Galdieria sulphuraria: An Extremophilic Alga as a Source of Antiviral Bioactive Compounds
Abstract
1. Introduction
2. Results
2.1. Chemical Characterization
2.2. Evaluation of Cytotoxicity Effects
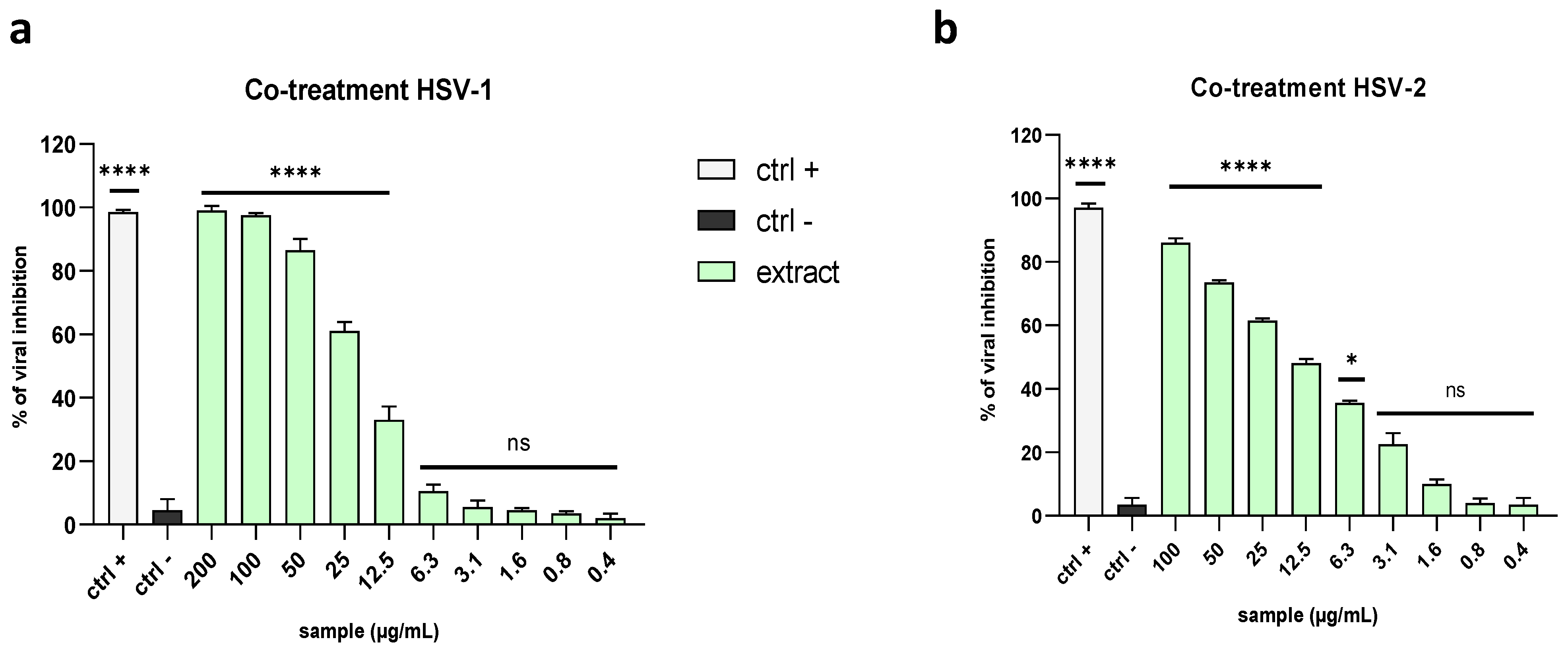
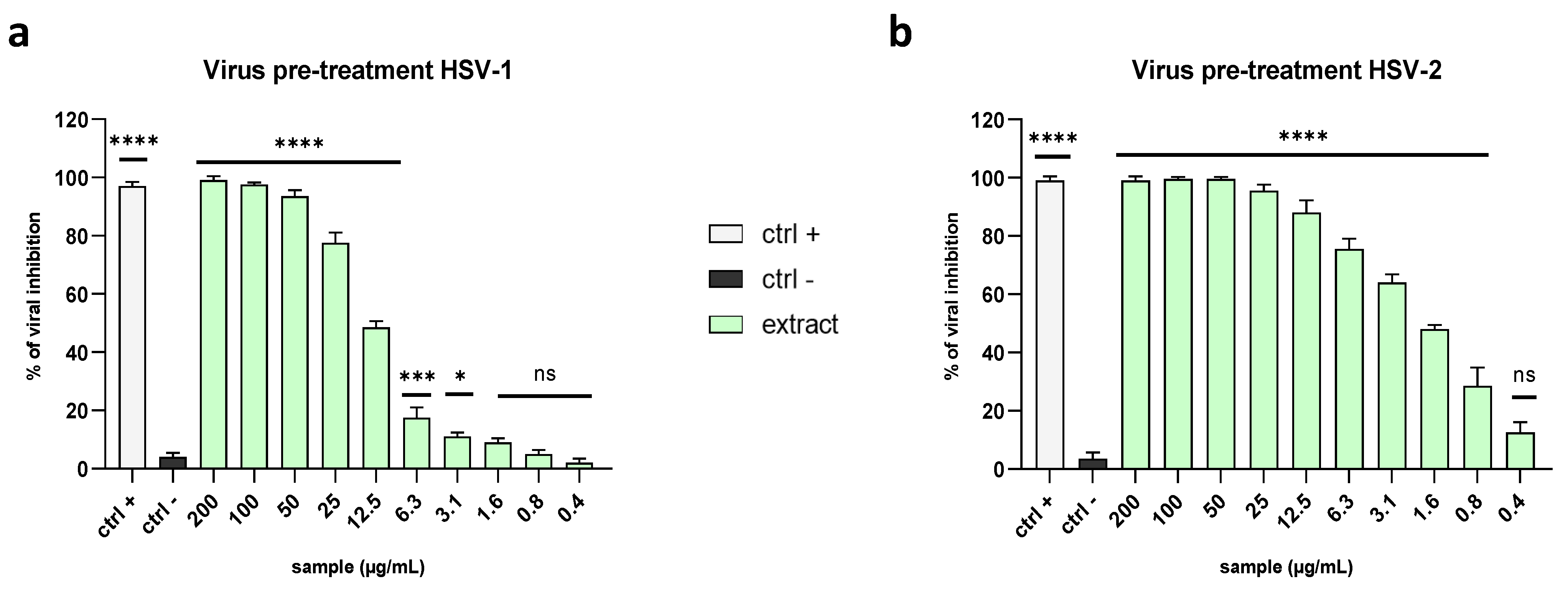
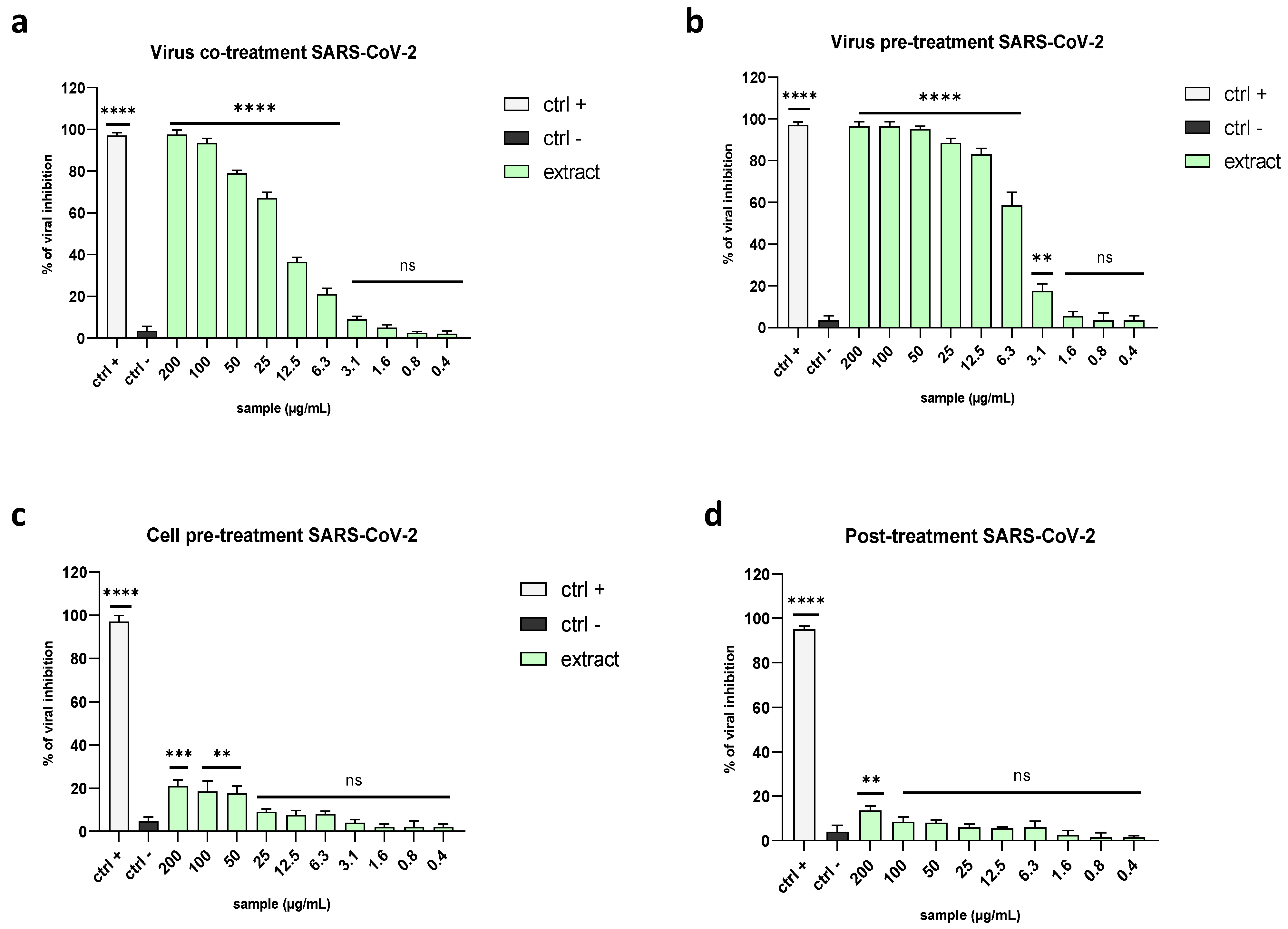
2.3. Evaluation of Antiviral Activity on Different Viruses
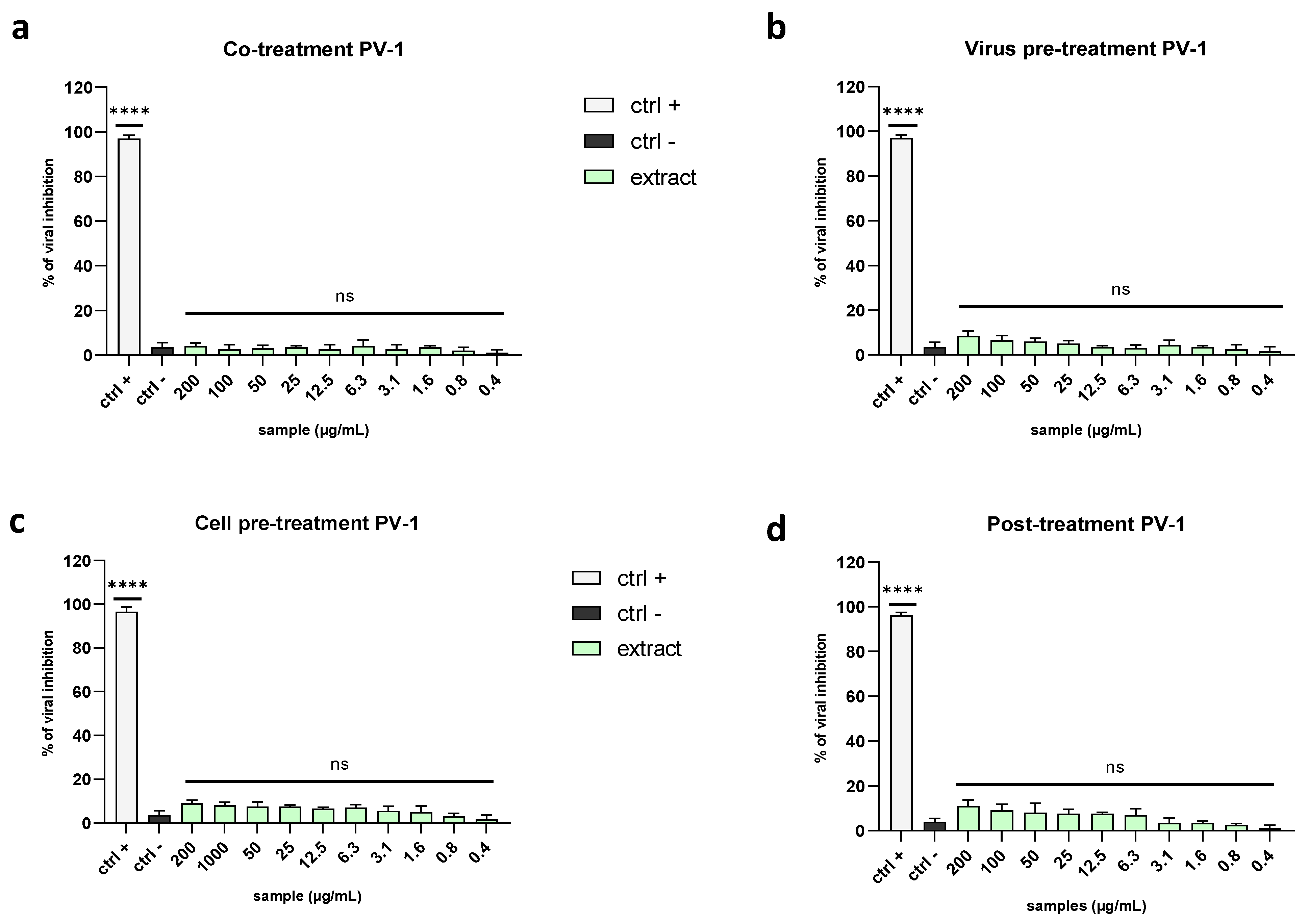
2.3.1. Enveloped Viruses
2.3.2. Non-Enveloped Viruses
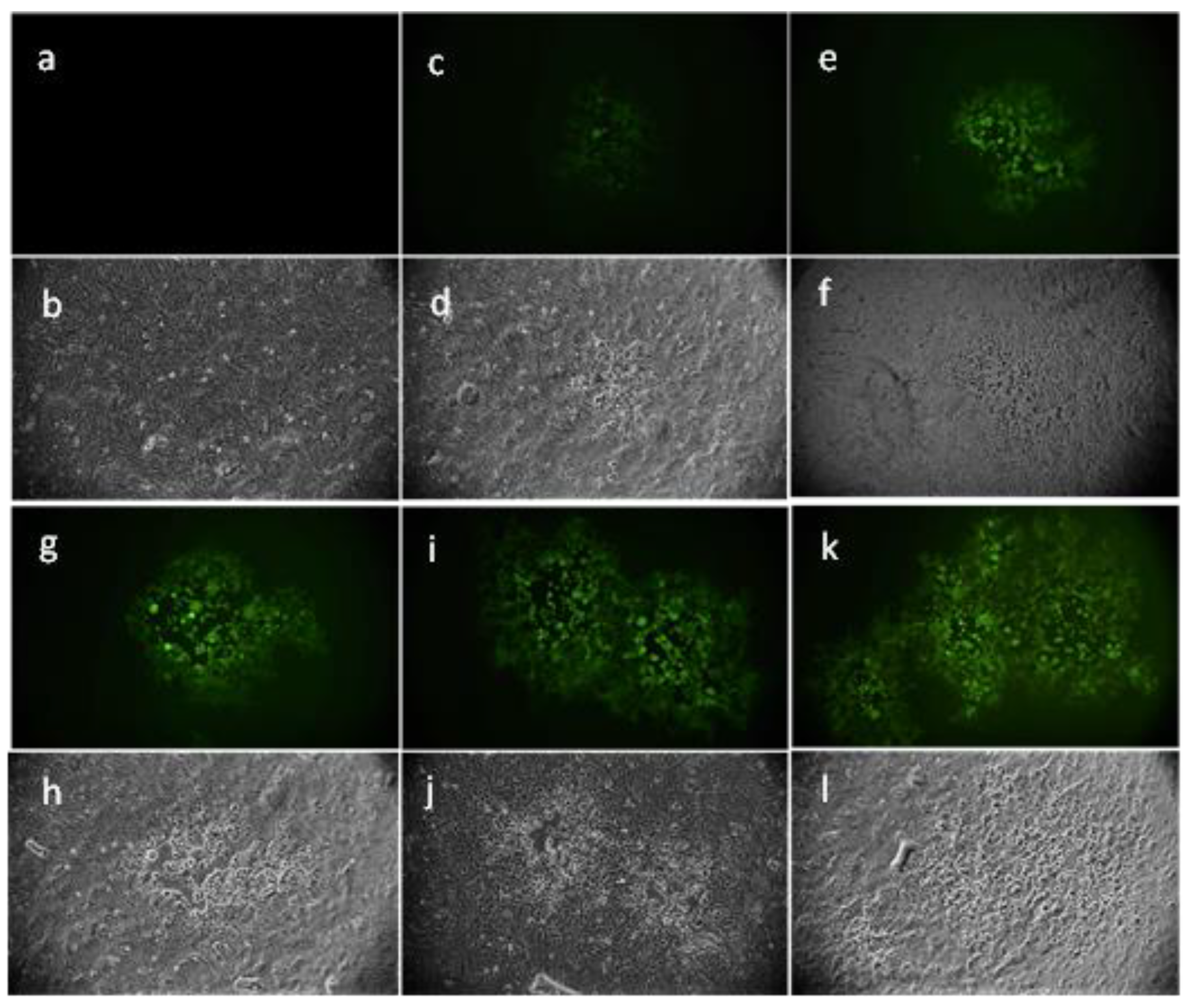
2.4. Evaluation of Viral Gene Expression
3. Discussion
4. Materials and Methods
4.1. Galdiera Sulphuraria Cultivation
4.2. Extract Preparation
4.3. Ultra-High Performance Liquid Chromatography–High-Resolution Mass Spectrometry Analyses
4.4. Cell Lines and Viral Strains
4.5. Cytotoxicity Assay
4.6. Antiviral Activity Assay
4.7. Fluorescence Microscopy
4.8. Gene Expression Analysis: Real-Time PCR
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tompa, D.R.; Immanuel, A.; Srikanth, S.; Kadhirvel, S. Trends and strategies to combat viral infections: A review on FDA approved antiviral drugs. Int. J. Biol. Macromol. 2021, 172, 524–541. [Google Scholar] [CrossRef] [PubMed]
- Carbone, D.A.; Pellone, P.; Lubritto, C.; Ciniglia, C. Evaluation of Microalgae Antiviral Activity and Their Bioactive Compounds. Antibiotics 2021, 10, 746. [Google Scholar] [CrossRef]
- Kim, S.K.; Vo, T.S.; Ngo, D.H. Potential application of marine algae as antiviral agents in medicinal foods. Adv. Food Nutr. Res. 2011, 64, 245–254. [Google Scholar] [PubMed]
- Raihan, T.; Rabbee, M.F.; Roy, P.; Choudhury, S.; Baek, K.H.; Azad, A.K. Microbial Metabolites: The Emerging Hotspot of Antiviral Compounds as Potential Candidates to Avert Viral Pandemic Alike COVID-19. Front. Mol. Biosci. 2021, 8, 732256. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.X.; Lee, Y.; Bae, M.; Hu, S.; Kang, H.; Kim, M.B.; Park, Y.K.; Lee, J.Y. Spirulina supplementation in a mouse model of diet-induced liver fibrosis reduced the pro-inflammatory response of splenocytes. Br. J. Nutr. 2019, 121, 748–755. [Google Scholar] [CrossRef]
- Samuels, R.; Mani, U.V.; Iyer, U.M.; Nayak, U.S. Hypocholesterolemic effect of spirulina in patients with hyperlipidemic nephrotic syndrome. J. Med. Food 2002, 5, 91–96. [Google Scholar] [CrossRef]
- Balasubramaniam, V.; Gunasegavan, R.D.; Mustar, S.; Lee, J.C.; Mohd Noh, M.F. Isolation of Industrial Important Bioactive Compounds from Microalgae. Molecules 2021, 26, 943. [Google Scholar] [CrossRef]
- Pulz, O.; Gross, W. Valuable products from biotechnology of microalgae. Appl. Microbiol. Biotechnol. 2004, 65, 635–648. [Google Scholar] [CrossRef]
- Lauritano, C.; Andersen, J.H.; Hansen, E.; Albrigtsen, M.; Escalera, L.; Esposito, F.; Helland, K.; Hanssen, K.Ø.; Romano, G.; Ianora, A. Bioactivity Screening of Microalgae for Antioxidant, Anti-Inflammatory, Anticancer, Anti-Diabetes, and Antibacterial Activities. Front. Mar. Sci. 2016, 3, 68. [Google Scholar] [CrossRef]
- Ku, C.S.; Pham, T.X.; Park, Y.; Kim, B.; Shin, M.S.; Kang, I.; Lee, J. Edible blue-green algae reduce the production of pro-inflammatory cytokines by inhibiting NF-κB pathway in macrophages and splenocytes. Biochim. Biophys. Acta 2013, 1830, 2981–2988. [Google Scholar] [CrossRef]
- Sami, N.; Ahmad, R.; Fatma, T. Exploring algae and cyanobacteria as a promising natural source of antiviral drug against SARS-CoV-2. Biomed. J. 2021, 44, 54–62. [Google Scholar] [CrossRef]
- Kasting, J.F.; Siefert, J.L. Life and the evolution of Earth’s atmosphere. Science 2002, 296, 1066–1068. [Google Scholar] [CrossRef]
- Kaushik, N.; Mitra, S.; Baek, E.J.; Nguyen, L.N.; Bhartiya, P.; Kim, J.H.; Choi, E.H.; Kaushik, N.K. The inactivation and destruction of viruses by reactive oxygen species generated through physical and cold atmospheric plasma techniques: Current status and perspectives. J. Adv. Res. 2023, 43, 59–71. [Google Scholar] [CrossRef]
- Suhail, S.; Zajac, J.; Fossum, C.; Lowater, H.; McCracken, C.; Severson, N.; Laatsch, B.; Narkiewicz-Jodko, A.; Johnson, B.; Liebau, J.; et al. Role of Oxidative Stress on SARS-CoV (SARS) and SARS-CoV-2 (COVID-19) Infection: A Review. Protein J. 2020, 39, 644–656. [Google Scholar] [CrossRef]
- Hayashi, K.; Asai, S.; Umezawa, K.; Kakizoe, H.; Miyachi, H.; Morita, M.; Akaike, T.; Kuno, H.; Komatsu, S.; Watanabe, T.; et al. Virucidal effect of monogalactosyl diacylglyceride from a green microalga, Coccomyxa sp. KJ, against clinical isolates of SARS-CoV-2 as assessed by a plaque assay. J. Clin. Lab. Anal. 2022, 36, e24146. [Google Scholar] [CrossRef]
- Hayashi, K.; Lee, J.B.; Atsumi, K.; Kanazashi, M.; Shibayama, T.; Okamoto, K.; Kawahara, T.; Hayashi, T. In vitro and in vivo anti-herpes simplex virus activity of monogalactosyl diacylglyceride from Coccomyxa sp. KJ (IPOD FERM BP-22254), a green microalga. PLoS ONE 2019, 14, e0219305. [Google Scholar] [CrossRef]
- Tang, L.; Qiu, L.; Liu, C.; Du, G.; Mo, Z.; Tang, X.; Mao, Y. Transcriptomic Insights into Innate Immunity Responding to Red Rot Disease in Red Alga Pyropia yezoensis. Int. J. Mol. Sci. 2019, 20, 5970. [Google Scholar] [CrossRef]
- Botos, I.; Wlodawer, A. Cyanovirin-N: A sugar-binding antiviral protein with a new twist. Cell. Mol. Life Sci. 2003, 60, 277–287. [Google Scholar] [CrossRef]
- Gustafson, K.R.; Sowder, R.C.; Henderson, L.E.; Cardellina, J.H.; McMahon, J.B.; Rajamani, U.; Pannell, L.K.; Boyd, M.R. Isolation, primary sequence determination, and disulfide bond structure of cyanovirin-N, an anti-HIV (human immunodeficiency virus) protein from the cyanobacterium Nostoc ellipsosporum. Biochem. Biophys. Res. Commun. 1997, 238, 223–228. [Google Scholar] [CrossRef]
- Bouslama, L.; Hayashi, K.; Lee, J.B.; Ghorbel, A.; Hayashi, T. Potent virucidal effect of pheophorbide a and pyropheophorbide a on enveloped viruses. J. Nat. Med. 2011, 65, 229–233. [Google Scholar] [CrossRef]
- Shih, S.R.; Tsai, K.N.; Li, Y.S.; Chueh, C.C.; Chan, E.C. Inhibition of enterovirus 71-induced apoptosis by allophycocyanin isolated from a blue-green alga Spirulina platensis. J. Med. Virol. 2003, 70, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Zainuddin, E.; Mundt, S.; Wegner, U.; Mentel, R. Cyanobacteria a potential source of antiviral substances against influenza virus. Med. Microbiol. Immunol. 2002, 191, 181–182. [Google Scholar]
- Li, H.C.; Yang, C.H.; Lo, S.Y. Hepatitis C Viral Replication Complex. Viruses 2021, 13, 520. [Google Scholar] [CrossRef] [PubMed]
- Stadnichuk, I.N.; Rakhimberdieva, M.G.; Bolychevtseva, Y.V.; Yurina, N.P.; Karapetyan, N.V.; Selyakh, I.O. Inhibition by glucose of chlorophyll and phycocyanobilin biosynthesis in the unicellular red alga Galdieria partita at the stage of coproporphyrinogen III formation. Plant Sci. 1998, 136, 11–23. [Google Scholar] [CrossRef]
- Xia, M.; Fu, D.; Chakraborty, R.; Singh, R.P.; Terry, N. Enhanced crude oil depletion by constructed bacterial consortium comprising bioemulsifier producer and petroleum hydrocarbon degraders. Bioresour. Technol. 2019, 282, 456–463. [Google Scholar] [CrossRef]
- Montes D’Oca, M.G.; Viêgas, C.V.; Lemões, J.S.; Miyasaki, E.K.; Morón-Villarreyes, J.A.; Primel, E.G.; Abreu, P.C. Production of FAMEs from several microalgal lipidic extracts and direct transesterification of the Chlorella pyrenoidosa. Biomass Bioenergy 2011, 35, 1533–1538. [Google Scholar] [CrossRef]
- Lopez, Y.; Soto, S.M. The Usefulness of Microalgae Compounds for Preventing Biofilm Infections. Antibiotics 2019, 9, 9. [Google Scholar] [CrossRef]
- Fernandes, A.S.; Petry, F.C.; Mercadante, A.Z.; Jacob-Lopes, E.; Zepka, L.Q. HPLC-PDA-MS/MS as a strategy to characterize and quantify natural pigments from microalgae. Curr. Res. Food Sci. 2020, 3, 100–112. [Google Scholar] [CrossRef]
- Ahmad, I.; Wilson, D.W. HSV-1 Cytoplasmic Envelopment and Egress. Int. J. Mol. Sci. 2020, 21, 5969. [Google Scholar] [CrossRef]
- Payne, S. Chapter 34—Family Herpesviridae. In Viruses; Payne, S., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 269–278. [Google Scholar]
- Giugliano, R.; Buonocore, C.; Zannella, C.; Chianese, A.; Palma Esposito, F.; Tedesco, P.; De Filippis, A.; Galdiero, M.; Franci, G.; de Pascale, D. Antiviral Activity of the Rhamnolipids Mixture from the Antarctic Bacterium Pseudomonas gessardii M15 against Herpes Simplex Viruses and Coronaviruses. Pharmaceutics 2021, 13, 2121. [Google Scholar] [CrossRef]
- Taylor, E.W. RNA Viruses vs. DNA Synthesis: A General Viral Strategy That May Contribute to the Protective Antiviral Effects of Selenium. Preprints 2020, 2020060069. [Google Scholar] [CrossRef]
- Fontaine-Rodriguez, E.C.; Knipe, D.M. Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J. Virol. 2008, 82, 3538–3545. [Google Scholar] [CrossRef]
- Weller, S.K.; Coen, D.M. Herpes simplex viruses: Mechanisms of DNA replication. Cold Spring Harb. Perspect. Biol. 2012, 4, a013011. [Google Scholar] [CrossRef]
- Baquero, E.; Albertini, A.A.V.; Gaudin, Y. Recent mechanistic and structural insights on class III viral fusion glycoproteins. Curr. Opin. Struct. Biol. 2015, 33, 52–60. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, J.; Nguyen, L.N.T.; Adkins, J.L.; Schank, M.; Khanal, S.; Nguyen, L.N.; Dang, X.; Cao, D.; Thakuri, B.K.C.; et al. Blockade of SARS-CoV-2 spike protein-mediated cell–cell fusion using COVID-19 convalescent plasma. Sci. Rep. 2021, 11, 5558. [Google Scholar] [CrossRef]
- Bai, Z.; Cao, Y.; Liu, W.; Li, J. The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses 2021, 13, 1115. [Google Scholar] [CrossRef]
- Monda, V.; Valenzano, A.; Moscatelli, F.; Messina, A.; Piombino, L.; Zannella, C.; Viggiano, E.; Monda, G.; De Luca, V.; Chieffi, S.; et al. Modifications of Activity of Autonomic Nervous System, and Resting Energy Expenditure in Women Using Hormone-Replacement Therapy. Biol. Med. (Aligarth) 2016, 8, 5. [Google Scholar]
- Stelitano, D.; Franci, G.; Chianese, A.; Galdiero, S.; Morelli, G.; Galdiero, M. HSV Membrane Glycoproteins, Their Function in Viral Entry and Their Use in Vaccine Studies, Amino Acids, Peptides and Proteins; Royal Society of Chemistry: London, UK, 2019; pp. 14–43. [Google Scholar]
- Selvaratnam, T.; Pegallapati, A.K.; Montelya, F.; Rodriguez, G.; Nirmalakhandan, N.; Van Voorhies, W.; Lammers, P.J. Evaluation of a thermo-tolerant acidophilic alga, Galdieria sulphuraria, for nutrient removal from urban wastewaters. Bioresour. Technol. 2014, 156, 395–399. [Google Scholar] [CrossRef]
- Sun, Y.; Shi, M.; Lu, T.; Ding, D.; Sun, Y.; Yuan, Y. Bio-removal of PtCl(6)(2-) complex by Galdieria sulphuraria. Sci. Total Environ. 2021, 796, 149021. [Google Scholar] [CrossRef]
- Scherhag, P.; Ackermann, J.U. Removal of sugars in wastewater from food production through heterotrophic growth of Galdieria sulphuraria. Eng. Life Sci. 2021, 21, 233–241. [Google Scholar] [CrossRef]
- Ju, X.; Igarashi, K.; Miyashita, S.; Mitsuhashi, H.; Inagaki, K.; Fujii, S.; Sawada, H.; Kuwabara, T.; Minoda, A. Effective and selective recovery of gold and palladium ions from metal wastewater using a sulfothermophilic red alga, Galdieria sulphuraria. Bioresour. Technol. 2016, 211, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Mallick, K.; Henkanatte Gedara, S.M.; Jarvis, J.M.; Schaub, T.; Jena, U.; Nirmalakhandan, N.; Brewer, C.E. Hydrothermal liquefaction of Galdieria sulphuraria grown on municipal wastewater. Bioresour. Technol. 2019, 292, 121884. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Cui, Z.; Mallick, K.; Nirmalakhandan, N.; Brewer, C.E. Hydrothermal liquefaction of high- and low-lipid algae: Mass and energy balances. Bioresour. Technol. 2018, 258, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Sanchez-Arcos, C.; Pohnert, G.; Wei, D. Untargeted Metabolomics Unveil Changes in Autotrophic and Mixotrophic Galdieria sulphuraria Exposed to High-Light Intensity. Int. J. Mol. Sci. 2021, 22, 1247. [Google Scholar] [CrossRef]
- Serive, B.; Nicolau, E.; Berard, J.B.; Kaas, R.; Pasquet, V.; Picot, L.; Cadoret, J.P. Community analysis of pigment patterns from 37 microalgae strains reveals new carotenoids and porphyrins characteristic of distinct strains and taxonomic groups. PLoS ONE 2017, 12, e0171872. [Google Scholar] [CrossRef]
- Getachew, P.; Getachew, M.; Joo, J.; Choi, Y.S.; Hwang, D.S.; Hong, Y.K. The slip agents oleamide and erucamide reduce biofouling by marine benthic organisms (diatoms, biofilms and abalones). Toxicol. Environ. Health Sci. 2016, 8, 341–348. [Google Scholar] [CrossRef]
- Çelik, P.A.; Manga, E.B.; Çabuk, A.; Banat, I.M. Biosurfactants’ Potential Role in Combating COVID-19 and Similar Future Microbial Threats. Appl. Sci. 2021, 11, 334. [Google Scholar] [CrossRef]
- Saide, A.; Lauritano, C.; Ianora, A. Pheophorbide a: State of the Art. Mar. Drugs 2020, 18, 257. [Google Scholar] [CrossRef]
- Meunier, T.; Desmarets, L.; Bordage, S.; Bamba, M.; Hervouet, K.; Rouille, Y.; Francois, N.; Decossas, M.; Sencio, V.; Trottein, F.; et al. A Photoactivable Natural Product with Broad Antiviral Activity against Enveloped Viruses, Including Highly Pathogenic Coronaviruses. Antimicrob. Agents Chemother. 2022, 66, e0158121. [Google Scholar] [CrossRef]
- Jimenez-Aleman, G.H.; Castro, V.; Londaitsbehere, A.; Gutierrez-Rodriguez, M.; Garaigorta, U.; Solano, R.; Gastaminza, P. SARS-CoV-2 Fears Green: The Chlorophyll Catabolite Pheophorbide A Is a Potent Antiviral. Pharmaceuticals 2021, 14, 1048. [Google Scholar] [CrossRef]
- Haarr, L.; Skulstad, S. The herpes simplex virus type 1 particle: Structure and molecular functions. Review article. APMIS 1994, 102, 321–346. [Google Scholar] [CrossRef]
- Johnston, C.; Gottlieb, S.L.; Wald, A. Status of vaccine research and development of vaccines for herpes simplex virus. Vaccine 2016, 34, 2948–2952. [Google Scholar] [CrossRef]
- Sanchez-Leon, E.; Bello-Morales, R.; Lopez-Guerrero, J.A.; Poveda, A.; Jimenez-Barbero, J.; Girones, N.; Abrusci, C. Isolation and characterization of an exopolymer produced by Bacillus licheniformis: In vitro antiviral activity against enveloped viruses. Carbohydr. Polym. 2020, 248, 116737. [Google Scholar] [CrossRef]
- Lamers, S.L.; Newman, R.M.; Laeyendecker, O.; Tobian, A.A.R.; Colgrove, R.C.; Ray, S.C.; Koelle, D.M.; Cohen, J.; Knipe, D.M.; Quinn, T.C. Global Diversity within and between Human Herpesvirus 1 and 2 Glycoproteins. J. Virol. 2015, 89, 8206–8218. [Google Scholar] [CrossRef]
- Wang, B.; Liu, Q.; Huang, Y.; Yuan, Y.; Ma, Q.; Du, M.; Cai, T.; Cai, Y. Extraction of Polysaccharide from Spirulina and Evaluation of Its Activities. Evid. Based Complement. Altern. Med. 2018, 2018, 3425615. [Google Scholar] [CrossRef]
- Brignati, M.J.; Loomis, J.S.; Wills, J.W.; Courtney, R.J. Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J. Virol. 2003, 77, 4888–4898. [Google Scholar] [CrossRef]
- Chianese, A.; Zannella, C.; Monti, A.; De Filippis, A.; Doti, N.; Franci, G.; Galdiero, M. The Broad-Spectrum Antiviral Potential of the Amphibian Peptide AR-23. Int. J. Mol. Sci. 2022, 23, 883. [Google Scholar] [CrossRef]




| Rt (min) | Tentative Identification | Molecular Formula | m/z Found | m/z Calc. | Error (ppm) | RDB |
|---|---|---|---|---|---|---|
| 17.934 | Octadecadienamide | C18H33NO | 280.2631 (+) | 280.2635 | −1.4 | 3 |
| 18.501 | Hydroxyoctadecadienamide | C18H33NO2 | 296.2579 (+) | 296.2584 | −1.7 | 3 |
| 22.076 | Palmitamide | C16H33NO | 256.2629 (+) | 256.2635 | −2.3 | 1 |
| 22.500 | Octadecenamide 1 | C18H35NO | 282.2783 (+) | 282.2791 | −2.8 | 2 |
| 22.831 | Octadecenamide 2 | C18H35NO | 282.2785 (+) | 282.2791 | −2.3 | 2 |
| 23.052 | Palmitic acid | C16H32O2 | 255.2337 (−) | 255.2330 | +2.9 | 1 |
| 23.308 | Pheophorbide a | C35H36N4O5 | 593.2731 (+) | 593.2758 | −4.6 | 20 |
| 24.200 | Stearamide | C18H37NO | 284.2939 (+) | 284.2948 | −3.1 | 1 |
| 29.638 | Docosatetraenoylethanolamine | C25H43NO2 | 390.3349 (+) | 390.3367 | −4.5 | 5 |
| 29.646 | Tetracosanoylethanolamine | C26H45NO2 | 404.3507 (+) | 404.3523 | −4.0 | 5 |
| 30.259 | hydroxypheophytin a | C55H74N4O6 | 887.5637 (+) | 887.5681 | −4.9 | 21 |
| Gene | Virus | Forward Sequence | Reverse Sequence |
|---|---|---|---|
|
UL54 UL52 UL27 | HSV-2 | TGGCGGACATTAAGGACATTG GACCGACGGGTGCGTTATT GCCTTCTTCGCCTTTCGC | TGGCCGTCAACTCGCAG GAAGGAGTCGCCATTTAGCC CGCTCGTGCCCTTCTTCTT |
|
S RdRp | HCoV-229E | CGTTGAACTTCAAACCTCAGA | ACCAACATTGGCATAAACAG |
|
S N | SARS-CoV-2 | AGGTTGATCACAGGCAGACT GGGGAACTTCTCCTGCTAGAAT | GCTGACTGAGGGAAGGAC CAGACATTTTGCTCTCAAGCTG |
| GAPDH | / | CCTTTCATTGAGCTCCAT | CGTACATGGGAGCGTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ambrosino, A.; Chianese, A.; Zannella, C.; Piccolella, S.; Pacifico, S.; Giugliano, R.; Franci, G.; De Natale, A.; Pollio, A.; Pinto, G.; et al. Galdieria sulphuraria: An Extremophilic Alga as a Source of Antiviral Bioactive Compounds. Mar. Drugs 2023, 21, 383. https://doi.org/10.3390/md21070383
Ambrosino A, Chianese A, Zannella C, Piccolella S, Pacifico S, Giugliano R, Franci G, De Natale A, Pollio A, Pinto G, et al. Galdieria sulphuraria: An Extremophilic Alga as a Source of Antiviral Bioactive Compounds. Marine Drugs. 2023; 21(7):383. https://doi.org/10.3390/md21070383
Chicago/Turabian StyleAmbrosino, Annalisa, Annalisa Chianese, Carla Zannella, Simona Piccolella, Severina Pacifico, Rosa Giugliano, Gianluigi Franci, Antonino De Natale, Antonino Pollio, Gabriele Pinto, and et al. 2023. "Galdieria sulphuraria: An Extremophilic Alga as a Source of Antiviral Bioactive Compounds" Marine Drugs 21, no. 7: 383. https://doi.org/10.3390/md21070383
APA StyleAmbrosino, A., Chianese, A., Zannella, C., Piccolella, S., Pacifico, S., Giugliano, R., Franci, G., De Natale, A., Pollio, A., Pinto, G., De Filippis, A., & Galdiero, M. (2023). Galdieria sulphuraria: An Extremophilic Alga as a Source of Antiviral Bioactive Compounds. Marine Drugs, 21(7), 383. https://doi.org/10.3390/md21070383











