Modulation of SREBP Expression and Fatty Acid Levels by Bacteria-Induced ER Stress Is Mediated by Hemocyanin in Penaeid Shrimp
Abstract
1. Introduction
2. Results
2.1. Bacteria Pathogens Induce Hemocyanin, SREBP, and ER Stress Proteins in Shrimp
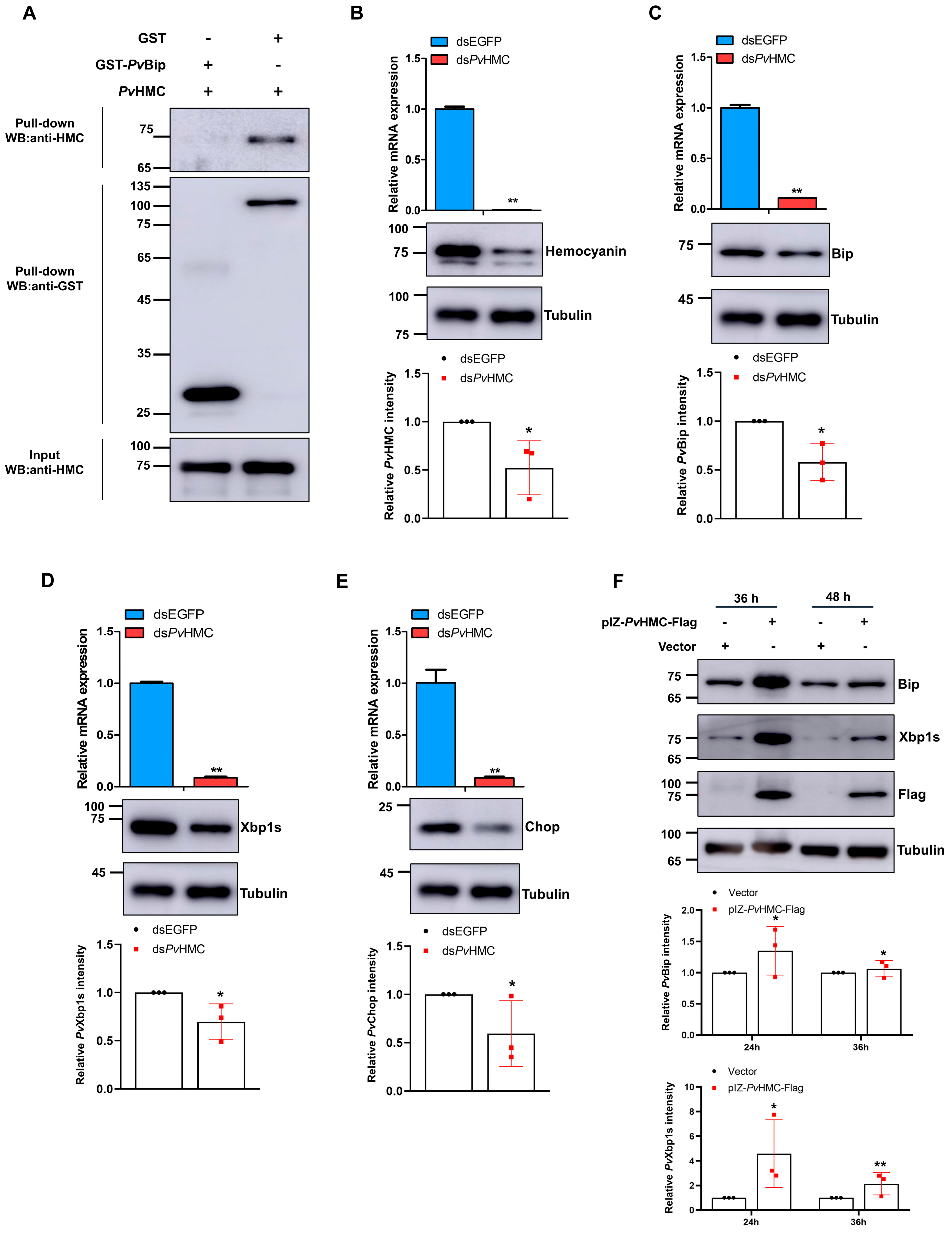
2.2. Hemocyanin Interacts with ER Stress Proteins
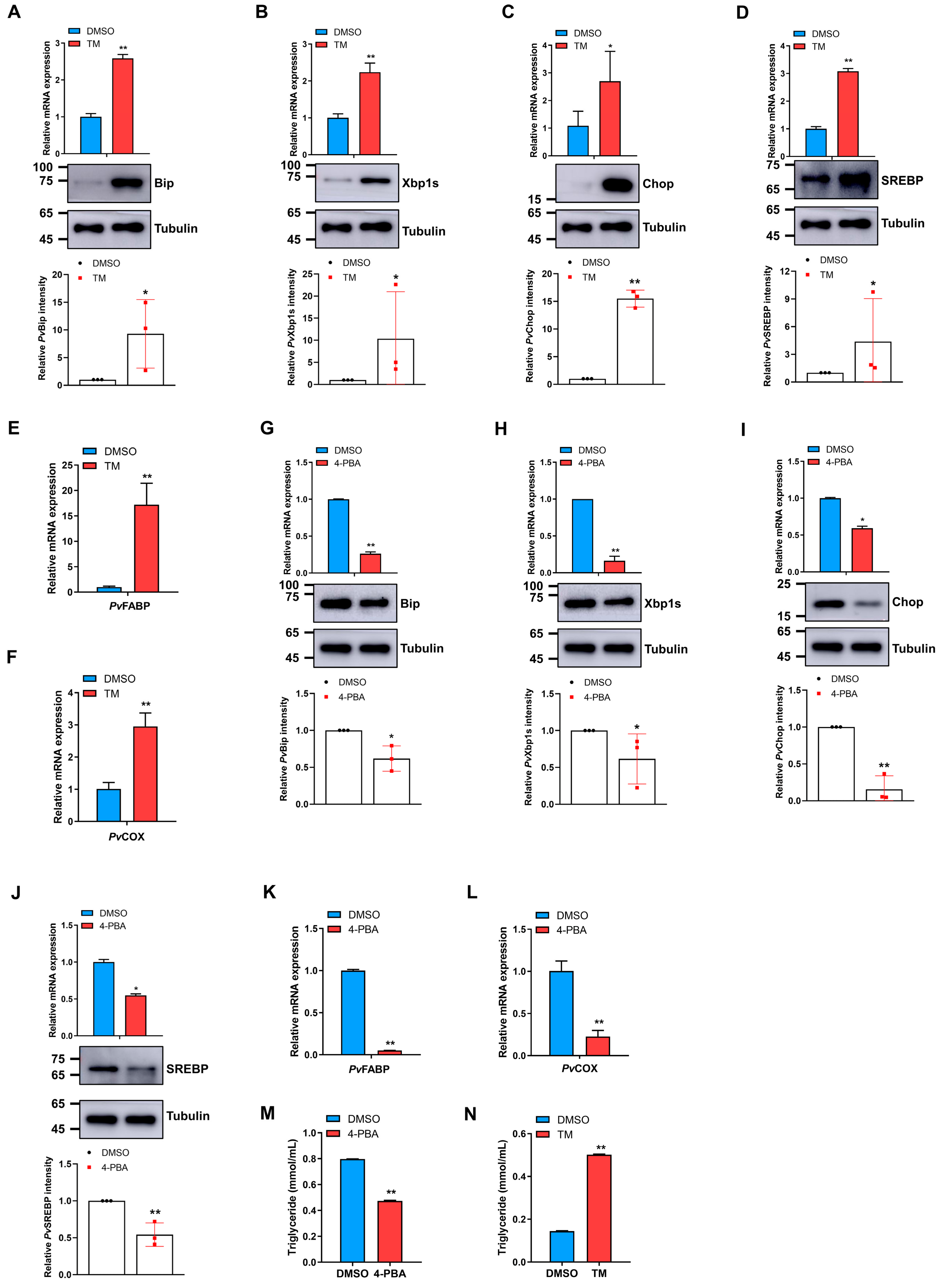
2.3. Bacteria-induced ER Stress Promotes SREBP Expression to Modulate Fatty Acid Levels in Shrimp
2.4. ER Stress Activation or Inhibition Modulates SREBP and Fatty Acid Levels in Shrimp
2.5. Hemocyanin Modulates ER Stress Proteins to Regulate SREBP Expression
2.6. Hemocyanin-Mediated ER Stress Alters Fatty Acid Profile in Shrimp Hepatopancreas
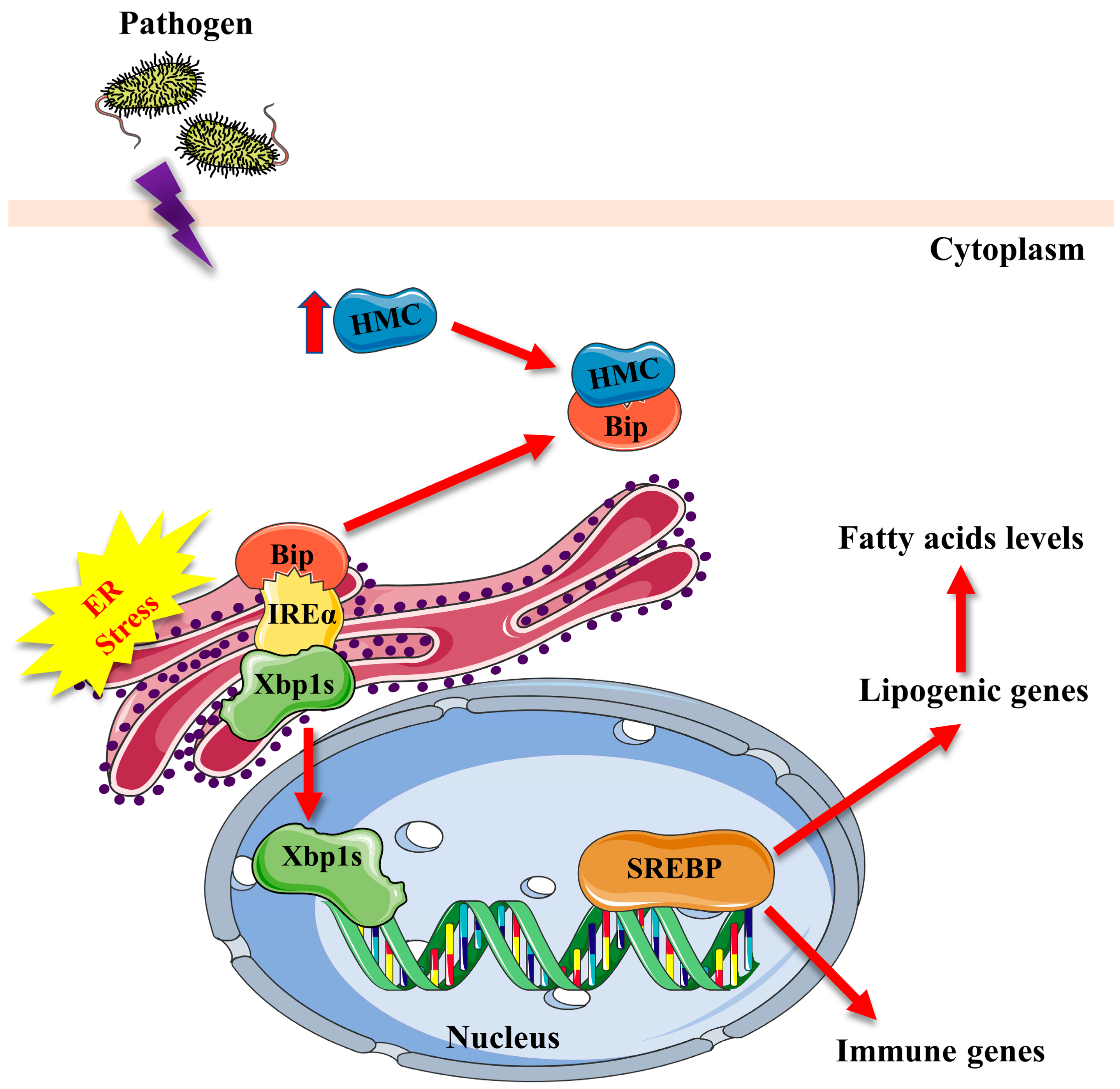
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. Reagents
4.3. Challenge Experiments and Samples Processing
4.4. Total RNA Extraction, cDNA Synthesis, and Quantitative RT-PCR
| Primers | Sequence (5′-3′) | Amplicon Size (bp) |
|---|---|---|
| Primers for protein expression | ||
| PvBip-F | ATGAGGTGTTGGACTGCA | 1926 |
| PvBip-R | CTACAATTCGTCCTTTTCATA | |
| pGEX-6P-1-PvBip-F | TCCAGGGGCCCCTGGGATCCATGAGGTGTTGGACTGCATTAG | 1950 |
| pGEX-6P-1-PvBip-R | CCCGGGAATTCCGGGGATCCCAATTCGTCCTTTTCATAATCT | |
| Primers for Real-time PCR | ||
| PvHMC-qF | CCTGGCCTCATAAAGACAACA | 104 |
| PvHMC-qR | TTTTCCACCCTTCAAAGATACC | |
| PvEF-1α-qF | TATGCTCCTTTTGGACGTTTTGC | 118 |
| PvEF-1α-qR | CCTTTTCTGCGGCCTTGGTAG | |
| PvBip-qF | GAGCGTCTGATTGGTGATT | 163 |
| PvBip-qR | GTGGCTTGTCGTTCTTGTT | |
| PvXbp1s-qF | AACTACGGGACCTGACATCTGC | 207 |
| PvXbp1s-qR | ACTGCCTTCTGCTGATCCACC | |
| PvChop-qF | TGACCCCCACCACCATCCC | 204 |
| PvChop-qR | ACTCGCTCCTCCGTCTCCC | |
| PvSREBP-qF | GGAGTTGTTGTTGCCGTGG | 134 |
| PvSREBP-qR | TGGCTGAGATGTTGGTAATGG | |
| PvFABP-qF | CGCTAAGCCCGTGCTGGAAGT | 103 |
| PvFABP-qR | CTCCTCGCCGAGCTTGATGGT | |
| PvCOX-qF | CCACAAGCGACTGATGACTTA | 103 |
| PvCOX-qR | GTAGGCATTGAGGGTGATGTAG | |
| Primers for dsRNA | ||
| dsPvHMC-T7F | GGATCCTAATACGACTCACTATAGGGTCCTCATCCACTGCAAA | 449 |
| dsPvHMC-R | TTGGACAGACGTTCAGCA | |
| dsPvHMC-T7R | GGATCCTAATACGACTCACTATAGGTTGGACAGACGTTCAGCA | 449 |
| dsPvHMC-F | GTCCTCATCCACTGCAAA | |
| dsEGFP-T7F | GGATCCTAATACGACTCACTATAGGCGTAAACGGCCACAAGTT | 429 |
| dsEGFP-R | TTCACCTTGATGCCGTTC | |
| dsEGFP-T7R | GGATCCTAATACGACTCACTATAGGTTCACCTTGATGCCGTTC | 429 |
| dsEGFP-F | CGTAAACGGCCACAAGTT |
4.5. Targeted RNA Interference (RNAi)
4.6. Plasmids
4.7. Cell Culture and Transfection
4.8. GST Pull-Down, SDS-PAGE, and Western Blot Analyses
4.9. Antibodies
4.10. Fatty Acid Profiling and Triglycerides Analysis
4.11. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gissi, E.; Manea, E.; Mazaris, A.D.; Fraschetti, S.; Almpanidou, V.; Bevilacqua, S.; Coll, M.; Guarnieri, G.; Lloret-Lloret, E.; Pascual, M. A review of the combined effects of climate change and other local human stressors on the marine environment. Sci. Total. Environ. 2021, 755, 142564. [Google Scholar] [CrossRef]
- Dong, F.; Zhu, X.; Qian, W.; Wang, P.; Wang, J. Combined effects of CO2-driven ocean acidification and Cd stress in the marine environment: Enhanced tolerance of Phaeodactylum tricornutum to Cd exposure. Mar. Pollut. Bull. 2020, 150, 110594. [Google Scholar] [CrossRef]
- Zhu, L.; Gao, N.; Wang, R.; Zhang, L. Proteomic and metabolomic analysis of marine medaka (Oryzias melastigma) after acute ammonia exposure. Ecotoxicology 2018, 27, 267–277. [Google Scholar] [CrossRef]
- Cohen, R.E.; James, C.C.; Lee, A.; Martinelli, M.M.; Muraoka, W.T.; Ortega, M.; Sadowski, R.; Starkey, L.; Szesciorka, A.R.; Timko, S.E. Marine Host-Pathogen Dynamics Influences of Global Climate Change. Oceanography 2018, 31, 182–193. [Google Scholar] [CrossRef]
- Hernandez-Llamas, A.; Cabanillas-Ramos, J.; Magallon-Barajas, F.J. Estimating impact of white spot disease on economic risk in semi-intensive shrimp farms in Mexico: The case of the State of Sinaloa. Rev. Aquac. 2016, 8, 111–120. [Google Scholar] [CrossRef]
- Tomanek, L. Proteomic responses to environmentally induced oxidative stress. J. Exp. Biol. 2015, 218, 1867–1879. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Zhang, Y.L.; Yao, D.F.; Zhao, Y.Z.; Tran, N.T.; Li, S.K.; Ma, H.Y.; Aweya, J.J. Metabolic reprogramming in crustaceans: A vital immune and environmental response strategy. Rev. Aquac. 2022, 14, 1094–1119. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Blower, M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell. Mol. Life Sci. 2016, 73, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Perkins, H.T.; Allan, V. Intertwined and Finely Balanced: Endoplasmic Reticulum Morphology, Dynamics, Function, and Diseases. Cells 2021, 10, 2341. [Google Scholar] [CrossRef] [PubMed]
- Chong, W.C.; Shastri, M.D.; Eri, R. Endoplasmic Reticulum Stress and Oxidative Stress: A Vicious Nexus Implicated in Bowel Disease Pathophysiology. Int. J. Mol. Sci. 2017, 18, 771. [Google Scholar] [CrossRef]
- Liang, Z.; Liu, R.; Zhao, D.; Wang, L.; Sun, M.; Wang, M.; Song, L. Ammonia exposure induces oxidative stress, endoplasmic reticulum stress and apoptosis in hepatopancreas of pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol. 2016, 54, 523–528. [Google Scholar] [CrossRef]
- Chipurupalli, S.; Kannan, E.; Tergaonkar, V.; D’Andrea, R.; Robinson, N. Hypoxia Induced ER Stress Response as an Adaptive Mechanism in Cancer. Int. J. Mol. Sci. 2019, 20, 749. [Google Scholar] [CrossRef]
- Carreras-Sureda, A.; Pihan, P.; Hetz, C. Calcium signaling at the endoplasmic reticulum: Fine-tuning stress responses. Cell Calcium 2018, 70, 24–31. [Google Scholar] [CrossRef]
- Grootjans, J.; Kaser, A.; Kaufman, R.J.; Blumberg, R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016, 16, 469–484. [Google Scholar] [CrossRef]
- Mogilenko, D.A.; Haas, J.T.; L’Homme, L.; Fleury, S.; Quemener, S.; Levavasseur, M.; Becquart, C.; Wartelle, J.; Bogomolova, A.; Pineau, L. Metabolic and Innate Immune Cues Merge into a Specific Inflammatory Response via the UPR. Cell 2019, 177, 1201–1216.e19. [Google Scholar] [CrossRef]
- Moncan, M.; Mnich, K.; Blomme, A.; Almanza, A.; Samali, A.; Gorman, A.M. Regulation of lipid metabolism by the unfolded protein response. J. Cell. Mol. Med. 2021, 25, 1359–1370. [Google Scholar] [CrossRef]
- Mohan, S.; Brown, L.; Ayyappan, P. Endoplasmic reticulum stress: A master regulator of metabolic syndrome. Eur. J. Pharmacol. 2019, 860, 172553. [Google Scholar] [CrossRef]
- Oyadomari, S.; Harding, H.P.; Zhang, Y.; Oyadomari, M.; Ron, D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008, 7, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Bobrovnikova-Marjon, E.; Hatzivassiliou, G.; Grigoriadou, C.; Romero, M.; Cavener, D.R.; Thompson, C.B.; Diehl, J.A. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc. Natl. Acad. Sci. USA 2008, 105, 16314–16319. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Ren, L.P.; Wang, C.; Zhu, Y.J.; Xing, H.Y.; Zhao, J.; Song, G.Y. Role of X-Box Binding Protein-1 in Fructose-Induced De Novo Lipogenesis in HepG2 Cells. Chin. Med. J. Engl. 2018, 131, 2310–2319. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Scapa, E.F.; Cohen, D.E.; Glimcher, L.H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 2008, 320, 1492–1496. [Google Scholar] [CrossRef] [PubMed]
- Henkel, A.S. Unfolded Protein Response Sensors in Hepatic Lipid Metabolism and Nonalcoholic Fatty Liver Disease. Semin. Liver Dis. 2018, 38, 320–332. [Google Scholar] [CrossRef]
- Lee, S.J.; Sekimoto, T.; Yamashita, E.; Nagoshi, E.; Nakagawa, A.; Imamoto, N.; Yoshimura, M.; Sakai, H.; Chong, K.T.; Tsukihara, T. The structure of importin-beta bound to SREBP-2: Nuclear import of a transcription factor. Science 2003, 302, 1571–1575. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.P.; Li, L.; Goldstein, J.L.; Brown, M.S. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 2005, 280, 26483–26490. [Google Scholar] [CrossRef]
- Nohturfft, A.; Yabe, D.; Goldstein, J.L.; Brown, M.S.; Espenshade, P.J. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell 2000, 102, 315–323. [Google Scholar] [CrossRef]
- DeBose-Boyd, R.A.; Ye, J. SREBPs in Lipid Metabolism, Insulin Signaling, and Beyond. Trends Biochem. Sci. 2018, 43, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Ferre, P.; Foufelle, F. Hepatic steatosis: A role for de novo lipogenesis and the transcription factor SREBP-1c. Diabetes Obes. Metab. 2010, 12 (Suppl. S2), 83–92. [Google Scholar] [CrossRef]
- Lee, J.N.; Ye, J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J. Biol. Chem. 2004, 279, 45257–45265. [Google Scholar] [CrossRef]
- Olivares, S.; Henkel, A.S. The role of X-box binding protein 1 in the hepatic response to refeeding in mice. J. Lipid. Res. 2019, 60, 353–359. [Google Scholar] [CrossRef]
- Aweya, J.J.; Zheng, X.; Zheng, Z.; Wang, W.; Fan, J.; Yao, D.; Li, S.; Zhang, Y. The sterol regulatory element binding protein homolog of Penaeus vannamei modulates fatty acid metabolism and immune response. Biochim. Biophys Acta Mol. Cell Biol. Lipids 2020, 1865, 158757. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Huang, Z.; Liu, Z.; Zheng, Z.; Zhang, Y.; Aweya, J.J. Transcriptome Analysis Reveals That SREBP Modulates a Large Repertoire of Genes Involved in Key Cellular Functions in Penaeus vannamei, although the Majority of the Dysregulated Genes Are Unannotated. Genes 2022, 13, 2057. [Google Scholar] [CrossRef] [PubMed]
- Marxen, J.C.; Pick, C.; Oakley, T.H.; Burmester, T. Occurrence of hemocyanin in ostracod crustaceans. J. Mol. Evol. 2014, 79, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Hoeger, U.; Harris, J.R. Vertebrate and Invertebrate Respiratory Proteins, Lipoproteins and other Body Fluid Proteins; Springer: Berlin/Heidelberg, Germany, 2020; Volume 94, pp. 233–250. [Google Scholar]
- Coates, C.J.; Decker, H. Immunological properties of oxygen-transport proteins: Hemoglobin, hemocyanin and hemerythrin. Cell. Mol. Life Sci. 2017, 74, 293–317. [Google Scholar] [CrossRef]
- Sun, S.; Xuan, F.; Fu, H.; Ge, X.; Zhu, J.; Qiao, H.; Jin, S.; Zhang, Y. Comparative proteomic study of the response to hypoxia in the muscle of oriental river prawn (Macrobrachium nipponense). J. Proteomics 2016, 138, 115–123. [Google Scholar] [CrossRef]
- Wei, K.; Yang, J. Oxidative damage of hepatopancreas induced by pollution depresses humoral immunity response in the freshwater crayfish Procambarus clarkii. Fish Shellfish Immunol. 2015, 43, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Li, J.; Lv, J.; Liu, P.; Li, J.; Li, S. Full-length transcriptome sequences of ridgetail white prawn Exopalaemon carinicauda provide insight into gene expression dynamics during thermal stress. Sci. Total Environ. 2020, 747, 141238. [Google Scholar] [CrossRef]
- Aweya, J.J.; Zhuang, K.; Liu, Y.; Fan, J.; Yao, D.; Wang, F.; Chen, X.; Li, S.; Ma, H.; Zhang, Y. The ARM repeat domain of hemocyanin interacts with MKK4 to modulate antimicrobial peptides expression. iScience 2022, 25, 103958. [Google Scholar] [CrossRef]
- Zhao, M.; Aweya, J.J.; Feng, Q.; Zheng, Z.; Yao, D.; Zhao, Y.; Chen, X.; Zhang, Y. Ammonia stress affects the structure and function of hemocyanin in Penaeus vannamei. Ecotoxicol. Environ. Saf. 2022, 241, 113827. [Google Scholar] [CrossRef]
- Zhao, M.M.; Yao, D.F.; Li, S.K.; Zhang, Y.L.; Aweya, J.J. Effects of ammonia on shrimp physiology and immunity: A review. Rev. Aquac. 2020, 12, 2194–2211. [Google Scholar] [CrossRef]
- Kim, J.Y.; Garcia-Carbonell, R.; Yamachika, S.; Zhao, P.; Dhar, D.; Loomba, R.; Kaufman, R.J.; Saltiel, A.R.; Karin, M. ER Stress Drives Lipogenesis and Steatohepatitis via Caspase-2 Activation of S1P. Cell 2018, 175, 133–145.e15. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, S.; Matsushita, Y.; Kurosaki, S.; Tange, M.; Fujiwara, N.; Hayata, Y.; Hayakawa, Y.; Suzuki, N.; Hata, M.; Tsuboi, M. Inhibiting SCAP/SREBP exacerbates liver injury and carcinogenesis in murine nonalcoholic steatohepatitis. J. Clin. Investig. 2022, 132, e151895. [Google Scholar] [CrossRef] [PubMed]
- Roszer, T. The invertebrate midintestinal gland (“hepatopancreas”) is an evolutionary forerunner in the integration of immunity and metabolism. Cell Tissue Res. 2014, 358, 685–695. [Google Scholar] [CrossRef]
- Kibenge, F.S.B.; Strange, R.J. Introduction to the anatomy and physiology of the major aquatic animal species in aquaculture. In Aquaculture Pharmacology; Kibenge, F.S.B., Baldisserotto, B., Chong, R.S.-M., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 1–111. [Google Scholar]
- Manubens, A.; Salazar, F.; Haussmann, D.; Figueroa, J.; Del Campo, M.; Pinto, J.M.; Huaquin, L.; Venegas, A.; Becker, M.I. Concholepas hemocyanin biosynthesis takes place in the hepatopancreas, with hemocytes being involved in its metabolism. Cell Tissue Res. 2010, 342, 423–435. [Google Scholar] [CrossRef]
- Zheng, Z.; Aweya, J.J.; Bao, S.; Yao, D.; Li, S.; Tran, N.T.; Ma, H.; Zhang, Y. The Microbial Composition of Penaeid Shrimps’ Hepatopancreas Is Modulated by Hemocyanin. J. Immunol. 2021, 207, 2733–2743. [Google Scholar] [CrossRef]
- Pobre, K.F.R.; Poet, G.J.; Hendershot, L.M. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J. Biol. Chem. 2019, 294, 2098–2108. [Google Scholar] [CrossRef]
- Jiang, Q.; Liu, G.; Chen, J.; Yao, K.; Yin, Y. Crosstalk Between Nuclear Glucose-Regulated Protein 78 and Tumor Protein 53 Contributes to the Lipopolysaccharide Aggravated Apoptosis of Endoplasmic Reticulum Stress-Responsive Porcine Intestinal Epithelial Cells. Cell. Physiol Biochem. 2018, 48, 2441–2455. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y.; Nawata, Y.; Sugimoto, Y. Lysozyme Mutants Accumulate in Cells while Associated at their N-terminal Alpha-domain with the Endoplasmic Reticulum Chaperone GRP78/BiP. Int. J. Biol. Sci. 2016, 12, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Ghemrawi, R.; Khair, M. Endoplasmic Reticulum Stress and Unfolded Protein Response in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 6127. [Google Scholar] [CrossRef]
- Lemmer, I.L.; Willemsen, N.; Hilal, N.; Bartelt, A. A guide to understanding endoplasmic reticulum stress in metabolic disorders. Mol. Metab. 2021, 47, 101169. [Google Scholar] [CrossRef]
- Cao, S.S.; Kaufman, R.J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox Signal. 2014, 21, 396–413. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, P.K.; Zhang, K.; Kumar, A. Toll-like receptor 2 (TLR2) engages endoplasmic reticulum stress sensor IRE1alpha to regulate retinal innate responses in Staphylococcus aureus endophthalmitis. FASEB J. 2020, 34, 13826–13838. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Mendez, R.; Heng, H.H.; Yang, Z.Q.; Zhang, K. Pharmacological ER stress promotes hepatic lipogenesis and lipid droplet formation. Am. J. Transl. Res. 2012, 4, 102–113. [Google Scholar] [PubMed]
- Park, S.M.; Kang, T.I.; So, J.S. Roles of XBP1s in Transcriptional Regulation of Target Genes. Biomedicines 2021, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Shimano, H.; Sato, R. SREBP-regulated lipid metabolism: Convergent physiology-divergent pathophysiology. Nat. Rev. Endocrinol. 2017, 13, 710–730. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Espenshade, P.J. Expanding roles for SREBP in metabolism. Cell Metab. 2012, 16, 414–419. [Google Scholar] [CrossRef]
- Casali, C.; Malvicini, R.; Erjavec, L.; Parra, L.; Artuch, A.; Fernandez Tome, M.C. X-box binding protein 1 (XBP1): A key protein for renal osmotic adaptation. Its role in lipogenic program regulation. Biochim. Biophys Acta Mol. Cell Biol. Lipids 2020, 1865, 158616. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, B.; Meng, M.; Zhao, W.; Wang, D.; Yuan, Y.; Zheng, Y.; Qiu, J.; Li, Y.; Li, G. FOXA3 induction under endoplasmic reticulum stress contributes to non-alcoholic fatty liver disease. J. Hepatol. 2021, 75, 150–162. [Google Scholar] [CrossRef]
- Raychaudhuri, S.; Prinz, W.A. The diverse functions of oxysterol-binding proteins. Annu. Rev. Cell Dev. Biol. 2010, 26, 157–177. [Google Scholar] [CrossRef]
- Akiyama, M.; Liew, C.W.; Lu, S.; Hu, J.; Martinez, R.; Hambro, B.; Kennedy, R.T.; Kulkarni, R.N. X-box binding protein 1 is essential for insulin regulation of pancreatic alpha-cell function. Diabetes 2013, 62, 2439–2449. [Google Scholar] [CrossRef]
- Das, U.N. Arachidonic acid and other unsaturated fatty acids and some of their metabolites function as endogenous antimicrobial molecules: A review. J. Adv. Res. 2018, 11, 57–66. [Google Scholar] [CrossRef]
- Pan, C.Y.; Liu, Y.H.; Gong, H.Y.; Chen, J.Y. Transcriptome analysis of the effect of polyunsaturated fatty acids against Vibrio vulnificus infection in Oreochromis niloticus. Fish Shellfish Immunol. 2017, 62, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




| Main Fatty Acids | Fatty Acid Composition (%) 1 | |
|---|---|---|
| Control (PBS) | V. parahaemolyticus | |
| Palmitic acid (C16:0) | 20.35 ± 0.58 | 21.64 ± 0.03 |
| Palmitoleic acid (C16:1) | 5.82 ± 0.20 | 6.82 ± 0.27 |
| Stearic acid (C18:0) | 0.11 ± 0.005 | 0.14 ± 0.002 * |
| Oleic acid (C18:1n-9c) | 5.14 ± 0.07 | 5.27 ± 0.72 |
| Linolelaidic acid (C18:2n-6c) | 6.44 ± 0.20 | 6.59 ± 0.16 |
| Arachidic acid (C20:0) | 20.23 ± 1.51 | 16.37 ± 0.46 |
| γ-Linolenic acid (C18:3n-6) | 3.95 ± 0.18 | 4.05 ± 0.09 |
| α-Linolenic acid (C18:3n-3) | 2.04 ± 0.06 | 2.18 ± 0.007 |
| Dihomo-γ-linolenic acid (C20:3n-6) | 1.58 ± 0.06 | 1.49 ± 0.009 |
| Arachidonic acid (C20:4n-6) | 3.90 ± 0.05 | 4.70 ± 0.024 ** |
| Eicosapentaenoic acid (C20:5n-3) | 6.72 ± 0.11 | 7.90 ± 0.06 ** |
| Docosapentaenoic acid (C22:5n-3) | 0.87 ± 0.02 | 1.0 ± 0.003 * |
| Docosahexaenoic acid (C22:6n-3) | 4.89 ± 0.16 | 5.18 ± 0.01 |
| ∑SFA | 40.70 ± 2.07 | 38.16 ± 0.43 |
| ∑MUFA | 10.96 ± 0.25 | 12.03 ± 0.20 * |
| ∑LC-PUFA | 30.40 ± 0.79 | 33.08 ± 0.07 |
| Main Fatty Acids | Fatty Acid Composition (%) 1 | |
|---|---|---|
| Control (PBS) | 4-Phenylbutyric Acid (4-PBA) | |
| Palmitic acid (C16:0) | 11.03 ± 1.18 | 7.33 ± 0.95 * |
| Palmitoleic acid (C16:1) | 1.44 ± 0.16 | 0.46 ± 0.06 |
| Stearic acid (C18:0) | 2.21 ± 0.17 | 2.02 ± 0.19 |
| Oleic acid (C18:1n-9c) | 11.93 ± 2.58 | 9.56 ± 1.39 |
| Linolelaidic acid (C18:2n-6c) | 4.31 ± 0.32 | 3.79 ± 0.37 |
| Arachidic acid (C20:0) | 16.09 ± 2.46 | 13.53 ± 1.53 |
| γ-Linolenic acid (C18:3n-6) | 0.1 ± 0.02 | 0.07 ± 0.008 |
| α-Linolenic acid (C18:3n-3) | 1.48 ± 0.29 | 0.97 ± 0.13 |
| Dihomo-γ-linolenic acid (C20:3n-6) | 1.07 ± 0.11 | 0.79 ± 0.09 |
| Arachidonic acid (C20:4n-6) | 2.01 ± 0.17 | 1.54 ± 0.07 |
| Eicosapentaenoic acid (C20:5n-3) | 2.03 ± 0.26 | 1.43 ± 0.39 |
| Docosapentaenoic acid (C22:5n-3) | 0.23 ± 0.03 | 0.09 ± 0.01 * |
| Docosahexaenoic acid (C22:6n-3) | 1.45 ± 0.02 | 0.87 ± 0.005 ** |
| ∑SFA | 29.33 ± 3.58 | 22.88 ± 2.67 |
| ∑MUFA | 13.37 ± 2.72 | 10.01 ± 1.45 |
| ∑LC-PUFA | 12.96 ± 0.96 | 9.57 ± 0.99 |
| Main Fatty Acids | Fatty Acid Composition (%) 1 | |
|---|---|---|
| Control PBS | Tunicamycin (TM) | |
| Palmitic acid (C16:0) | 11.43 ± 0.03 | 13.23 ± 0.28 ** |
| Palmitoleic acid (C16:1) | 2.11 ± 0.25 | 1.06 ± 0.05 |
| Stearic acid (C18:0) | 2.06 ± 0.09 | 3.84 ± 0.22 ** |
| Oleic acid (C18:1n-9c) | 15.11 ± 0.83 | 14.75 ± 1.39 |
| Linolelaidic acid (C18:2n-6c) | 4.26 ± 0.10 | 6.22 ± 0.31 ** |
| Arachidic acid (C20:0) | 20.37 ± 0.64 | 22.59 ± 2.39 |
| γ-Linolenic acid (C18:3n-6) | 0.06 ± 0.003 | 0.16 ± 0.02 ** |
| α-Linolenic acid (C18:3n-3) | 2.17 ± 0.12 | 1.54 ± 0.11 |
| Dihomo-γ-linolenic acid (C20:3n-6) | 1.12 ± 0.12 | 1.37 ± 0.09 |
| Arachidonic acid (C20:4n-6) | 1.79 ± 0.16 | 3.32 ± 0.31 * |
| Eicosapentaenoic acid (C20:5n-3) | 2.25 ± 0.17 | 4.14 ± 0.42 * |
| Docosapentaenoic acid (C22:5n-3) | 0.23 ± 0.08 | 0.31 ± 0.11 |
| Docosahexaenoic acid (C22:6n-3) | 1.37 ± 0.17 | 2.06 ± 0.42 |
| ∑SFA | 33.86 ± 0.69 | 39.66 ± 2.87 |
| ∑MUFA | 17.22 ± 1.05 | 15.81 ± 1.34 |
| ∑LC-PUFA | 13.25 ± 0.70 | 19.12 ± 1.15 * |
| Main Fatty Acid | Fatty Acid Composition (%) 1 | |||
|---|---|---|---|---|
| dsEGFP | dsPvHMC | |||
| DMSO | Tunicamycin (TM) | DMSO | Tunicamycin (TM) | |
| Palmitic acid (C16:0) | 29.17 ± 0.27 | 33.49 ± 1.02 * | 22.38 ± 0.33 ** | 22.45 ± 0.33 |
| Palmitoleic acid(C16:1) | 2.86 ± 0.03 | 3.87 ± 0.19 ** | 2.11 ± 0.56 ** | 2.17 ± 0.09 |
| Stearic acid (C18:0) | 5.64 ± 0.05 | 6.35 ± 0.24 | 5.12 ± 0.18 | 4.93 ± 0.64 |
| Oleic acid (C18:1n-9c) | 40.59 ± 0.45 | 45.57 ± 1.31 * | 29.38 ± 0.57 ** | 28.99 ± 0.18 |
| Linolelaidic acid (C18:2n-6c) | 10.65 ± 0.34 | 12.39 ± 0.66 | 8.17 ± 0.12 ** | 8.08 ± 0.36 |
| Arachidic acid (C20:0) | 56.48 ± 0.80 | 62.41 ± 1.70 * | 41.27 ± 0.80 ** | 40.57 ± 0.22 |
| γ-Linolenic acid (C18:3n-6) | 4.37 ± 0.30 | 5.29 ± 0.2 * | 3.14 ± 0.06 ** | 3.18 ± 0.07 |
| α-Linolenic acid (C18:3n-3) | 2.69 ± 0.03 | 3.00 ± 0.04 ** | 2.01 ± 0.03 ** | 2.05 ± 0.01 |
| Dihomo-γ-linolenic acid (C20:3n-6) | 2.98 ± 0.05 | 3.18 ± 0.11 | 2.09 ± 0.04 ** | 2.01 ± 0.01 |
| Arachidonic acid (C20:4n-6) | 5.14 ± 0.03 | 5.84 ± 0.21 * | 4.74 ± 0.05 ** | 4.93 ± 0.02 † |
| Eicosapentaenoic acid (C20:5n-3) | 5.46 ± 0.04 | 6.31 ± 0.19* | 5.50 ± 0.06 | 6.01 ± 0.02 †† |
| Docosapentaenoic acid (C22:5n-3) | 0.63 ± 0.01 | 0.73 ± 0.02 * | 0.52 ± 0.005 ** | 0.52 ± 0.01 |
| Docosahexaenoic acid (C22:6n-3) | 6.74 ± 0.02 | 8.09 ± 0.23 ** | 6.08 ± 0.07 ** | 6.63 ± 0.03 †† |
| ∑SFA | 91.3 ± 1.12 | 102.25 ± 2.92 * | 68.77 ± 1.02 ** | 67.95 ± 0.42 |
| ∑MUFA | 43.45 ± 0.49 | 49.44 ± 1.51 * | 31.49 ± 0.62 ** | 31.13 ± 0.27 |
| ∑LC-PUFA | 38.65 ± 0.50 | 44.83 ± 1.60 * | 32.27 ± 0.38 ** | 33.41 ± 0.48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Zheng, X.; Chen, Z.; Zheng, Z.; Yao, D.; Yang, S.; Zhang, Y.; Aweya, J.J. Modulation of SREBP Expression and Fatty Acid Levels by Bacteria-Induced ER Stress Is Mediated by Hemocyanin in Penaeid Shrimp. Mar. Drugs 2023, 21, 164. https://doi.org/10.3390/md21030164
Huang Z, Zheng X, Chen Z, Zheng Z, Yao D, Yang S, Zhang Y, Aweya JJ. Modulation of SREBP Expression and Fatty Acid Levels by Bacteria-Induced ER Stress Is Mediated by Hemocyanin in Penaeid Shrimp. Marine Drugs. 2023; 21(3):164. https://doi.org/10.3390/md21030164
Chicago/Turabian StyleHuang, Zishu, Xiaoyu Zheng, Zeyan Chen, Zhihong Zheng, Defu Yao, Shen Yang, Yueling Zhang, and Jude Juventus Aweya. 2023. "Modulation of SREBP Expression and Fatty Acid Levels by Bacteria-Induced ER Stress Is Mediated by Hemocyanin in Penaeid Shrimp" Marine Drugs 21, no. 3: 164. https://doi.org/10.3390/md21030164
APA StyleHuang, Z., Zheng, X., Chen, Z., Zheng, Z., Yao, D., Yang, S., Zhang, Y., & Aweya, J. J. (2023). Modulation of SREBP Expression and Fatty Acid Levels by Bacteria-Induced ER Stress Is Mediated by Hemocyanin in Penaeid Shrimp. Marine Drugs, 21(3), 164. https://doi.org/10.3390/md21030164









