Citrinin Is a Potential Quorum Sensing Inhibitor against Pseudomonas aeruginosa
Abstract
1. Introduction
2. Results
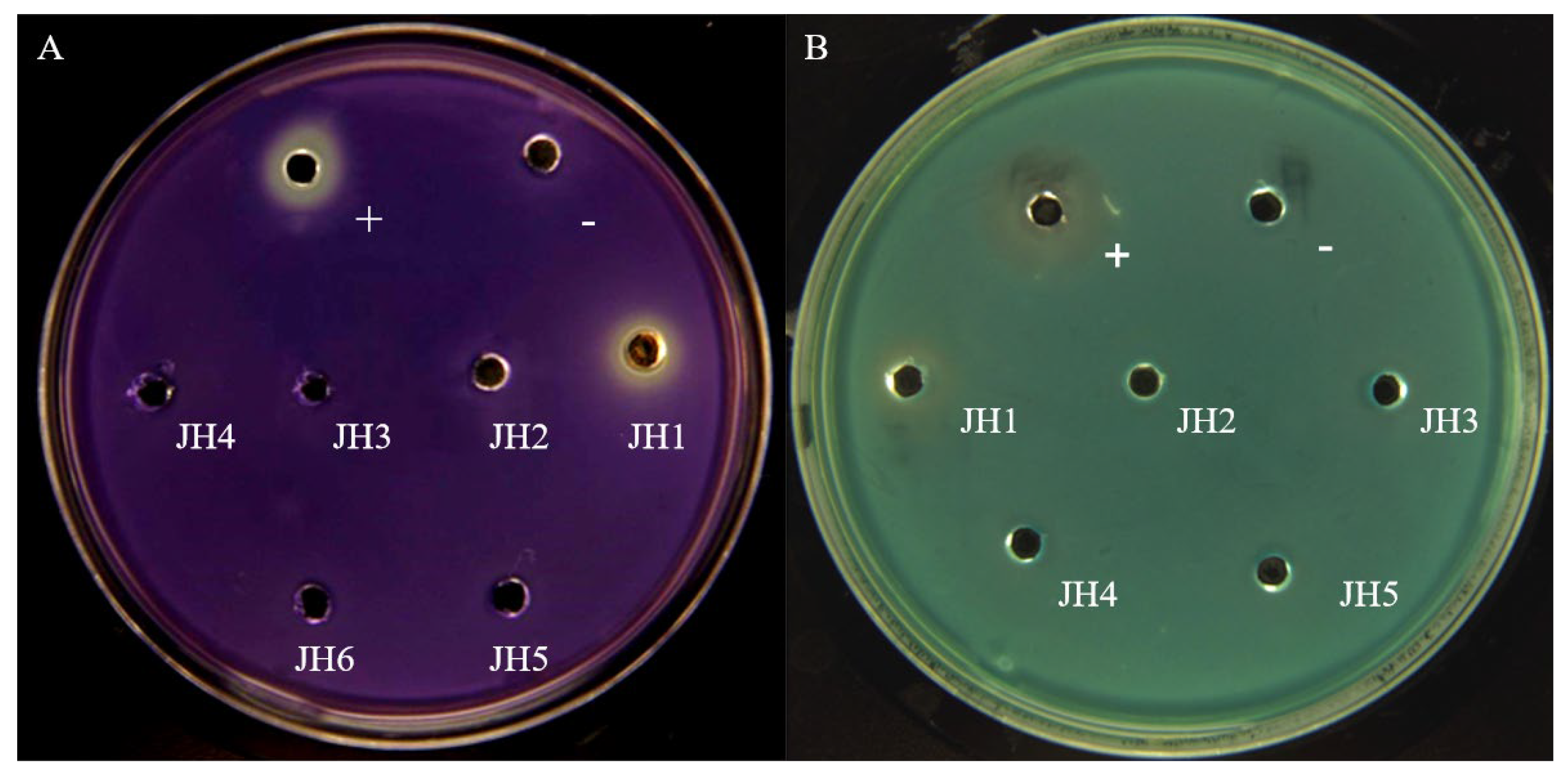
2.1. Screening and Identification of Active Compounds
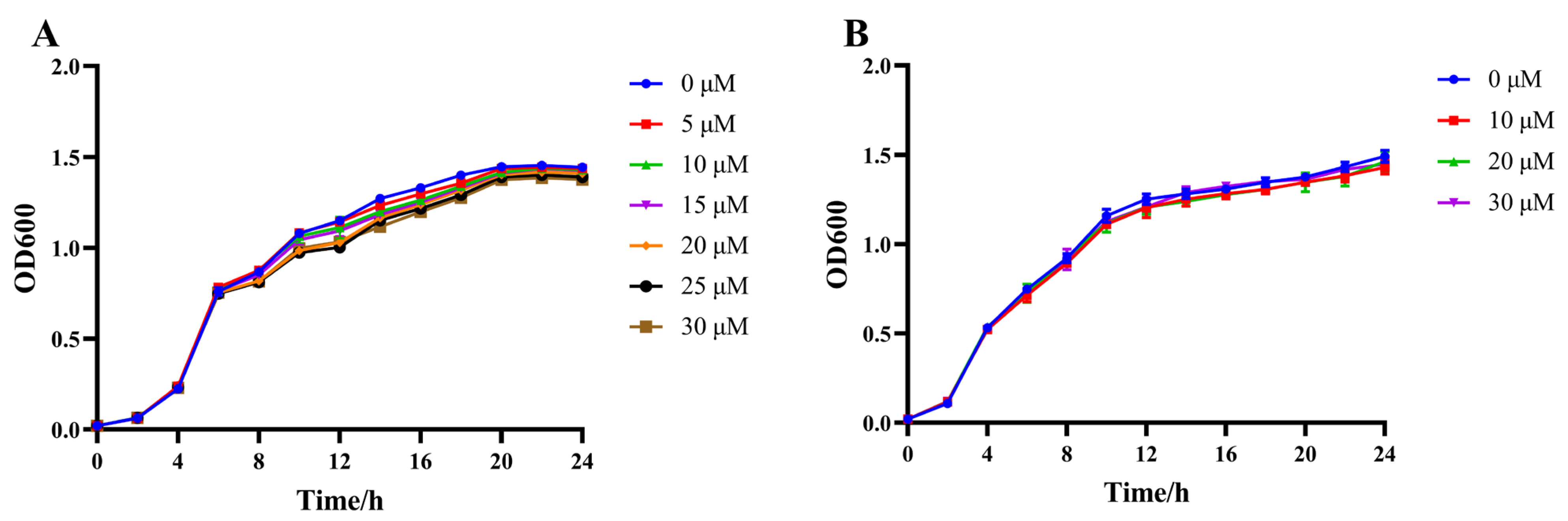
2.2. Effect of Citrinin on Bacterial Growth
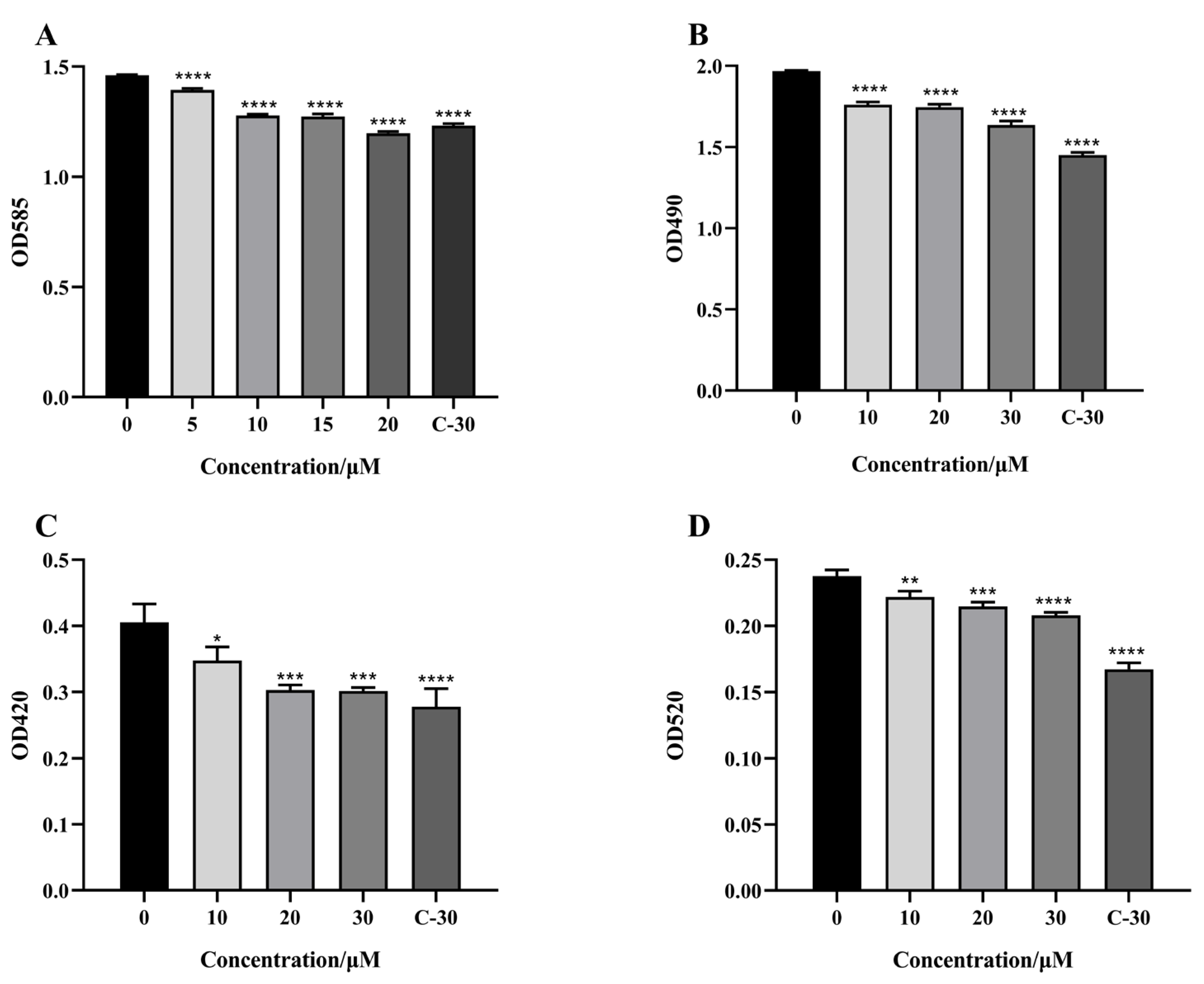
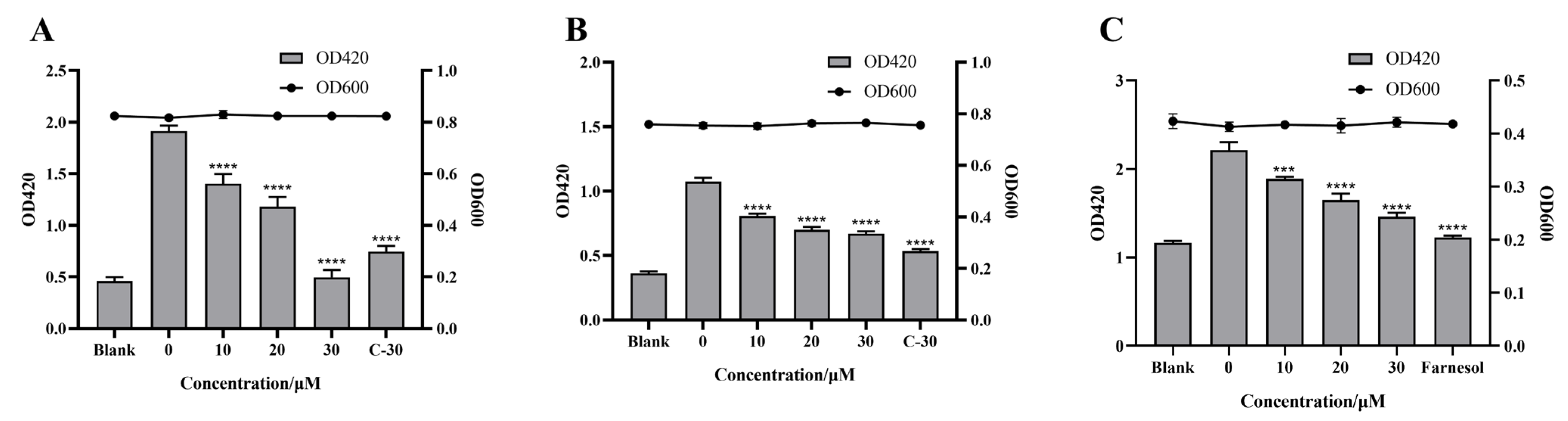
2.3. Effect of Citrinin on Virulence Factors
2.4. Effect of Citrinin on the Motility of P. aeruginosa
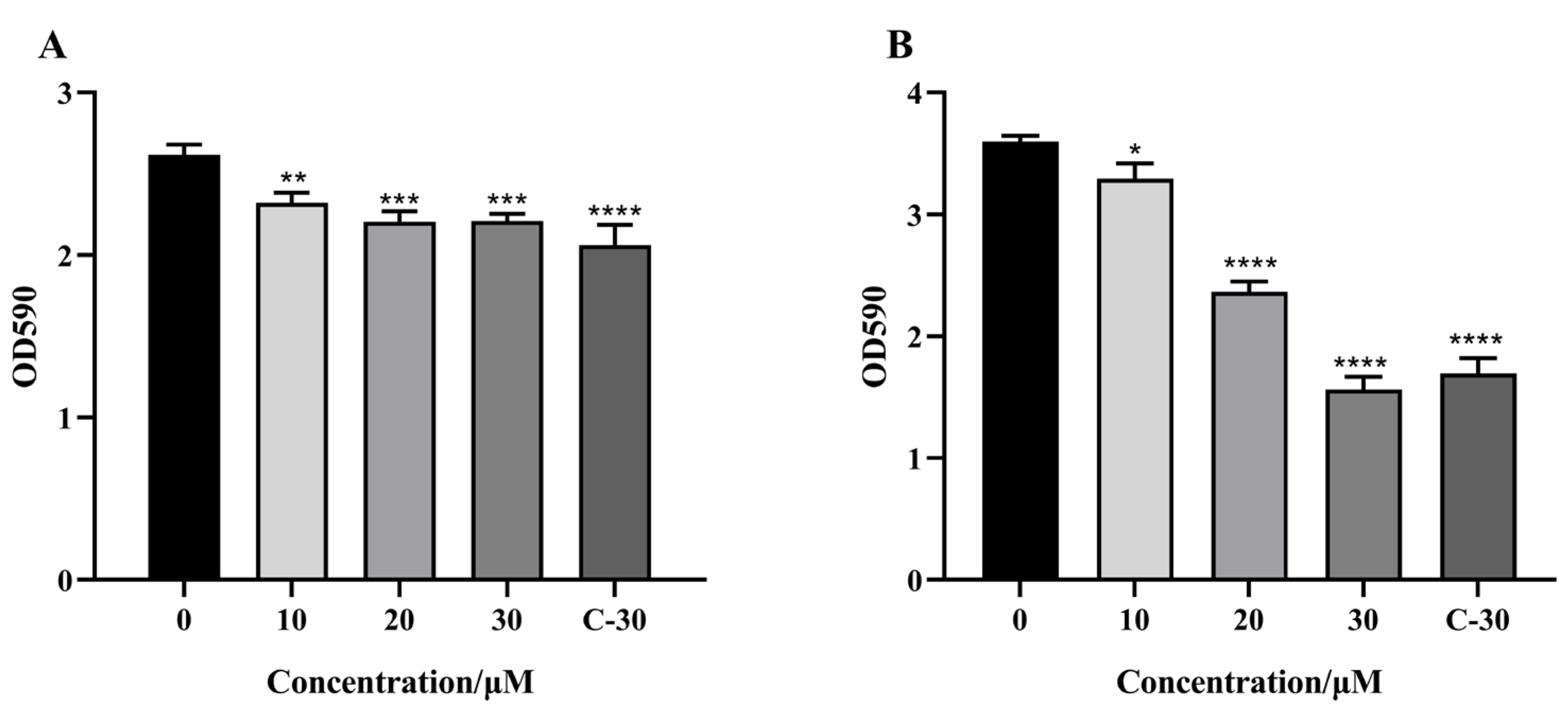
2.5. Effect of Citrinin on the Biofilm Formation of P. aeruginosa
2.6. Effect of Citrinin on the Promoter Activity of the Quorum Sensing System
2.7. Effect of Citrinin on the mRNA Transcription Level of Quorum Sensing-Related Genes
2.8. Molecular Docking Analysis
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Isolation of Marine Fungi and Preparation of the Crude Extracts
4.3. Screening for Quorum Sensing Inhibitors
4.4. Identification of Fungal Species
4.5. Purification Process of Quorum Sensing Inhibitors
4.6. Growth Curve Measurement
4.7. Effect of Citrinin on Virulence Factors
4.7.1. Elastase Activity Assay
4.7.2. Pyocyanin Assay
4.7.3. Rhamnolipid Assay
4.8. Motility Assay
4.9. Biofilm Assay
4.10. β-Galactosidase Assay
4.11. Real-Time RT-PCR
4.12. Molecular Docking
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahator, S.D.; Zhang, L. Small is mighty-chemical communication systems in Pseudomonas aeruginosa. Annu. Rev. Microbiol. 2019, 73, 559–578. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Parkins, M.D.; Somayaji, R.; Waters, V.J. Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin. Microbiol. Rev. 2018, 31, e00019-18. [Google Scholar] [CrossRef]
- Thi, M.T.; Wibowo, D.; Rehm, B.H. Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Gallagher, L.A.; McKnight, S.L.; Kuznetsova, M.S.; Pesci, E.C.; Manoil, C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 2002, 184, 6472–6480. [Google Scholar] [CrossRef] [PubMed]
- Schauder, S.; Bassler, B.L. The languages of bacteria. Genes Dev. 2001, 15, 1468–1480. [Google Scholar] [CrossRef]
- Pearson, J.P.; Gray, K.M.; Passador, L.; Tucker, K.D.; Eberhard, A.; Iglewski, B.H.; Greenberg, E.P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 1994, 91, 197–201. [Google Scholar] [CrossRef]
- Pearson, J.P.; Passador, L.; Iglewski, B.H.; Greenberg, E.P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 1490–1494. [Google Scholar] [CrossRef]
- Coleman, J.P.; Hudson, L.L.; McKnight, S.L.; Farrow, J.M.; Calfee, M.W.; Lindsey, C.A.; Pesci, E.C. Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme a ligase. J. Bacteriol. 2008, 190, 1247–1255. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Bonsall, R.F.; Delaney, S.M.; Soule, M.J.; Phillips, G.; Thomashow, L.S. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001, 183, 6454–6465. [Google Scholar] [CrossRef]
- Shih, P.C.; Huang, C.T. Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J. Antimicrob. Chemother. 2002, 49, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.E.; Caiazza, N.C.; O’Toole, G.A. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2003, 185, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Moustafa, D.A.; Stergioula, V.; Smith, C.D.; Goldberg, J.B.; Bassler, B.L. The PqsE and RhlR proteins are an autoinducer synthase-receptor pair that control virulence and biofilm development in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2018, 115, E9411–E9418. [Google Scholar] [CrossRef] [PubMed]
- Chanda, W.; Joseph, T.P.; Padhiar, A.A.; Guo, X.; Min, L.; Wang, W.; Lolokote, S.; Ning, A.; Cao, J.; Huang, M.; et al. Combined effect of linolenic acid and tobramycin on Pseudomonas aeruginosa biofilm formation and quorum sensing. Exp. Ther. Med. 2017, 14, 4328–4338. [Google Scholar] [CrossRef]
- Fong, J.; Mortensen, K.T.; Nørskov, A.; Qvortrup, K.; Yang, L.; Tan, C.H.; Nielsen, T.E.; Givskov, M. Itaconimides as novel quorum sensing inhibitors of Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2019, 8, 443. [Google Scholar] [CrossRef]
- Cheng, W.J.; Zhou, J.W.; Zhang, P.P.; Luo, H.Z.; Tang, S.; Li, J.J.; Deng, S.M.; Jia, A.Q. Quorum sensing inhibition and tobramycin acceleration in Chromobacterium violaceum by two natural cinnamic acid derivatives. Appl. Microbiol. Biotechnol. 2020, 104, 5025–5037. [Google Scholar] [CrossRef]
- Murray, E.J.; Dubern, J.F.; Chan, W.C.; Chhabra, S.R.; Williams, P. A Pseudomonas aeruginosa PQS quorum-sensing system inhibitor with anti-staphylococcal activity sensitizes polymicrobial biofilms to tobramycin. Cell Chem. Biol. 2022, 29, 1187–1199. [Google Scholar] [CrossRef]
- Kalia, V.C.; Patel, S.K.; Kang, Y.C.; Lee, J.K. Quorum sensing inhibitors as antipathogens: Biotechnological applications. Biotechnol. Adv. 2019, 37, 68–90. [Google Scholar] [CrossRef]
- Certner, R.H.; Vollmer, S.V. Inhibiting bacterial quorum sensing arrests coral disease development and disease-associated microbes. Environ. Microbiol. 2018, 20, 645–657. [Google Scholar] [CrossRef]
- Ding, T.; Li, T.; Wang, Z.; Li, J. Curcumin liposomes interfere with quorum sensing system of Aeromonas sobria and in silico analysis. Sci. Rep. 2017, 7, 8612. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, L.; Wu, H.; Zhao, C.; Gong, Q.; Yu, W. Cladodionen is a potential quorum sensing inhibitor against Pseudomonas aeruginosa. Mar. Drugs 2020, 18, 205. [Google Scholar] [CrossRef] [PubMed]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, M.; Zhu, X.; Yu, W.; Gong, Q. Equisetin as potential quorum sensing inhibitor of Pseudomonas aeruginosa. Biotechnol. Lett. 2018, 40, 865–870. [Google Scholar] [CrossRef]
- Fux, C.A.; Costerton, J.W.; Stewart, P.S.; Stoodley, P. Survival strategies of infectious biofilms. Trends Microbiol. 2005, 13, 34–40. [Google Scholar] [CrossRef]
- Zhao, C.; Zheng, H.; Zhou, L.; Ji, H.; Zhao, L.; Yu, W.; Gong, Q. Falcarindiol isolated from Notopterygium incisum inhibits the quorum sensing of Pseudomonas aeruginosa. Molecules 2021, 26, 5896. [Google Scholar] [CrossRef] [PubMed]
- Cugini, C.; Calfee, M.W.; Farrow, J.M.; Morales, D.K.; Pesci, E.C.; Hogan, D.A. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol. Microbiol. 2007, 65, 896–906. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Rudden, M.; Elias, S.M.; Smyth, T.J.; Marchant, R.; Banat, I.M.; Dooley, J.S. Pseudomonas aeruginosa PA80 is a cystic fibrosis isolate deficient in RhIRI quorum sensing. Sci. Rep. 2021, 11, 5729. [Google Scholar] [CrossRef] [PubMed]
- Mould, D.L.; Botelho, N.J.; Hogan, D.A. Intraspecies signaling between common variants of Pseudomonas aeruginosa increases production of quorum-sensing-controlled virulence factors. MBio 2020, 11, e01865-20. [Google Scholar] [CrossRef]
- He, Y.; Cox, R.J. The molecular steps of citrinin biosynthesis in fungi. Chem. Sci. 2016, 7, 2119–2127. [Google Scholar] [CrossRef]
- Lee, C.H.; Lee, C.L.; Pan, T.M. A 90-d toxicity study of monascus-fermented products including high citrinin level. J. Food Sci. 2010, 75, T91–T97. [Google Scholar] [CrossRef]
- Liu, B.H.; Yu, F.Y.; Wu, T.S.; Li, S.Y.; Su, M.C.; Wang, M.C.; Shih, S.M. Evaluation of genotoxic risk and oxidative DNA damage in mammalian cells exposed to mycotoxins, patulin and citrinin. Toxicol. Appl. Pharmacol. 2003, 191, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, Y.; Iguchi, H.; Kamisuki, S.; Sugawara, F.; Furuichi, T.; Shinoda, Y. Low doses of the mycotoxin citrinin protect cortical neurons against glutamate-induced excitotoxicity. J. Toxicol. Sci. 2016, 41, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, K.; Yamazaki, R.; Kinoshita, M.; Kamata, Y.; Tani, F.; Minai, Y.; Sugita-Konishi, Y. Inhibitory effect of citrinin on lipopolisaccharide-induced nitric oxide production by mouse macrophage cells. Mycotoxin Res. 2013, 29, 229–234. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Filho, J.W.; Islam, M.T.; Ali, E.S.; Uddin, S.J.; Santos, J.V.; de Alencar, M.V.; Júnior, A.L.; Paz, M.F.; de Brito, M.D.; Sousa, J.M.E.; et al. A comprehensive review on biological properties of citrinin. Food Chem. Toxicol. 2017, 110, 130–141. [Google Scholar] [CrossRef]
- Gyawali, R.; Ibrahim, S.A. Natural products as antimicrobial agents. Food Control 2014, 46, 412–429. [Google Scholar] [CrossRef]
- Ding, X.; Yin, B.; Qian, L.; Zeng, Z.; Yang, Z.; Li, H.; Lu, Y.; Zhou, S. Screening for novel quorum-sensing inhibitors to interfere with the formation of Pseudomonas aeruginosa biofilm. J. Med. Microbiol. 2011, 60, 1827–1834. [Google Scholar] [CrossRef]
- Tsang, P.W.; Bandara, H.M.; Fong, W.P. Purpurin suppresses Candida albicans biofilm formation and hyphal development. PLoS ONE 2012, 7, e50866. [Google Scholar] [CrossRef]
- Ahmed Adam, M.A.; Tabana, Y.M.; Musa, K.B.; Sandai, D.A. Effects of different mycotoxins on humans, cell genome and their involvement in cancer (Review). Oncol. Rep. 2017, 37, 1321–1336. [Google Scholar] [CrossRef]
- Ohman, D.E.; Chakrabarty, A.M. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect. Immun. 1981, 33, 142–148. [Google Scholar] [CrossRef]
- Pedersen, S.S. Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis. APMIS Suppl. 1992, 28, 1–79. [Google Scholar]
- Høiby, N. Recent advances in the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. BMC Med. 2011, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Dai, X.; Sun, J.; Bu, X.; Weng, C.; Li, H.; Zhu, H. A diketopiperazine factor from Rheinheimera aquimaris QSI02 exhibits anti-quorum sensing activity. Sci. Rep. 2016, 6, 39637. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Lee, S.J.; Park, N.H.; Mechesso, A.F.; Birhanu, B.T.; Kang, J.; Reza, M.A.; Suh, J.W.; Park, S.C. Impact of phenolic compounds in the acyl homoserine lactone-mediated quorum sensing regulatory pathways. Sci. Rep. 2017, 7, 10618. [Google Scholar] [CrossRef]
- Fu, T.K.; Ng, S.K.; Chen, Y.E.; Lee, Y.C.; Demeter, F.; Herczeg, M.; Borbás, A.; Chiu, C.H.; Lan, C.Y.; Chen, C.L.; et al. Rhamnose binding protein as an anti-bacterial agent-targeting biofilm of Pseudomonas aeruginosa. Mar. Drugs 2019, 17, 355. [Google Scholar] [CrossRef]


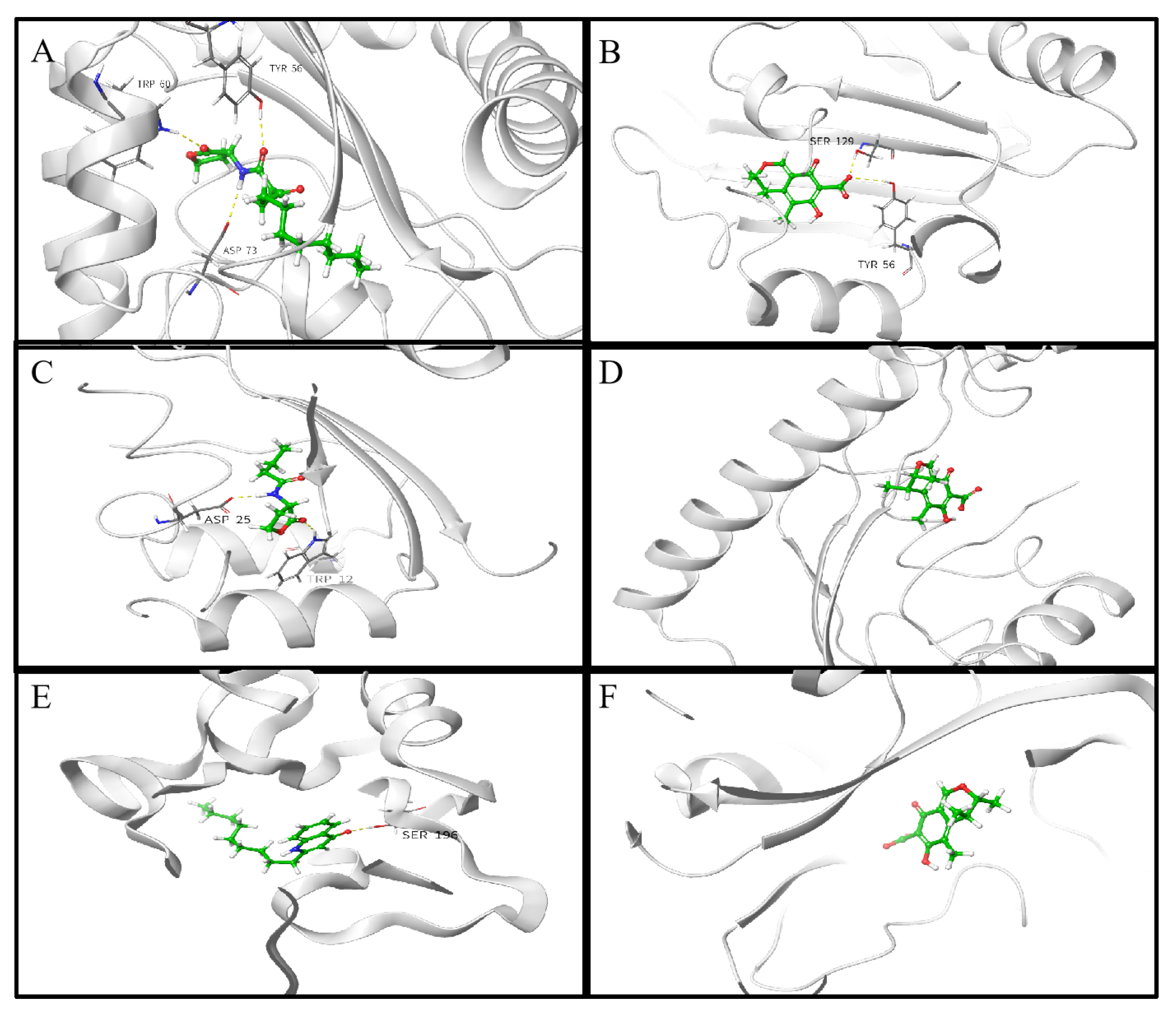

| Molecule | Docking Energy (kcal/mol) | Hydrogen Bonding Interactions | Key Hydrophobic Interactions |
|---|---|---|---|
| 3-oxo-C12-HSL | −6.168 | Tyr56, Trp60, Asp73 | Tyr64, Tyr56, Trp60, Ala105, Leu110, Phe101, Tyr93, Ala70, Trp88, Ile52, Ala50, Tyr47, Val76, Cys79, Leu36, Leu39, Leu40, Ala127, Leu125 |
| Citrinin | −6.300 | Tyr56, Ser129 | Leu36, Leu39, Leu40, Tyr64, Ile52, Ala50, Tyr47, Val76, Ala70, Tyr56, Ala127 |
| Molecule | Docking Energy (kcal/mol) | Hydrogen Bonding Interactions | Key Hydrophobic Interactions |
|---|---|---|---|
| 3-oxo-C12-HSL | −6.168 | Tyr56, Trp60, Asp73 | Tyr64, Tyr56, Trp60, Ala105, Leu110, Phe101, Tyr93, Ala70, Trp88, Ile52, Ala50, Tyr47, Val76, Cys79, Leu36, Leu39, Leu40, Ala127, Leu125 |
| Citrinin | −6.300 | Tyr56, Ser129 | Leu36, Leu39, Leu40, Tyr64, Ile52, Ala50, Tyr47, Val76, Ala70, Tyr56, Ala127 |
| Molecule | Docking Energy (kcal/mol) | Hydrogen Bonding Interactions | Key Hydrophobic Interactions |
|---|---|---|---|
| NHQ | −4.357 | Ser196 | Phe221, Met224, Leu197, Ile149, Ile236, Trp234, Tyr258, Ile186, Leu189, Ile263, Val170, Val211, Pro210, Leu208, Leu207 |
| Citrinin | −6.716 | / | Ala102, Ala168, Ile149, Ile263, Leu207, Leu208, Phe221, Met224, Leu197, Ile236 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, H.; Zhao, L.; Lv, K.; Zhang, Y.; Gao, H.; Gong, Q.; Yu, W. Citrinin Is a Potential Quorum Sensing Inhibitor against Pseudomonas aeruginosa. Mar. Drugs 2023, 21, 296. https://doi.org/10.3390/md21050296
Ji H, Zhao L, Lv K, Zhang Y, Gao H, Gong Q, Yu W. Citrinin Is a Potential Quorum Sensing Inhibitor against Pseudomonas aeruginosa. Marine Drugs. 2023; 21(5):296. https://doi.org/10.3390/md21050296
Chicago/Turabian StyleJi, Hongrui, Lu Zhao, Kaiwen Lv, Yuzhu Zhang, Haibo Gao, Qianhong Gong, and Wengong Yu. 2023. "Citrinin Is a Potential Quorum Sensing Inhibitor against Pseudomonas aeruginosa" Marine Drugs 21, no. 5: 296. https://doi.org/10.3390/md21050296
APA StyleJi, H., Zhao, L., Lv, K., Zhang, Y., Gao, H., Gong, Q., & Yu, W. (2023). Citrinin Is a Potential Quorum Sensing Inhibitor against Pseudomonas aeruginosa. Marine Drugs, 21(5), 296. https://doi.org/10.3390/md21050296






