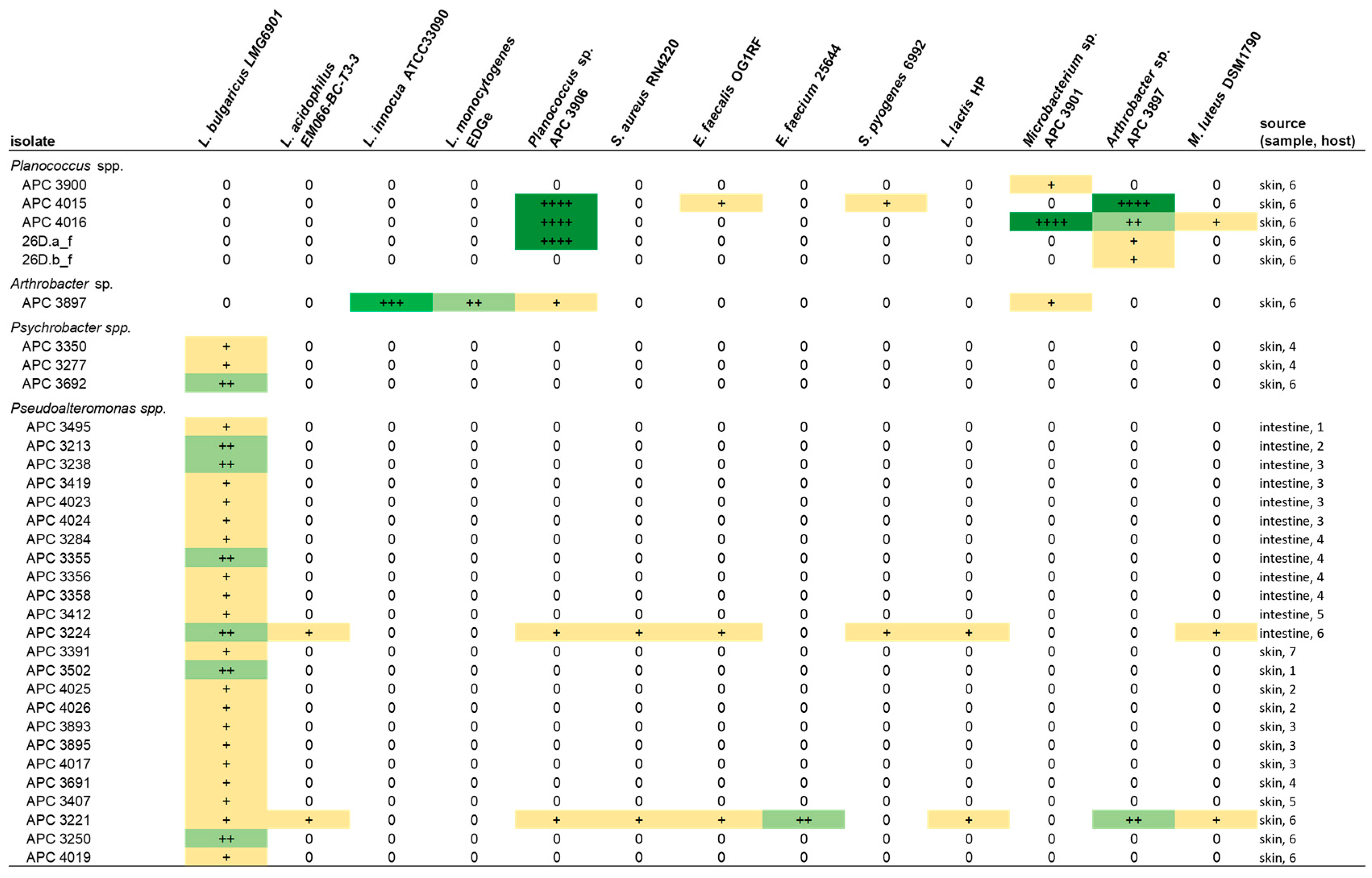
Following the initial screen against L. bulgaricus, the isolates were tested against a range of different target strains, including foodborne pathogens, food spoilage bacteria and marine bacteria. MA was chosen as the culture medium for the spectrum of activity screen as it supported the growth of the widest range of the marine isolates as compared to BHI and mTSA. Representative zones of inhibition are shown in Figure 4. Overall, 13 of the 22 target strains were inhibited (Figure 5). Four of the deep-sea strains that initially showed little or no activity against L. bulgaricus after culturing on marine agar were able to inhibit several of the other strains. Isolates of the genus Planococcus (strains APC 3900, APC 4015, APC 4016, 26D.a_f and 26D.b_f) were most effective at inhibiting the more closely related indicators: Planococcus sp. APC 3906 (of the same genus) and those of the order Micrococcales: Arthrobacter sp. APC 3897, Microbacterium sp. APC 3901 and Micrococcus luteus. Planococcus sp. APC 4016 additionally showed limited activity against E. faecalis and S. pyogenes. Arthrobacter sp. APC 3897 showed the greatest activity against L. innocua and L. monocytogenes. This isolate also showed activity against the Planococcus sp. and Microbacterium sp. indicators. Pseudoalteromonas sp. APC 4017 had the broadest spectrum of activity, inhibiting nine of the twenty-two indicators. None of the isolates inhibited B. cereus, S. intermedius, Rhodococcus sp. APC 3903 or Kocuria sp. APC 4018. None of the Gram-negative target strains (E. coli, P. aeruginosa, S. Typhumurium, V. fischerii, Psychrobacter sp. APC 3276) were inhibited. Thirteen of the isolates had no inhibitory effects against any of the indicators under these conditions. It was observed that several strains from the same sample site and the same host produced identical inhibition patterns and are likely to be identical strains. For example, Psychrobacter sp. strains APC 3350 and APC 3277, both from the skin of Centroscymnus coelolepis, only inhibited L. bulgaricus; similarly, Pseudoalteromonas sp. strains APC 4020 and APC 4023 from the intestine of deep-sea fish Bathysaurus ferox both also inhibited only L. bulgaricus.
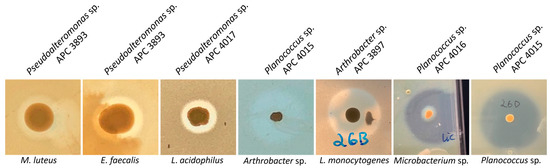
Figure 4.
Representative zones of inhibition of the bioactive marine isolates. The strongest inhibition was demonstrated against other marine isolates, Planococcus sp. and Microbacterium sp. and Listeria, including L. monocytogenes. The producer IDs are given above the images, and the indicator strains are below.
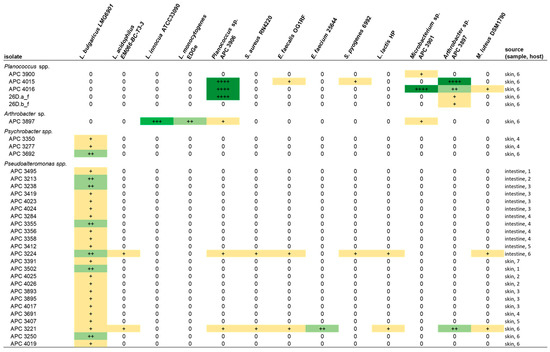
Figure 5.
The spectrum of inhibition of the bioactive deep-sea isolates against the target strains by colony overlay assays on marine agar. The overall broadest range of activity was exhibited by Pseudoalteromonas strains APC 4017 and APC 3893. Inhibition is indicated by + (0.5–2.5 mm), ++ (2.6–5 mm) and +++ (5.1–10 mm) and is coloured to highlight inhibition patterns. Only the results of strains that demonstrated inhibitory activity under these conditions are reported in the table.
2.4. In Silico Antimicrobial Screening
A selection of isolates that demonstrated in vitro antimicrobial activity and a representative of the identified taxonomic genera were chosen for whole genome sequencing. A total of 36 genomes were sequenced to enable genome-based in silico screening to identify antimicrobial-encoding genes and to identify potential bacteriocin producers that may not have shown in vitro activity under our tested conditions. The genome assemblies were cross-checked against the MAG sequence data (from [22]), and it was confirmed that the isolate sequences were not present in the MAGs.
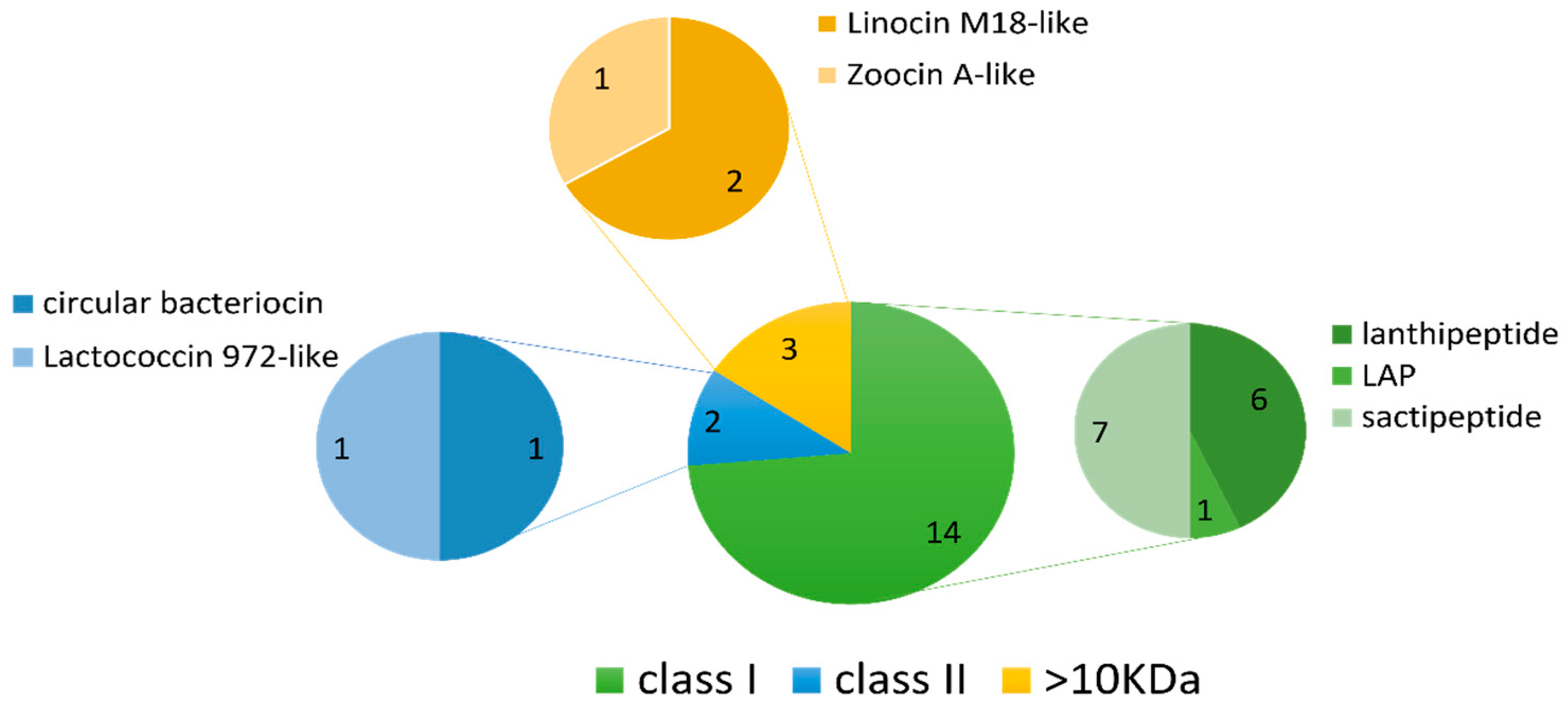
The BAGEL4 mining software searches for potential motifs and core peptides of ribosomally synthesised and post-translationally modified peptides (RiPPs), such as bacteriocins, as well as larger antimicrobial peptides (>10 kDa), and presents each output as an area of interest (AOI) [26]. Nineteen AOIs were predicted by BAGEL4 across 13 of the 36 genomes consisting of fourteen class I bacteriocins, two class II bacteriocins and three larger antimicrobial proteins (>10 kDa) (Table 2 and Figure 6). Sactipeptides were the most abundant sub-type (seven hits), followed by lanthipeptides (six hits). Of the lanthipeptide hits, three were of the lanthipeptide subclass II, sharing 50–56% similarity to the bacteriocins mersacidin and cerecidin (Table 2). The remaining three lanthipeptides were subclass III; however, they lacked an identifiable core peptide gene within the predicted gene cluster. The remaining bacteriocin/RiPP sub-types included one linear-azol(in)e-containing peptide (LAP), one class II circular bacteriocin with similarity to circularin A and one class II bacteriocin with similarity to lactococcin 972. Four AOIs each belonged to Planococcus strains, Pseudoalteromonas strains and the Rhodococcus sp. strain APC 3903. The remaining AOIs belonged to Arthrobacter sp. APC 3897, Curtobacterium sp. APC 3903, Kocuria sp. APC 4018, Psychrobacter sp. APC 3350 and Winogradskyella sp. APC 3343.

Table 2.
Summary of the potential bacteriocin/RiPP gene clusters and their annotations predicted by BAGEL4 in the deep-sea bacterial genomes. The predicted bacteriocins included subclass II lanthipeptides (with similarity to mersacidin/cerecidin), subclass III lanthipeptides and a circularin A-like class II bacteriocin. Other predicted bacteriocin subtypes included sactipeptides, a LAP and a lactococcin 972-like peptide. Predicted AOIs which lacked an identifiable core peptide gene and could not be annotated by BAGEL4 are given as “undefined”. ×2 = two clusters found; %sim = % similarity.
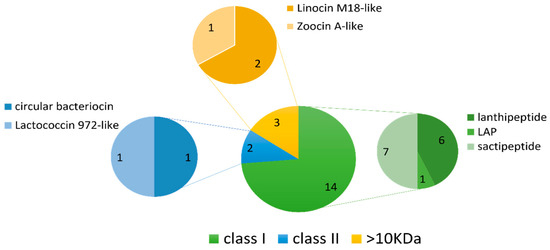
Figure 6.
Summary of the bacteriocin classes and their subtypes identified using BAGEL4. Nineteen AOIs were predicted in total across the 36 deep-sea fish gut isolate genomes. Sactipeptides and lanthipeptides (class I bacteriocins) were the most frequently predicted bacteriocin subtypes.
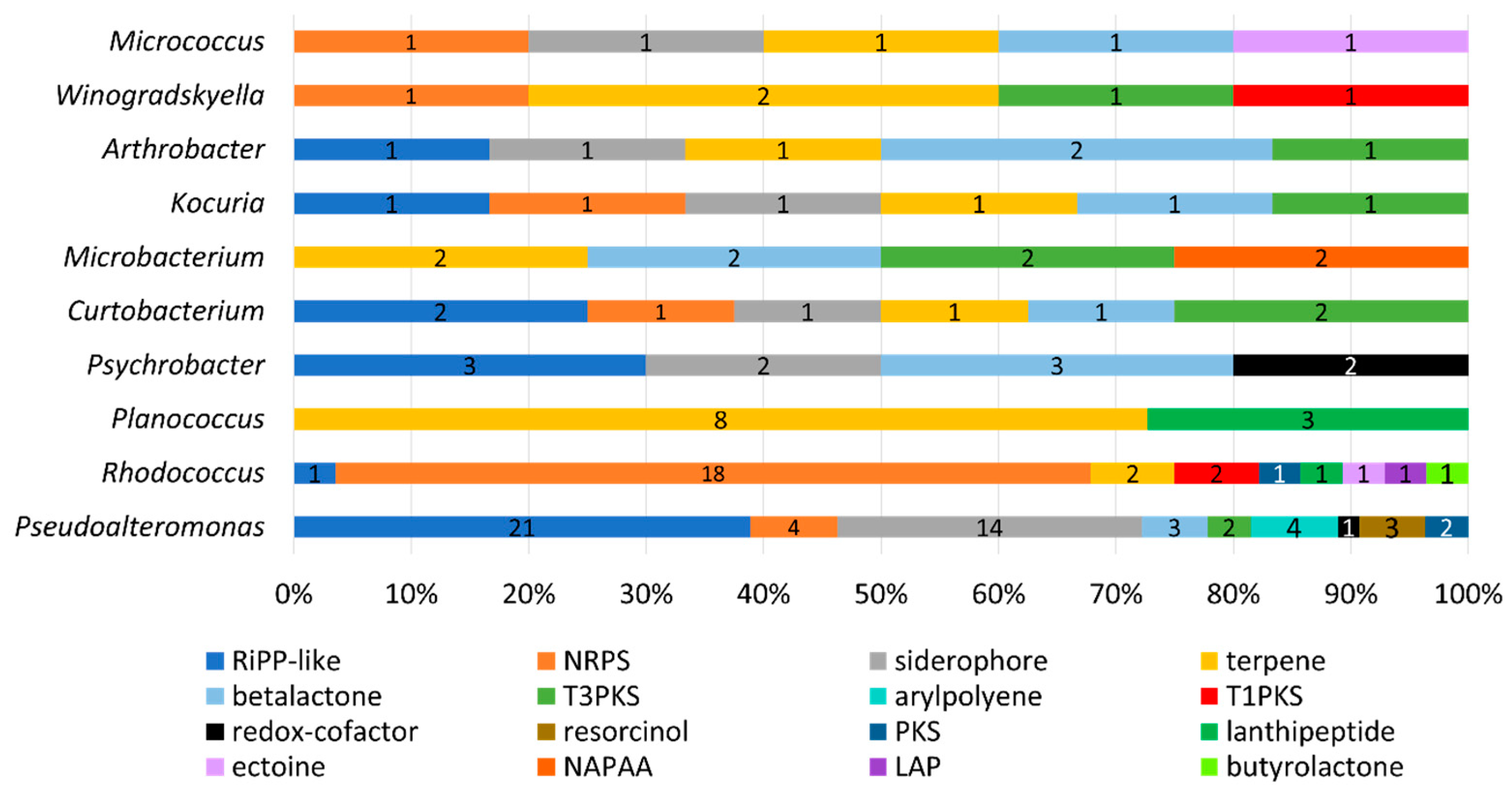
AntiSMASH identified 141 putative biosynthetic gene clusters (PBGCs) encoding secondary metabolites, with PBGCs being found in all 36 genomes (Table 3 and Figure 7). The most frequent PBGC types identified were RiPP-like (29 hits), non-ribosomal peptide synthase (NRPS, 26 hits), siderophore (20 hits), terpene (18 hits) and betalactone (13 hits). Additionally, PBGCs encoding resorcinol, redox-cofactors, type 3 and type 1 polyketide synthases (PKSs), non-alpha poly-amino acids (NAPAAs), linear azol(in)e-containing peptides (LAPs), ectoine, butyrolactone and aryl polyene were also found. Proportionally, the Rhodococcus strain APC 3903 had the greatest number of predicted secondary metabolite gene clusters at 28 (18% of the total). The presence of terpene BGCs is an indication that the orange colour observed in several of the colonies (Figure 4)., for example, the Planococcus spp. is the result of terpene/carotenoid productionAs with the BAGEL4 findings (Table 2), many of the predicted RiPP-like hits lacked an identifiable core biosynthetic gene, in which case the PBGC prediction may have been based on the presence of bacteriocin or RiPP-associated motifs or genes; this is indicated in where either no annotation was provided, or no known cluster hit was identified (Table 3).

Table 3.
In silico predictions by AntiSMASH of bacteriocin/RiPP-like PBGCs and other secondary metabolite gene clusters. Where no core peptide or no similar cluster was found, the result is given as “none.” ×2 = two clusters found; % sim = similarity; nd = no data.
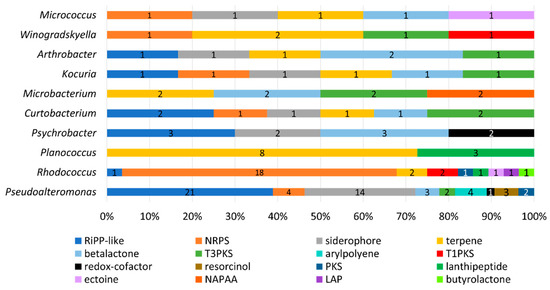
Figure 7.
PBGC hits predicted by the AntiSMASH database of secondary metabolites and antibiotics. Results are given as a percentage of the total per genus, and counts of each PBGC type are given. Default search strictness parameters were used. The most frequent hits included RiPPs and lanthipeptides, which suggests these isolates encode bioactive compounds, such as bacteriocins. Other gene cluster hits include polyketide synthases, siderophores, terpenes and non-ribosomal peptide synthases (NRPS).
The most promising candidates for further analysis, based on the presence of an identified core peptide/biosynthetic gene, were the putative cyclic bacteriocin in Arthrobacter APC 3897, the Planococcus spp. cerecidin-like lanthipeptides and the lactococcin 972-like peptides of Curtobacterium sp. APC 4022.
2.4.1. Class I LanM-Group Bacteriocins (Lanthipeptides)
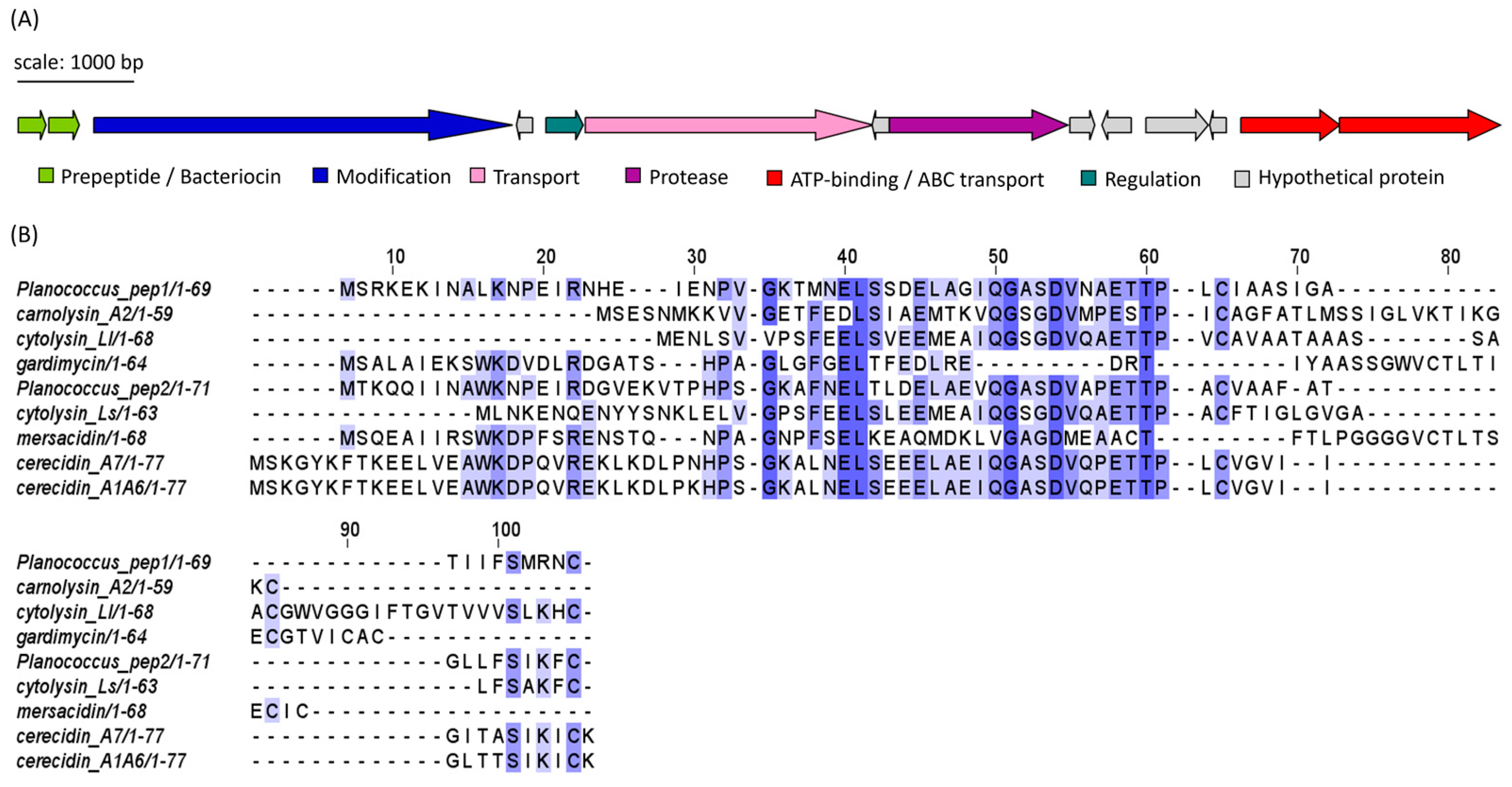
Isolates APC 3900, 4015 and 4016 of the genus Planococcus all encoded identical sequences, which corresponded to a cerecidin-like lanthipeptide (Figure 8A). The gene cluster encoded two premature/pro-peptides, a large modification protein (LanM-like), a transporter, a regulation protein, a protease and ATP-binding/ABC transport proteins. Several small open reading frames (sORFs) were also identified, but their function could not be determined. The two peptide precursors, denoted Pep1 and Pep2 for clarity, shared 52.94% identity with each other. When queried with RiPPMiner’s sequence similarity search tool, the top hits were cerecidin A1-6 and cerecidin A7 (B. cereus). Pep1 shared 46.27% and 44.78% identity with cerA7 and cerA1-6, respectively. Pep2 shared 54.29% and 55.71% id to cerA7 and cerA1-6, respectively. Other similar peptides from the database were cytolysin Ll (E. faecalis), carnolysin A2 (Carnobacterium maltaromaticum), gardimycin (Actinoplanes liguriae), mersacidin (B. amyloliquefaciens) and cytolysin Ls (E. faecalis). The multiple sequence alignment (MSA) of the Planococcus and RiPP hit sequences in Figure 8B shows only a few conserved residues within the region between residues 50 and 60. As with cerecidin, the Pep1 and Pep2 peptide sequences appear to possess a leader sequence, denoted by the GG/GA motif, followed by six additional amino acids which precede a potential core peptide (initiated by TT) [27]. The putative Planococcus modification gene shared 45.49% identity to LanM of cerecidin (Genbank accession: AHJ59542.1), indicating that the Planococcus peptides are class II lanthipeptides [28]. The statistics generated using EMBOSS Needle of the pairwise alignments of the precursor peptides, and the reference peptides are given in Table S3. Figures of the pairwise alignments are given in Figures S1 and S2.
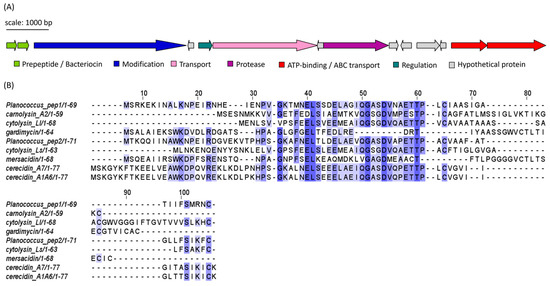
Figure 8.
(A) Putative lanthipeptide (subclass II) gene cluster identified in Planococcus spp. isolates (Planococcus peptides 1 and 2). Identical clusters were found in APC 3900, APC 4015 and APC 4017. Genes are coloured according to predicted function. (B) MSA of the predicted Planococcus peptide precursor/bacteriocin sequence with most similar bacteriocin sequences identified using RiPPMiner: cytolysin Ll (E. faecalis), carnolysin A2 (Carnobacterium maltaromaticum), mersacidin (B. amyloliquefaciens), cytolysin Ls (E. faecalis), cerecidin A1-6 and cerecidin A7 (Bacillus cereus), gardimycin (Actinoplanes liguriae). The MSA is coloured according to shared identity of the aligned amino acid residues at a given position, with the colour shade deepening as the percentage of sequences sharing the same residue increases. No colour indicates <40% of sequences share the residue.
2.4.2. Class IIc (Circular) Bacteriocin
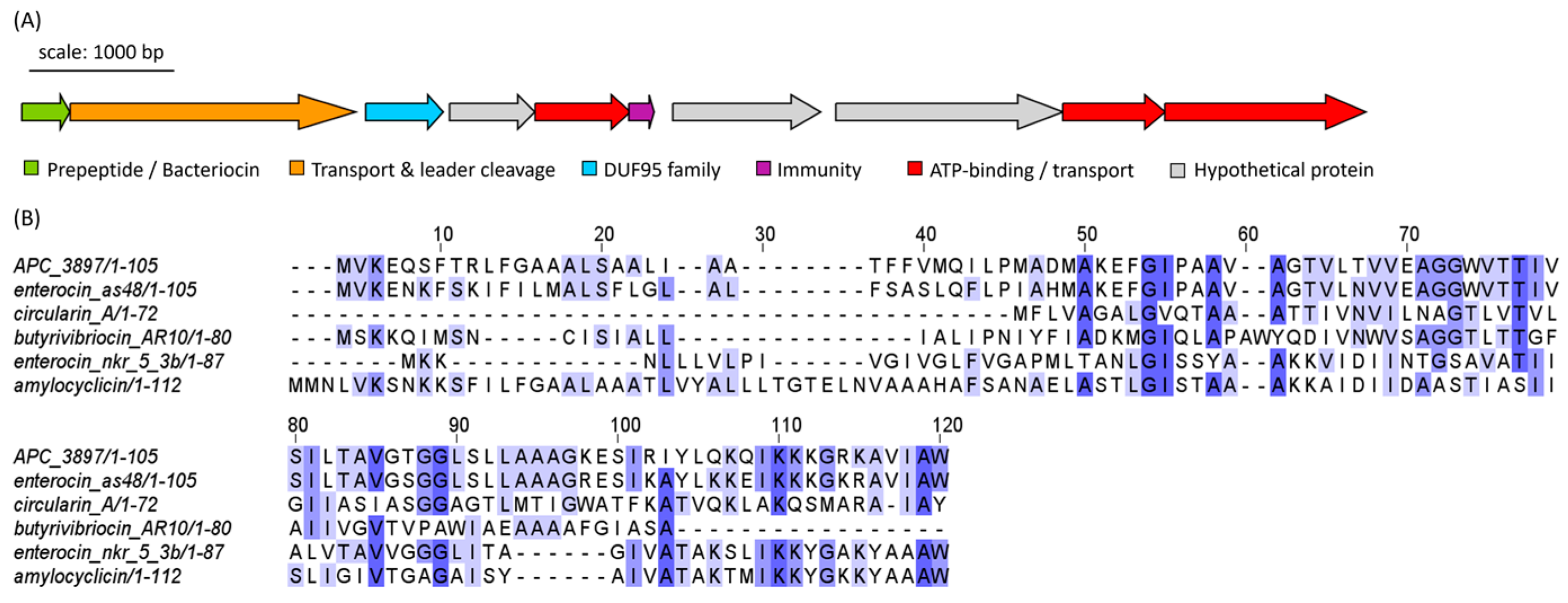
The cyclic bacteriocin gene cluster predicted in Arthrobacter isolate APC 3897 was annotated using BLASTp. The gene cluster consists of ten genes: a peptide precursor, a transporter/cleavage protein, a DUF95 family protein, three ATP-binding/transport proteins, an immunity protein and three hypothetical proteins of unknown function (Figure 9A). The predicted core precursor peptide sequence was queried using RiPPMiner: The closest related sequence was enterocin AS-48, a circular bacteriocin from Enterococcus faecalis, with which the peptide precursor sequences shared 71.3% identity. The other top bacteriocin/RiPP hits were butyrivibriocin (30.56%ID; Butyrivibrio fibrisolvens), circularin A (25.37%; Clostridium beijerinckii), enterocin NKR-5-3b (32.47%; Enterococcus faecium) and amylocyclicin (28.32%; Bacillus amyloliquefaciens). The peptide sequences were aligned and are illustrated in Figure 9B, which is coloured according to shared identity of the aligned amino acid residues and sequences are arranged according to pairwise identity. The pairwise alignment statistics and figures generated using EMBOSS Needle are provided in Table S3 and Figure S3, respectively.
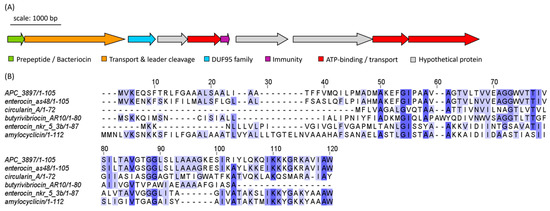
Figure 9.
(A) Biosynthetic gene cluster (BGC) of a putative class IIc bacteriocin from Arthrobacter sp. isolate APC 3897. Genes are coloured according to predicted function. (B) Multiple sequence alignment (MSA) of APC 3897 BGC with similar bacteriocin sequences identified using RiPPMiner: butyrivibriocin (Butyrivibrio fibrisolvens), circularin A (Clostridium beijerinckii), enterocin NKR-5-3b (Enterococcus faecium), enterocin AS-48 (E. faecalis) and amylocyclicin (Bacillus amyloliquefaciens). The MSA is coloured according to shared identity of the aligned amino acid residues at a given position, with the colour shade deepening as the percentage of sequences sharing the same residue increases. No colour indicates <40% of sequences share the residue.
2.4.3. Class IId Bacteriocins
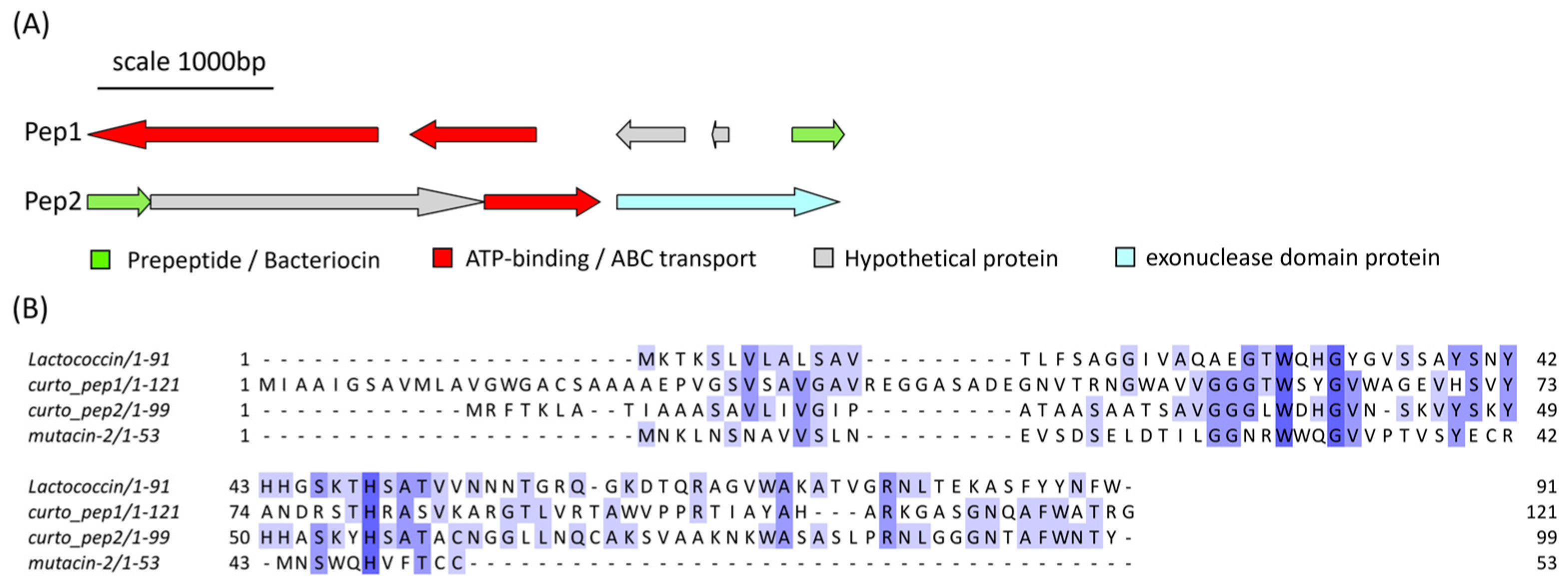
Curtobacterium sp. APC 4022 was found to contain two PBGCs located on two separate contigs that each encoded a lactococcin 972-like peptide (Figure 10A). The predicted gene cluster of the first peptide, denoted Pep1, consists of a core peptide gene, two ATP-binding/ABC-transport genes and two hypothetical genes of unknown function. The second gene cluster consists of four genes: a core peptide (Pep2) gene, a hypothetical gene of unknown function, an ATP-binding/ABC-transporter gene and a gene encoding an exonuclease family protein. When queried with RiPPMiner’s sequence similarity tool, the most similar hit to both core peptide sequences was mutacin II (Streptococcus mutans). The two Curtobacterium peptide sequences were aligned with mutacin II and lactococcin 972 (Figure 10B). Pep1 and Pep2 share 20.97% identity with each other. Pep1 shared 15.32% and 12.4% identity with lactococcin 972 and mutacin II, respectively. Pep2 shared 26% and 15% identity with lactococcin 972 and mutacin II, respectively. The pairwise alignment statistics generated using EMBOSS Needle are provided in Table S3. Figures of the pairwise alignments of Curtobacterium precursor peptides with the reference sequences are given in Figures S4 and S5. The pairwise alignment of Pep1 and Pep2 is also shown in Figure S4.
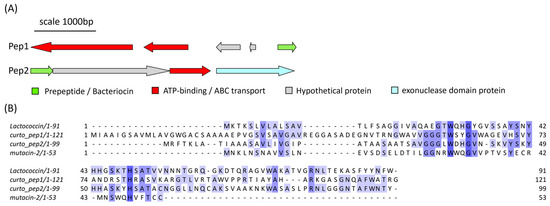
Figure 10.
(A) The predicted gene clusters of the two lactococcin 972-like class IId bacteriocins from Curtobacterium sp. APC 4022, labelled pep1 and pep2. (B) The MSA of the peptide precursor sequences with mutacin II (Streptococcus mutans) and lactococcin 972 (L. lactis subsp. lactis) shows some conserved residues between the sequences The MSA is coloured according to shared identity of the aligned amino acid residues at a given position, with the colour shade deepening as the percentage of sequences sharing the same residue increases. No colour indicates <40% of sequences share the residue.
2.5. Antimicrobial Resistance Genes
The 36 deep-sea genomes were screened for AMR genes using ABRicate. Hits to resistance genes were only detected in the genome of Rhodococcus sp. APC 4022 (Table 4). Two genes corresponding to rifamycin/rifampin resistance were detected: (rif)iri, which encodes a rifampin (inactivating) monooxygenase and rpoB2, which encodes a resistant variant of the beta-subunit of RNA polymerase. A gene encoding a multidrug efflux protein (mtrA) was also detected.

Table 4.
The output of ABRicate genome screening for antimicrobial resistance (AMR) genes. All hits came from the genome sequence of Rhodococcus sp. APC 4022.
3. Discussion
This study aimed to determine the bacteriocinogenic potential of bacterial isolates from the microbiomes of deep-sea fish. A combined approach of in vitro and in silico methods was used to identify antimicrobial-producing isolates and to further establish the potential source of this antimicrobial activity, with a focus on finding bacteriocin gene clusters.
The initial repeated screening against L. bulgaricus demonstrated that 44 of these isolates retained their bioactive activity (Table 1). The loss of activity by the remaining 21 isolates could be explained by the fact that antimicrobial production is often a tightly regulated process, dependent on the induction by external factors, such as other microbial metabolites present in the natural environment. Purification and removal of these isolates from the microbiome community may have affected their activity. Analysis of the spectrum of activity of these deep-sea isolates revealed that most strains had a narrow spectrum of activity. The only exceptions were Pseudoalteromonas strains APC 4017 and APC 3893 which were able to inhibit nine and eight of the indicator strains, respectively, with APC 4017 being particularly active against E. faecium 25644 and Arthrobacter sp. APC 3897 (Figure 6). Overall, just over half (11 out of 20) of the indicator strains were inhibited under the conditions tested. None of the Gram-negative indicator strains were impacted. Some inhibition patterns were noted amongst the taxonomic groups. The Planococcus strains were most effective against the Planococcus indicator strain APC 3906 and against the Micrococcales strains: Arthrobacter sp. APC 3897 and Microbacterium sp. APC 3901. The Gram-negative Pseudoalteromonas and Psychrobacter strains were only active against L. bulgaricus (with the aforementioned exceptions). The Arthrobacter strain APC 3897 was almost exclusively active against the Listeria strains. Based on the in vitro screening and taxonomic results, a number of isolates were chosen for whole-genome sequencing and in silico screening.
Interestingly, the whole-genome sequence data from the isolates was not found within the MAGs. This is a reflection of the difference in sample sizes between the two studies but also indicates that the isolates in this study may only represent a very small sample of the deep-sea fish microbiome, many of which may, in fact, be unculturable.
Through in silico screening using BAGEL4, we identified 19 AOIs, including lanthipeptide PBGCs and a cyclic bacteriocin PBGC. Though eleven of these AOIs lacked an identifiable core biosynthetic bacteriocin gene, the predictions by BAGEL4 may be based on similarities of the AOIs with bacteriocin-associated proteins. For example, in four of the predicted sactipeptide clusters, a gene was identified which encoded a putative GTP 3’,8-cyclase that shared similarity with MoaA, a member of the radical S-adenosylmethionine (rSAM) enzyme superfamily [29]. Specific rSAM enzymes (sactisynthases) are essential for forming the characteristic sactionine linkages of sactipeptides [30]. It is also possible that the core peptide genes are located elsewhere on these genomes, especially given that the in silico screening was carried out on contigs rather than full genomes. The lack of identifiable core peptides could also be due to the presence of unique motifs that are not found in the database.
The Planococcus strains APC 4015 and APC 4016, as mentioned above, demonstrated potent antimicrobial activity against several of the Gram-positive indicator strains, particularly against Planococcus sp. APC 3906, Microbacterium sp. APC 3901 and Arthrobacter sp. APC 3897. The Planococcus genomes were subsequently found to have identical gene clusters that encoded two putative bacteriocins. Of the known bacteriocin/RiPP sequences, the Planococcus peptides were most similar to the cerecidin peptides of B.cereus. Cerecidins are class II lanthipeptides that occur as two variants, CerA1 (of which there are six identical encoding genes, CerA1-6) and CerA7 [27]. The sequences also shared some similarities with the haemolytic enterotoxin of E. faecalis, cytolysin, which is composed of two structural subunits, CylLL and CylLs [31], and the two-component lantibiotic carnolysin (C. maltaromaticum) [32]. Cerecidin and cytolysin both contain two cleavage sites. The first occurs at a GA motif site after the first leader sequence. The second cleavage site occurs after a secondary leader sequence of six amino acids, leaving behind the core peptide [27,31]. These processing sites are also potentially present in the Planococcus peptides, as indicated by the area of homology seen in the region between residues 50 and 60 (Figure 8B). This suggests that these peptides undergo multistep processing.
Arthrobacter isolate APC 3897, which was observed to have anti-listerial activity in vitro, was found to contain genes encoding a class IIc cyclic bacteriocin. The predicted bacteriocin peptide precursor was found to be most closely related to enterocin AS-48, a circular bacteriocin from Enterococcus faecalis. AS-48 is most active against Gram-positive bacteria [33], particularly Listeria species [34], which is consistent with the in vitro activity we observed for Arthrobacter sp. APC 3897. The structure of the PBGC was also consistent with that of enterocin AS-48, each consisting of ten genes, including a structural peptide precursor, a transporter protein, ATP-binding/transport proteins and an immunity protein gene [35]. The Arthrobacter gene cluster was also found to encode a putative DUF95 family protein which is characteristic of circular bacteriocin gene clusters and is believed to be involved in the transport and secretion of these peptides [36]. To date, the identification of an AS-48-like circular bacteriocin has not been reported in an Arthrobacter strain; in fact, some Arthrobacter strains have been used as sensitive indicators in studies of AS-48 activity [37,38].
Curtobacterium sp. APC 4022 displayed very little in vitro antimicrobial activity, having only shown inhibition of L. bulgaricus when cultured on mTSA (Table 1). The genome of this strain was found to contain two separate gene clusters encoding predicted bacteriocins with sequence similarities, albeit low, to the class IId bacteriocins lactococcin 972 and mutacin II. Lactococcin 972, first isolated from Lactococcus lactis subsp. lactis IPLA972, inhibits peptidoglycan synthesis by targeting lipid II and is almost exclusively active against lactococci [39]. Members of the lactococcin 972 protein family (Pfam IPR006540) are usually associated with transmembrane putative immunity proteins. Mutacin II, from S. mutans NTCC 10449, is active against a range of Gram-positive species, including streptococci, and targets cell membrane functions [40].
The observed in vitro antimicrobial activity of the Planococcus, Arthrobacter and Curtobacterium strains may be attributed to their respective putatively identified bacteriocins; however, we cannot rule out the possibility that these strains also are producing other bioactive secondary metabolites. The production of bacteriocins can also be a tightly regulated process, which we expect holds true for these deep-sea strains. For example, the cerecidins of B. cereus are predicted to be under the control of competence quorum-sensing system genes (comQXPA), as well as the CerR regulatory protein, the stimulus of which is still unknown. Production of cerecidin in its native host has only been possible by constitutive expression of cerR using plasmids [41]. Further genomic and molecular analysis is needed to understand the regulatory systems behind the predicted bacteriocins of this study.
Lastly, we screened all of the genomes for the presence of AMR genes using ABRicate. Only three AMR genes were detected, and all three were from a single genome, that of Rhodococcus sp. APC 4022. Firstly, a resistant variant of the RNA polymerase beta subunit (RpoB2) was detected. Mutations to the RNA polymerase beta subunit (RpoB) can result in a gene duplication whereby one gene produces the rifamycin-sensitive RpoB and the second produces the resistant variant [42]. Possessing rpoB paralogs can be indicative of secondary metabolism and antibiotic production in Actinobacteria, though in vitro antimicrobial production was not detected in APC 4022 [43]. Secondly, a (rif)iri gene, which encodes a rifampin-inactivating monooxygenase, was identified. Production of rifamycin-inactivating or -modifying enzymes and mutations of RpoB are common resistance mechanisms and have previously been reported in Rhodococcus strains and related taxa; they are particularly well-studied in pathogenic bacteria such as Mycobacteria [44,45]. The third AMR gene detected was a multidrug efflux pump gene. The identification of multiple AMR genes indicates a potential multiple resistance mechanism utilised by Rhodococcus sp. APC 4022. The overall relative lack of detectable AMR genes across all the genomes is consistent with other reports of the low incidence of AMR genes in deep-sea fish-associated microbial genomes [22,46]. AMR genes have been detected in deep-sea water and sediment, but the incidence may be low compared to sample sites heavily impacted by anthropogenic activities [47,48]. There have been suggestions that AMR genes found in marine environments are largely unclassified [49]; therefore, the accurate identification of AMR genes in deep-sea samples may be even more challenging.
It is particularly notable that we were able to successfully isolate strains capable of targeting food-borne pathogens. Foodborne disease is a huge threat to public health and safety. The World Health Organisation (WHO) has estimated that globally up to 1 in 10 people fall ill to foodborne disease every year, leading to almost half a million premature deaths and huge economic losses [50]. It is important, now more than ever, to be able to find novel compounds for combatting foodborne pathogens, particularly when treatments are being compounded by the increasing incidence of antimicrobial resistance. The aquaculture industry is another potential application of deep-sea bacteriocins, which we have not explored here in detail. For example, the overuse of antibiotics and the development of antimicrobial resistance is also a great concern in the fish farming industry, with disease outbreaks having significant economic and health impacts [51].
4. Materials and Methods
4.1. Isolation and Cultivation of Antimicrobial-Producing Isolates
Fish were collected from sample sites in international waters off the Grand Banks of Newfoundland at depths ranging from 850 m to 1000 m, as previously described [22]. An example of one the fish specimens and the sampling location is shown in Figure 11. Swabs were taken from the skin and intestines of the fish, plated onto BD DifcoTM Marine Agar 2216 (MA, Becton Drive, NJ, USA) and incubated at 4 °C for three weeks. Individual colonies (n = 374) were picked and stocked in 35% glycerol and stored at −80 °C.

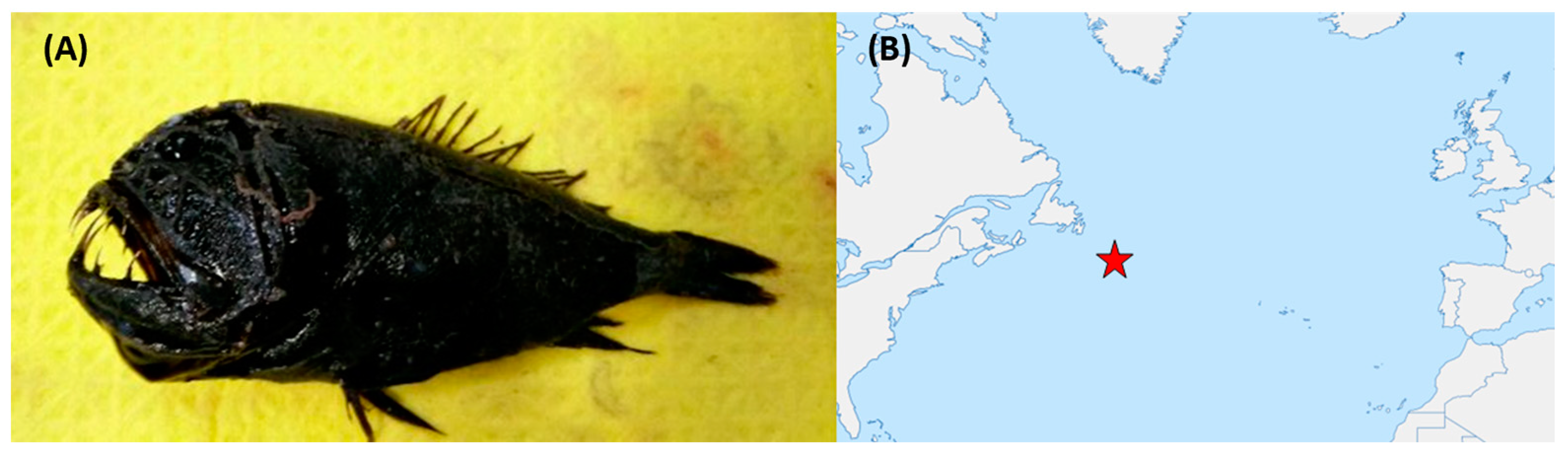
Figure 11.
(A) Anoplogaster cornuta—an example of one of the fish species from which bioactive bacteria were isolated. (B) Approximate sampling location (indicated by the red star icon) in the Atlantic Ocean. Deep-sea fish specimens were collected from depths of approximately 1000 m.
4.2. In Vitro Screening
Previous work in our laboratory (not published) conducted preliminary screening of a cohort of 374 stocked isolates against the indicator strain Lactobacillus delbrueckii subsp. bulgaricus LMG 6901 and identified 65 strains with antimicrobial activity. In this study, we reassessed the activity against L. bulgaricus by the 65 strains and examined the spectrum of antimicrobial activity of these strains against various target bacteria strains (listed in Table 5).

Table 5.
The target strains and respective growth conditions used in this study.
For analysis of activity against L. bulgaricus, isolates were plated on MA, brain heart infusion (BHI, Oxoid, Basingstoke, UK) agar and modified tryptic soy agar (mTSA, +0.5 g/L NaCl, Oxoid) and incubated at 5 °C (±1 °C) for two weeks. The plates were then overlaid with sloppy media containing 0.8% agar and 0.25% of an overnight culture of L. bulgaricus in modified Difco MRS (mMRS, +0.5 g/L cysteine). Following incubation overnight, we measured the distance from the edge of the producing colony to the edge of the area of growth of the indicator organism and recorded the results qualitatively as follows: + (0.5–2.5 mm), ++ (2.6–5 mm), +++ (5.1–10 mm) and ++++ (>10 mm).
For subsequent colony overlay screening to assess the spectrum of activity, isolates were plated on MA and incubated at 5 °C (±1 °C) for 2 weeks. The plates were then overlaid with indicator organisms in the relevant sloppy media, as listed in Table 5, using the same method as mentioned above. Inhibition distances were measured after overnight incubation or after 48 h for indicators cultured at 20 °C.
4.3. 16S rRNA Gene Sequencing and Taxonomy
Colony PCR was performed on all of the isolates. Cells were lysed by resuspending in 10 µL lysis buffer (20 mM Tris-HCL, pH 8.0, 2 mM EDTA, 1.2% Triton X-100; reagents from Sigma-Aldrich/Merck, Burlington, Massachusetts, USA) and incubatingat 95 °C for 10 min. PCR was performed in a total volume of 50 μL using 25 μL of BioMixTM Red (Meridian Bioscience®, London, UK), 16 μL of sterile molecular-grade water, 2 μL (at 10 pmol/µL) of the non-specific universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5’-GGTTACCTTGTTACGACTT-3’) and 5 μL of DNA template from lysed cells. The following amplification conditions were used: 95 °C for 1 min, followed by 32 cycles of 95 °C for 15 s, 55 °C for 15 s and 72 °C for 32 s with a final extension at 72 °C for 10 min. PCR products were run on a 1.5% agarose gel, stained with MIDORI Green Advance and subject to gel electrophoresis. The gel was visualised using a UV-transilluminator to confirm the presence of amplified DNA. PCR products were purified using the GeneJET PCR purification Kit (Thermo Scientific, Waltham, MA, USA). DNA sequencing of both the forward and reverse strands was performed by GENEWIZ (Azenta Life Sciences, Leipzig, Germany) or Source BioScience (Waterford, Ireland). Where possible, sequence pairs were concatenated before analysis. Where available, 16S rRNA gene sequences were also extracted from subsequent gDNA sequencing data and used for inference. Taxonomy was inferred by searching the resulting 16S rRNA gene sequences for sequence similarity with NCBI’s 16S rRNA database using the BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 3 November 2021).
4.4. Genomic DNA Extraction and Sequencing
Genomic DNA (gDNA) was extracted from 36 of the isolates using the GeneJET Genomic DNA Purification Kit (Thermo Scientific) and the relevant protocol, Gram-negative or Gram-positive, for the isolate. DNA quality was checked using agarose (1%) gel electrophoresis. An overview of the sequencing and assembly information of each sample is given in Table S1. In brief, the first set of gDNA samples were sequenced on an Illumina MiSeq platform (2 × 250 bp paired-end reads) at GenProbio s.r.l. (Parma, Italy). The second set of samples were sequenced on the Illumina MiSeq platform (2 × 250 bp paired-end reads) by MicrobesNG (http://www.microbesng.com), which is supported by the BBSRC, (grant number BB/L024209/1). APC 4015 was sequenced by GENEWIZ (Azenta Life Sciences, Burlington, MA, USA) on the Illumina NovaSeq platform (2 × 150 bp paired-end reads).
4.5. Genome Assembly and Annotation
Reads provided by MicrobesNG were processed and assembled by the sequencing company. For all other samples, read quality was assessed with FastQC (v0.11.9; https://www.bioinformatics.babraham.ac.uk/projects/fastqc), trimmed with Trimmomatic (v0.36) [52] to a Phred score of 20 across a 4 bp sliding window. Reads less than 50 bp were discarded. Contigs (>500 bp) were assembled using SPAdes (v3.15.3) [53], and assembly quality was assessed using QUAST (v4.4) [54] and CheckM (v1.0.18) [55]. Assemblies were submitted to Genbank and annotated using the NCBI Prokaryotic Annotation Pipeline (PGAP) upon submission [56]. An overview of the assembly information, QUAST and CheckM statistics is given in Table S1. Additionally, the 16S rRNA gene sequences from the WGS data were searched against the SILVA rRNA gene project database [57]. The resulting taxonomic descriptions and closest related reference strain(s) are reported in Table S2.
The datasets generated in this study are available under the BioProject accession number PRJNA883941.
4.6. In Silico Antimicrobial Screening
Assembled genomes were screened for putative bacteriocin and secondary metabolite gene clusters using the genome mining tools BAGEL4 (BAGEL v4) [26] and AntiSMASH (v.5.1.2.) [58], respectively.
4.7. Putative Biosynthetic Gene Cluster (PBGC) Analysis
Putative core precursor peptide genes were analysed using RiPPMiner’s sequence similarity search [59] to query their similarity to known RiPPs from the database. Multiple alignments of the core peptide sequences and the RiPPMiner hits were made using Clustal Omega [60] and visualised using Jalview [61]. Pairwise alignments between the precursor peptide sequences and the RiPPMiner hits/reference sequences were generated using EMBOSS Needle [62] using default parameters. The matrix used for each pairwise alignment is provided along with the results in Table S3. Genes within the PBGCs adjacent to the core peptide genes were queried and annotated using BLASTp to determine their potential function. The annotated PBGCs were visualised using Gene Graphics [63].
4.8. Antimicrobial Resistance Gene Screening
Genomes were screened for the presence of antimicrobial resistance genes using ABRicate (https://github.com/tseemann/abricate). The databases used were ARG-ANNOT [64], CARD [65], EcOH [66], MEGARES [67], NCBI [68], PlasmidFinder [69], Resfinder [70] and VFDb [71].
5. Conclusions
This study has shown that not only are bacteria from the skin and gut microbiomes of deep-sea fish cultivable, but they are a potentially rich source of antimicrobial agents. The majority of the antimicrobial producers in this study were Gram-negative Pseudoalteromonas strains of the phylum Pseudomonadota, though some of the most potent producers included Planococcus and Arthrobacter strains of the phyla Bacillota and Actinomycetota, respectively. Whilst initially selected based on their inhibitory activity against Lactobacillus bulgaricus, it was demonstrated that these deep-sea isolates could inhibit foodborne pathogens, such as Listeria monocytogenes and Enterococcus spp. Overall, the most susceptible indicators were other native marine strains, or more closely related organisms, a characteristic of the narrow spectrum of activity of many bacteriocins. In silico screening found that these isolates encode a wide variety of PBGCs encoding antimicrobial molecules from type I and II bacteriocins to LAPs and sactipeptides as well as PKSs, NRPSs, betalactones and terpenes. Included in these are potentially novel bacteriocins and variants of known bacteriocins. This provides great scope for future work in exploring applications of targeting pathogenic bacteria with antimicrobials that are produced, and potentially active at low temperatures. The dual in vitro and in silico approach allowed for a more comprehensive understanding of the potential targets of deep-sea bacteriocins and an insight into the antimicrobial molecules that they may produce.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/md21080444/s1; Table S1: Overview of the whole-genome sequencing data, QUAST and CheckM metrics; Table S2: Taxonomic descriptions, based on the 16S rRNA gene sequence, of the strains selected for whole-genome sequencing. Sequences were searched against the SILVA rRNA gene project database; Table S3: Resulting statistics from the pairwise alignments of precursor peptide sequences using EMBOSS Needle. Figure S1: Pairwise alignment of the Planococcus precursor peptide, Pep1, with (A) Pep2 (Planococcus spp.); (B) cerecidin A1A6 (B. cereus); (C) cerecidin A7 (B. cereus); (D) cytolysin Ls (E. faecalis); (E) carnolysin A2 (C. maltaromaticum); (F) carnolysin A1 (C. maltaromaticum); (G) cytolysin Ll (E.faecalis); (H) gardimycin (A. liguriae); (I) mersacidin (B. amyloliquifaciens). Figure S2: Pairwise alignment of the Planococcus precursor peptide, Pep2, with (A) cerecidin A7 (B. cereus); (B) cerecidin A1A6 (B. cereus); (C) cytolysin Ls (E. faecalis); (D) carnolysin A1 (C. maltaromaticum); (E) carnolysin A2 (C. maltaromaticum); (F) cytolysin Ll (E.faecalis); (G) gardimycin (A. liguriae); (H) mersacidin (B. amyloliquifaciens). Figure S3: Pairwise alignment of the Arthrobacter sp. APC 3897 precursor peptide with (A) enterocin AS48 (E. faecalis); (B) amylocyclicin (B. amyloliquifaciens); (C) circularin A (C. beierinckii); (D) enterocin nkr_5_3b (E. faecium); (E) butyrivibriocin AR10 (B. fibriosolvens). Figure S4: Pairwise alignment of the Curtobacterium sp. APC 4022 precursor peptide curto_Lct1 with (A) curto_Lct2); (B) lactococcin 972 (L. lactis); (C) mutacin 2 (S. mutans). Figure S5: Pairwise alignment of the Curtobacterium sp. APC 4022 precursor peptide curto_Lct2 with (A) lactococcin 972 (L. lactis) and (B) mutacin 2 (S. mutans).
Author Contributions
Conceptualisation, C.H. and R.P.R.; data curation, S.U.-L.; funding acquisition, R.P.R.; investigation, S.U.-L.; methodology, S.U.-L. and F.W.J.C.; resources, S.U.-L., F.W.J.C. and R.P.R.; supervision, C.H. and R.P.R.; visualisation, S.U.-L.; writing—original draft, S.U.-L.; writing—review and editing, C.H. and R.P.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was co-funded by a Teagasc Walsh Fellowship (grant number 2017218), Science Foundation Ireland (SFI) under Grant Number SFI/12/RC/2273_P2 and the European Union (ERC, BACtheWINNER, 101054719). Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council. Neither the European Union nor the granting authority can be held responsible for them.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The datasets generated in this study are available under the BioProject accession number PRJNA883941.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Greenaway, S.F.; Sullivan, K.D.; Umfress, S.H.; Beittel, A.B.; Wagner, K.D. Revised depth of the Challenger Deep from submersible transects; including a general method for precise, pressure-derived depths in the ocean. Deep Sea Res. Part I Oceanogr. Res. Pap. 2021, 178, 103644. [Google Scholar] [CrossRef]
- Danovaro, R.; Corinaldesi, C.; Dell’Anno, A.; Snelgrove, P.V.R. The deep-sea under global change. Curr. Biol. 2017, 27, R461–R465. [Google Scholar] [CrossRef] [PubMed]
- Somero, G.N. Biochemical ecology of deep-sea animals. Experientia 1992, 48, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Pernice, M.C.; Giner, C.R.; Logares, R.; Perera-Bel, J.; Acinas, S.G.; Duarte, C.M.; Gasol, J.M.; Massana, R. Large variability of bathypelagic microbial eukaryotic communities across the world’s oceans. ISME J. 2016, 10, 945–958. (In English) [Google Scholar] [CrossRef]
- Zhang, J.-W.; Zeng, R.-Y. Molecular Cloning and Expression of a Cold-Adapted Lipase Gene from an Antarctic Deep Sea Psychrotrophic Bacterium Pseudomonas sp. 7323. Mar. Biotechnol. 2008, 10, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Liu, G.; Zheng, R.; Sun, C.; Wu, S. Structural and Functional Insights into Iturin W, a Novel Lipopeptide Produced by the Deep-Sea Bacterium Bacillus sp. Strain wsm-1. Appl. Environ. Microbiol. 2020, 86, e01597-20. [Google Scholar] [CrossRef]
- Penhallurick, R.W.; Ichiye, T. Pressure Adaptations in Deep-Sea Moritella Dihydrofolate Reductases: Compressibility versus Stability. Biology 2021, 10, 11. (In English) [Google Scholar] [CrossRef]
- Yan, B.-Q.; Chen, X.-L.; Hou, X.-Y.; He, H.; Zhou, B.-C.; Zhang, Y.-Z. Molecular analysis of the gene encoding a cold-adapted halophilic subtilase from deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913: Cloning, expression, characterization and function analysis of the C-terminal PPC domains. Extremophiles 2009, 13, 725–733. [Google Scholar] [CrossRef]
- Zhou, X.; Huang, H.; Chen, Y.; Tan, J.; Song, Y.; Zou, J.; Tian, X.; Hua, Y.; Ju, J. Marthiapeptide A, an Anti-infective and Cytotoxic Polythiazole Cyclopeptide from a 60 L Scale Fermentation of the Deep Sea-Derived Marinactinospora thermotolerans SCSIO 00652. J. Nat. Prod. 2012, 75, 2251–2255. [Google Scholar] [CrossRef]
- Niu, S.; Li, S.; Chen, Y.; Tian, X.; Zhang, H.; Zhang, G.; Zhang, W.; Yang, X.; Zhang, S.; Ju, J.; et al. Lobophorins E and F, new spirotetronate antibiotics from a South China Sea-derived Streptomyces sp. SCSIO 01127. J. Antibiot. 2011, 64, 711–716. [Google Scholar] [CrossRef]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Osset, M.; Pérez-Victoria, I.; Martín, J.; De Pedro, N.; De la Cruz, M.; Díaz, C.; Vicente, F.; Reyes, F.; et al. Lobophorin K, a New Natural Product with Cytotoxic Activity Produced by Streptomyces sp. M-207 Associated with the Deep-Sea Coral Lophelia pertusa. Marine Drugs 2017, 15, 144. [Google Scholar] [CrossRef]
- Jang, K.H.; Nam, S.J.; Locke, J.B.; Kauffman, C.A.; Beatty, D.S.; Paul, L.A.; Fenical, W. Anthracimycin, a Potent Anthrax Antibiotic from a Marine-Derived Actinomycete. Angew. Chem. Int. Ed. 2013, 30, 7822–7824. [Google Scholar] [CrossRef]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Pérez-Victoria, I.; Otero, L.; Fernández, J.; Palacios, J.J.; Martín, J.; de la Cruz, M.; Díaz, C.; Vicente, F.; et al. Branimycins B and C, Antibiotics Produced by the Abyssal Actinobacterium Pseudonocardia carboxydivorans M-227. J. Nat. Prod. 2017, 80, 569–573. [Google Scholar] [CrossRef]
- Back, C.R.; Stennett, H.L.; Williams, S.E.; Wang, L.; Ojeda Gomez, J.; Abdulle, O.M.; Duffy, T.; Neal, C.; Mantell, J.; Jepson, M.A.; et al. A New Micromonospora Strain with Antibiotic Activity Isolated from the Microbiome of a Mid-Atlantic Deep-Sea Sponge. Mar. Drugs 2021, 19, 105. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Montalbán-López, M.; Scott, T.A.; Ramesh, S.; Rahman, I.R.; Van Heel, A.J.; Viel, J.H.; Bandarian, V.; Dittmann, E.; Genilloud, O.; Goto, Y.; et al. New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep. 2021, 38, 130–239. [Google Scholar] [CrossRef] [PubMed]
- Nissen-Meyer, J.; Rogne, P.; Oppegard, C.; Haugen, S.H.; Kristiansen, E.P. Structure-Function Relationships of the Non-Lanthionine-Containing Peptide (class II) Bacteriocins Produced by Gram-Positive Bacteria. Curr. Pharm. Biotechnol. 2009, 10, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States, 2019 ed.; U.S. Department of Health and Human Services, CDC: Washington, DC, USA, 2019.
- Bindiya, E.S.; Tina, K.J.; Sasidharan, R.S.; Bhat, S.G. BaCf3: Highly thermostable bacteriocin from Bacillus amyloliquefaciens BTSS3 antagonistic on food-borne pathogens. 3 Biotech 2019, 9, 136. [Google Scholar] [CrossRef]
- Bindiya, E.S.; Tina, K.J.; Raghul, S.S.; Bhat, S.G. Characterization of Deep Sea Fish Gut Bacteria with Antagonistic Potential, from Centroscyllium fabricii (Deep Sea Shark). Probiotics Antimicrob. Proteins 2015, 7, 157–163. (In English) [Google Scholar] [CrossRef] [PubMed]
- Collins, F.W.; Walsh, C.J.; Gomez-Sala, B.; Guijarro-García, E.; Stokes, D.; Jakobsdóttir, K.B.; Kristjánsson, K.; Burns, F.; Cotter, P.D.; Rea, M.C.; et al. The microbiome of deep-sea fish reveals new microbial species and a sparsity of antibiotic resistance genes. Gut Microbes 2021, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.N.; O’Connor, P.M.; Rea, M.C.; O’Sullivan, O.; Walsh, C.J.; Healy, B.; Mathur, H.; Field, D.; Hill, C.; Ross, R.P. Nisin J, a Novel Natural Nisin Variant, Is Produced by Staphylococcus capitis Sourced from the Human Skin Microbiota. J. Bacteriol. 2020, 202, e00639-19. [Google Scholar] [CrossRef] [PubMed]
- Sugrue, I.; O’Connor, P.M.; Hill, C.; Stanton, C.; Ross, R.P. Actinomyces Produces Defensin-Like Bacteriocins (Actifensins) with a Highly Degenerate Structure and Broad Antimicrobial Activity. J. Bacteriol. 2020, 202, e00529-19. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, E.F.; Gardiner, G.E.; O’Connor, P.M.; Mills, S.; Ross, R.P.; Hill, C. Characterization of enterocin- and salivaricin-producing lactic acid bacteria from the mammalian gastrointestinal tract. FEMS Microbiol. Lett. 2009, 291, 24–34. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, L.; Teng, K.; Sun, S.; Sun, Z.; Zhong, J. Cerecidins, novel lantibiotics from Bacillus cereus with potent antimicrobial activity. Appl. Environ. Microbiol. 2014, 80, 2633–2643. (In English) [Google Scholar] [CrossRef]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef]
- Balo, A.R.; Tao, L.; Britt, R.D. Characterizing SPASM/twitch Domain-Containing Radical SAM Enzymes by EPR Spectroscopy. Appl. Magn. Reson. 2022, 53, 809–820. [Google Scholar] [CrossRef]
- Grell, T.A.; Kincannon, W.M.; Bruender, N.A.; Blaesi, E.J.; Krebs, C.; Bandarian, V.; Drennan, C.L. Structural and spectroscopic analyses of the sporulation killing factor biosynthetic enzyme SkfB, a bacterial AdoMet radical sactisynthase. J. Biol. Chem. 2018, 293, 17349–17361. [Google Scholar] [CrossRef]
- Booth, M.C.; Bogie, C.P.; Sahl, H.-G.; Siezen, R.J.; Hatter, K.L.; Gilmore, M.S. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol. Microbiol. 1996, 21, 1175–1184. [Google Scholar] [CrossRef]
- Lohans, C.T.; Li, J.L.; Vederas, J.C. Structure and Biosynthesis of Carnolysin, a Homologue of Enterococcal Cytolysin with d-Amino Acids. J. Am. Chem. Soc. 2014, 136, 13150–13153. [Google Scholar] [CrossRef] [PubMed]
- Gálvez, A.; Maqueda, M.; Martínez-Bueno, M.; Valdivia, E. Bactericidal and bacteriolytic action of peptide antibiotic AS-48 against gram-positive and gram-negative bacteria and other organisms. Res. Microbiol. 1989, 140, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, F.; Maqueda, M.; Gálvez, A.; Martínez-Bueno, M.; Valdivia, E. Antilisterial activity of peptide AS-48 and study of changes induced in the cell envelope properties of an AS-48-adapted strain of Listeria monocytogenes. Appl. Environ. Microbiol. 1999, 65, 618–625. (In English) [Google Scholar] [CrossRef]
- Grande Burgos, M.J.; Pulido, R.P.; Del Carmen López Aguayo, M.; Gálvez, A.; Lucas, R. The Cyclic Antibacterial Peptide Enterocin AS-48: Isolation, Mode of Action, and Possible Food Applications. Int. J. Mol. Sci. 2014, 15, 22706–22727. [Google Scholar] [CrossRef]
- Mu, F.; Masuda, Y.; Zendo, T.; Ono, H.; Kitagawa, H.; Ito, H.; Nakayama, J.; Sonomoto, K. Biological function of a DUF95 superfamily protein involved in the biosynthesis of a circular bacteriocin, leucocyclicin Q. J. Biosci. Bioeng. 2014, 117, 158–164. [Google Scholar] [CrossRef]
- Cebrián, R.; Maqueda, M.; Neira José, L.; Valdivia, E.; Martínez-Bueno, M.; Montalbán-López, M. Insights into the Functionality of the Putative Residues Involved in Enterocin AS-48 Maturation. Appl. Environ. Microbiol. 2010, 76, 7268–7276. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cebrián, R.; Martínez-Bueno, M.; Valdivia, E.; Albert, A.; Maqueda, M.; Sánchez-Barrena, M.J. The bacteriocin AS-48 requires dimer dissociation followed by hydrophobic interactions with the membrane for antibacterial activity. J. Struct. Biol. 2015, 190, 162–172. [Google Scholar] [CrossRef]
- Martínez, B.; Suárez, J.E.; Rodríguez, A. Lactococcin 972: A homodimeric lactococcal bacteriocin whose primary target is not the plasma membrane. Microbiology 1996, 142, 2393–2398. (In English) [Google Scholar] [CrossRef]
- Chikindas, M.L.; Novák, J.; Driessen, A.J.; Konings, W.N.; Schilling, K.M.; Caufield, P.W. Mutacin II, a bactericidal antibiotic from Streptococcus mutans. Antimicrob. Agents Chemother. 1995, 39, 2656–2660. (In English) [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Teng, K.; Wang, J.; Zhang, Z.; Zhang, J.; Sun, S.; Li, L.; Yang, X.; Zhong, J. CerR, a Single-Domain Regulatory Protein of the LuxR Family, Promotes Cerecidin Production and Immunity in Bacillus cereus. Appl. Environ. Microbiol. 2018, 84, e02245-17. [Google Scholar] [CrossRef]
- Ishikawa, J.; Chiba, K.; Kurita, H.; Satoh, H. Contribution of rpoB2 RNA polymerase beta subunit gene to rifampin resistance in Nocardia species. Antimicrob. Agents Chemother. 2006, 50, 1342–1346. (In English) [Google Scholar] [CrossRef] [PubMed]
- Talà, A.; Wang, G.; Zemanova, M.; Okamoto, S.; Ochi, K.; Alifano, P. Activation of Dormant Bacterial Genes by Nonomuraea sp. Strain ATCC 39727 Mutant-Type RNA Polymerase. J. Bacteriol. 2009, 191, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Yazawa, K.; Dabbs, E.R.; Nishikawa, K.; Komaki, H.; Mikami, Y.; Miyaji, M.; Morisaki, N.; Iwasaki, S. Different Rifampicin Inactivation Mechanisms in Nocardia and Related Taxa. Microbiol. Immunol. 1996, 40, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.P. Resistance to rifampicin: A review. J. Antibiot. 2014, 67, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Z.; Wang, Y.; Liao, M.; Rong, X.; Li, B.; Wang, K.; Chen, J.; Zhang, H. Complete Genome Sequence of Photobacterium damselae Subsp. damselae Strain SSPD1601 Isolated from Deep-Sea Cage-Cultured Sebastes schlegelii with Septic Skin Ulcer. Int. J. Genom. 2019, 4242653, 2019. [Google Scholar] [CrossRef]
- Chen, H.; Bai, X.; Jing, L.; Chen, R.; Teng, Y. Characterization of antibiotic resistance genes in the sediments of an urban river revealed by comparative metagenomics analysis. Sci. Total Environ. 2019, 653, 1513–1521. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Liu, P.; Sun, Y.; Dong, X.; Hu, X. Unveiling the occurrence, hosts and mobility potential of antibiotic resistance genes in the deep ocean. Sci. Total Environ. 2022, 816, 151539. [Google Scholar] [CrossRef]
- Hatosy, S.M.; Martiny, A.C. The Ocean as a Global Reservoir of Antibiotic Resistance Genes. Appl. Environ. Microbiol. 2015, 81, 7593–7599. [Google Scholar] [CrossRef]
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015. (In English) [Google Scholar]
- Rather, I.A.; Galope, R.; Bajpai, V.K.; Lim, J.; Paek, W.K.; Park, Y.H. Diversity of Marine Bacteria and Their Bacteriocins: Applications in Aquaculture. Rev. Fish. Sci. Aquac. 2017, 25, 257–269. (In English) [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. (In English) [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. (In English) [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. (In English) [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 2016, 44, 6614–6624. (In English) [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Agrawal, P.; Khater, S.; Gupta, M.; Sain, N.; Mohanty, D. RiPPMiner: A bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res 2017, 45, W80–W88. (In English) [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. (In English) [Google Scholar] [CrossRef]
- Harrison, K.J.; Crécy-Lagard, V.; Zallot, R. Gene Graphics: A genomic neighborhood data visualization web application. Bioinformatics 2018, 34, 1406–1408. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. (In English) [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017, 45, D566–D573. (In English) [Google Scholar] [CrossRef] [PubMed]
- Ingle, D.J.; Valcanis, M.; Kuzevski, A.; Tauschek, M.; Inouye, M.; Stinear, T.; Levine, M.M.; Robins-Browne, R.M.; Holt, K.E. In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genom. 2016, 2, e000064. (In English) [Google Scholar] [CrossRef] [PubMed]
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2020, 48, D561–D569. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, 11. (In English) [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014, 58, 3895–3903. (In English) [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. (In English) [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 2016, 44, D694–D697. (In English) [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).