Bioactivity Profiling and Untargeted Metabolomics of Microbiota Associated with Mesopelagic Jellyfish Periphylla periphylla
Abstract
1. Introduction
2. Results
2.1. Isolation and Characterization of Isolates
2.2. Antimicrobial and Anticancer Activity
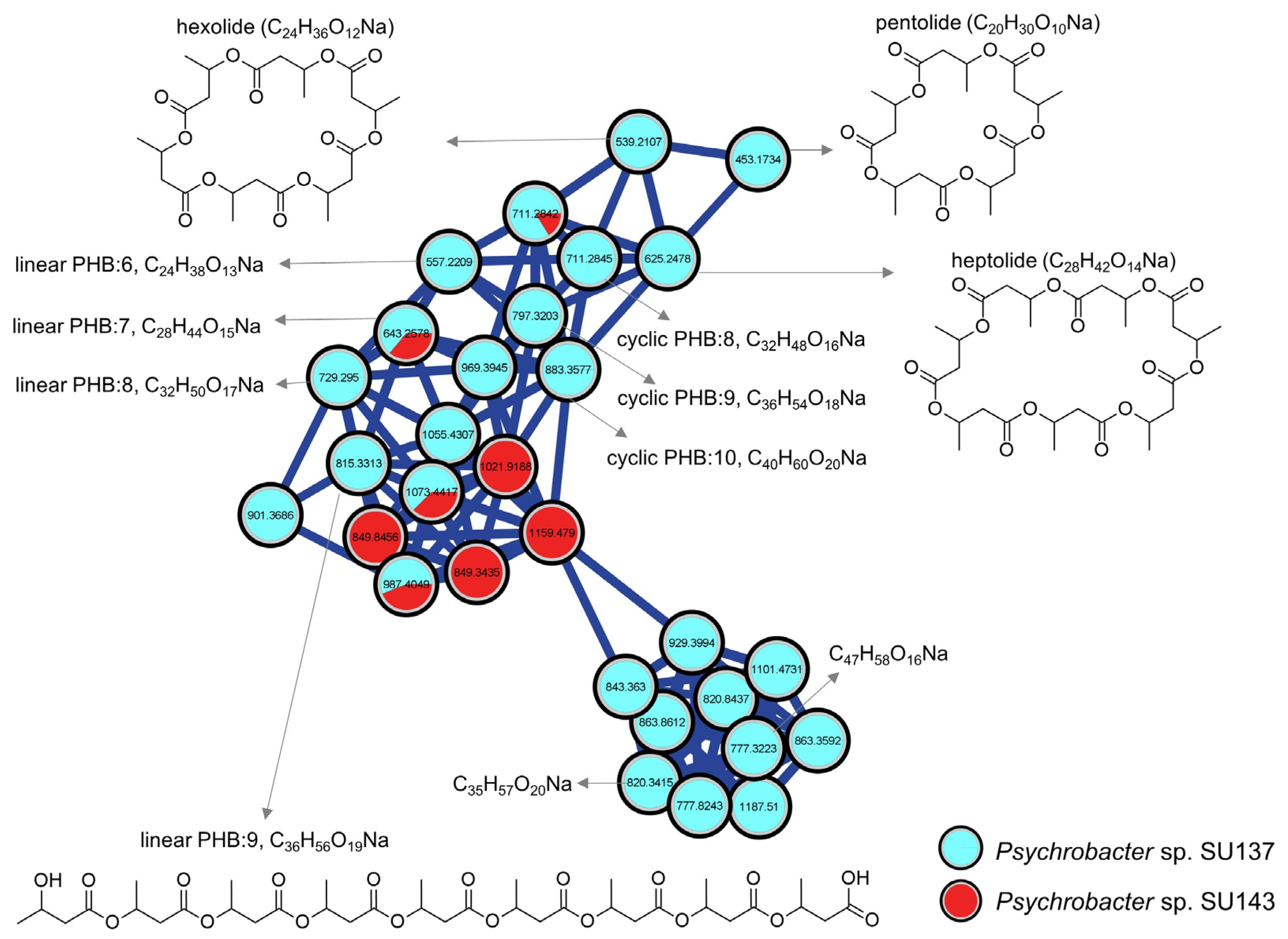
2.3. Metabolomics
3. Discussion
4. Materials and Methods
4.1. Sampling and Microbial Isolation
4.2. Identification of Microorganisms
4.3. Cultivation and Extraction
4.4. LC–MS/MS Sample Analysis
4.5. Molecular Networking and Spectral Library Search
4.6. Antimicrobial and Anticancer Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ICES. Working Group on Zooplankton Ecology (WGZE, Outputs from 2020 Meeting); ICES Scientific Reports; ICES: Copenhagen, Denmark, 2021. [Google Scholar]
- Watanuki, Y.; Thiebot, J.-B. Factors affecting the importance of myctophids in the diet of the world’s seabirds. Mar. Biol. 2018, 165, 79. [Google Scholar] [CrossRef]
- del Giorgio, P.A.; Duarte, C.M. Respiration in the open ocean. Nature 2002, 420, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Dipper, F. Chapter 5—Open water lifestyles: Marine nekton. In Elements of Marine Ecology, 5th ed.; Dipper, F., Ed.; Butterworth-Heinemann: Oxford, UK, 2022; pp. 229–255. [Google Scholar]
- Sutton, T.T.; Milligan, R.J. Deep-Sea ecology. In Encyclopedia of Ecology, 2nd ed.; Fath, B., Ed.; Elsevier: Oxford, UK, 2019; pp. 35–45. [Google Scholar]
- Katsuki, T.; Greenspan, R.J. Jellyfish nervous systems. Curr. Biol. 2013, 23, R592–R594. [Google Scholar] [CrossRef] [PubMed]
- Zapata, F.; Goetz, F.E.; Smith, S.A.; Howison, M.; Siebert, S.; Church, S.H.; Sanders, S.M.; Ames, C.L.; McFadden, C.S.; France, S.C.; et al. Phylogenomic analyses support traditional relationships within Cnidaria. PLoS ONE 2015, 10, e0139068. [Google Scholar] [CrossRef] [PubMed]
- Leone, A.; Lecci, R.M.; Milisenda, G.; Piraino, S. Mediterranean jellyfish as novel food: Effects of thermal processing on antioxidant, phenolic, and protein contents. Eur. Food Res. Technol. 2019, 245, 1611–1627. [Google Scholar] [CrossRef]
- Doyle, T.K.; Hays, G.C.; Harrod, C.; Houghton, J.D. Ecological and societal benefits of jellyfish. In Jellyfish Blooms; Springer: Berlin/Heidelberg, Germany, 2014; pp. 105–127. [Google Scholar]
- Hsieh, Y.; Leong, F.-M.; Rudloe, J. Jellyfish as food. In Jellyfish Blooms: Ecological and Societal Importance; Springer: Berlin/Heidelberg, Germany, 2001; pp. 11–17. [Google Scholar]
- Wright, R.M.; Le Quéré, C.; Buitenhuis, E.; Pitois, S.; Gibbons, M.J. Role of jellyfish in the plankton ecosystem revealed using a global ocean biogeochemical model. Biogeosciences 2021, 18, 1291–1320. [Google Scholar] [CrossRef]
- Cunha, S.A.; Dinis-Oliveira, R.J. Raising awareness on the clinical and forensic aspects of jellyfish stings: A worldwide increasing threat. Int. J. Environ. Res. Public Health 2022, 19, 8430. [Google Scholar] [CrossRef]
- Amreen Nisa, S.; Vinu, D.; Krupakar, P.; Govindaraju, K.; Sharma, D.; Vivek, R. Jellyfish venom proteins and their pharmacological potentials: A review. Int. J. Biol. Macromol. 2021, 176, 424–436. [Google Scholar] [CrossRef]
- Stabili, L.; Schirosi, R.; Parisi, M.G.; Piraino, S.; Cammarata, M. The mucus of Actinia equina (Anthozoa, Cnidaria): An unexplored resource for potential applicative purposes. Mar. Drugs 2015, 13, 5276–5296. [Google Scholar] [CrossRef]
- Tinta, T.; Kogovšek, T.; Klun, K.; Malej, A.; Herndl, G.J.; Turk, V. Jellyfish-associated microbiome in the marine environment: Exploring its biotechnological potential. Mar. Drugs 2019, 17, 94. [Google Scholar] [CrossRef]
- Schmahl, G. Induction of stolon settlement in the scyphopolyps of Aurelia aurita (Cnidaria, Scyphozoa, Semaeostomeae) by glycolipids of marine bacteria. Helgol. Mar. Res. 1985, 39, 117–127. [Google Scholar] [CrossRef]
- Viver, T.; Orellana, L.H.; Hatt, J.K.; Urdiain, M.; Díaz, S.; Richter, M.; Antón, J.; Avian, M.; Amann, R.; Konstantinidis, K.T.; et al. The low diverse gastric microbiome of the jellyfish Cotylorhiza tuberculata is dominated by four novel taxa. Environ. Microbiol. 2017, 19, 3039–3058. [Google Scholar] [CrossRef] [PubMed]
- Schuett, C.; Doepke, H. Endobiotic bacteria and their pathogenic potential in cnidarian tentacles. Helgol. Mar. Res. 2010, 64, 205–212. [Google Scholar] [CrossRef]
- Rath, C.M.; Janto, B.; Earl, J.; Ahmed, A.; Hu, F.Z.; Hiller, L.; Dahlgren, M.; Kreft, R.; Yu, F.; Wolff, J.J.; et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol. 2011, 6, 1244–1256. [Google Scholar] [CrossRef]
- Casertano, M.; Menna, M.; Imperatore, C. The Ascidian-derived metabolites with antimicrobial properties. Antibiotics 2020, 9, 510. [Google Scholar] [CrossRef]
- Piel, J. Bacterial symbionts: Prospects for the sustainable production of invertebrate-derived pharmaceuticals. Curr. Med. Chem. 2006, 13, 39–50. [Google Scholar] [CrossRef]
- Moore, B.S.; Trischman, J.A.; Seng, D.; Kho, D.; Jensen, P.R.; Fenical, W. Salinamides, antiinflammatory depsipeptides from a marine Streptomycete. J. Org. Chem. 1999, 64, 1145–1150. [Google Scholar] [CrossRef]
- Trischman, J.A.; Tapiolas, D.M.; Jensen, P.R.; Dwight, R.; Fenical, W.; McKee, T.C.; Ireland, C.M.; Stout, T.J.; Clardy, J. Salinamides A and B: Anti-inflammatory depsipeptides from a marine streptomycete. J. Am. Chem. Soc. 1994, 116, 757–758. [Google Scholar] [CrossRef]
- Hanif, N.; Ohno, O.; Kitamura, M.; Yamada, K.; Uemura, D. Symbiopolyol, a VCAM-1 Inhibitor from a symbiotic dinoflagellate of the Jellyfish Mastigias papua. J. Nat. Prod. 2010, 73, 1318–1322. [Google Scholar] [CrossRef]
- Liu, J.; Li, F.; Kim, E.L.; Li, J.L.; Hong, J.; Bae, K.S.; Chung, H.Y.; Kim, H.S.; Jung, J.H. Antibacterial polyketides from the jellyfish-derived fungus Paecilomyces variotii. J. Nat. Prod. 2011, 74, 1826–1829. [Google Scholar] [CrossRef]
- Hassan, H.M.; Degen, D.; Jang, K.H.; Ebright, R.H.; Fenical, W. Salinamide F, new depsipeptide antibiotic and inhibitor of bacterial RNA polymerase from a marine-derived Streptomyces sp. J. Antibiot. 2015, 68, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.L.; Li, J.L.; Xiao, B.; Hong, J.; Yoo, E.S.; Yoon, W.D.; Jung, J.H. A new cyclic tetrapeptide from the Jellyfish-derived fungus Phoma sp. Chem. Pharm. Bull. 2012, 60, 1590–1593. [Google Scholar] [CrossRef]
- Lucas, C.H.; Reed, A.J. Gonad morphology and gametogenesis in the deep-sea jellyfish Atolla wyvillei and Periphylla periphylla (Scyphozoa: Coronatae) collected from Cape Hatteras and the Gulf of Mexico. J. Mar. Biol. Assoc. UK 2009, 90, 1095–1104. [Google Scholar] [CrossRef]
- Båmstedt, U. Life History of A Deep-Water Medusa: First Records of Size, Longevity, Growth and Recruitment at Varying Ages. SSRN 2022, 1–17. Available online: https://ssrn.com/abstract=4137382 (accessed on 1 January 2023). [CrossRef]
- Delroisse, J.; Duchatelet, L.; Flammang, P.; Mallefet, J. Leaving the dark side? Insights Into the evolution of luciferases. Front. Mar. Sci. 2021, 8, 673620. [Google Scholar] [CrossRef]
- Lucas, C.H. Biochemical composition of the mesopelagic coronate jellyfish Periphylla periphylla from the Gulf of Mexico. J. Mar. Biol. Assoc. UK 2008, 89, 77–81. [Google Scholar] [CrossRef]
- Sötje, I.; Tiemann, H.; Båmstedt, U. Trophic ecology and the related functional morphology of the deepwater medusa Periphylla periphylla (Scyphozoa, Coronata). Mar. Biol. 2007, 150, 329–343. [Google Scholar] [CrossRef]
- Dziurzynski, M.; Gorecki, A.; Pawlowska, J.; Istel, L.; Decewicz, P.; Golec, P.; Styczynski, M.; Poszytek, K.; Rokowska, A.; Gorniak, D. Revealing the diversity of bacteria and fungi in the active layer of permafrost at Spitsbergen island (Arctic)–combining classical microbiology and metabarcoding for ecological and bioprospecting exploration. Sci. Total Environ. 2023, 856, 159072. [Google Scholar] [CrossRef]
- Wu, B.; Wiese, J.; Labes, A.; Kramer, A.; Schmaljohann, R.; Imhoff, J.F. Lindgomycin, an unusual antibiotic polyketide from a marine fungus of the Lindgomycetaceae. Mar. Drugs 2015, 13, 4617–4632. [Google Scholar] [CrossRef]
- Oppong-Danquah, E.; Parrot, D.; Blümel, M.; Labes, A.; Tasdemir, D. Molecular networking-based metabolome and bioactivity analyses of marine-adapted fungi co-cultivated with phytopathogens. Front. Microbiol. 2018, 9, 2072. [Google Scholar] [CrossRef]
- Utermann, C.; Echelmeyer, V.A.; Oppong-Danquah, E.; Blümel, M.; Tasdemir, D. Diversity, bioactivity profiling and untargeted metabolomics of the cultivable gut microbiota of Ciona intestinalis. Mar. Drugs 2021, 19, 6. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Park, S.J.; Li, X.H.; Choi, Y.S.; Im, D.S.; Lee, J.H. Bacterial ornithine lipid, a surrogate membrane lipid under phosphate-limiting conditions, plays important roles in bacterial persistence and interaction with host. Environ. Microbiol. 2018, 20, 3992–4008. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2015, 40, 133–159. [Google Scholar] [CrossRef]
- da Costa, M.S.; Albuquerque, L.; Nobre, M.F.; Wait, R. The identification of fatty acids in bacteria. In Methods in Microbiology; Rainey, F., Oren, A., Eds.; Academic Press: Cambridge, MA, USA, 2011; Volume 38, pp. 183–196. [Google Scholar]
- Vanounou, S.; Parola, A.H.; Fishov, I. Phosphatidylethanolamine and phosphatidylglycerol are segregated into different domains in bacterial membrane. A study with pyrene-labelled phospholipids. Mol. Microbiol. 2003, 49, 1067–1079. [Google Scholar] [CrossRef]
- Murai, N.; Skoog, F.; Doyle, M.E.; Hanson, R.S. Relationships between cytokinin production, presence of plasmids, and fasciation caused by strains of Corynebacterium fascians. Proc. Natl. Acad. Sci. USA 1980, 77, 619–623. [Google Scholar] [CrossRef]
- Li, H.; Shinde, P.B.; Lee, H.J.; Yoo, E.S.; Lee, C.-O.; Hong, J.; Choi, S.H.; Jung, J.H. Bile acid derivatives from a sponge-associated bacterium Psychrobacter sp. Arch. Pharmacal Res. 2009, 32, 857–862. [Google Scholar] [CrossRef]
- Petta, T.; Raichardt, L.; Melo, I.S.; Moraes, L.A.B. Bioassay-guided isolation of a low molecular weight PHB from Burkholderia sp. with phytotoxic activity. Appl. Biochem. Biotechnol. 2013, 170, 1689–1701. [Google Scholar] [CrossRef]
- Esposito, F.P.; Vecchiato, V.; Buonocore, C.; Tedesco, P.; Noble, B.; Basnett, P.; de Pascale, D. Enhanced production of biobased, biodegradable, Poly (3-hydroxybutyrate) using an unexplored marine bacterium Pseudohalocynthiibacter aestuariivivens, isolated from highly polluted coastal environment. Bioresour. Technol. 2023, 368, 128287. [Google Scholar] [CrossRef] [PubMed]
- Seebach, D.; Brändli, U.; Müller, H.-M.; Dobler, M.; Egli, M.; Przybylski, M.; Schneider, K. On the macrolactonization of β-hydroxy acids. Crystal structures of the pentolide and the hexolide from (R)-3-hydroxybutanoic acid. Molecular modeling studies of the tetrolide. Helv. Chim. Acta 1989, 72, 1704–1717. [Google Scholar] [CrossRef]
- Seebach, D.; Brändli, U.; Schnurrenberger, P.; Przybylski, M. High-yield synthesis of 20-, 24-, and 28-membered macropentolide, -hexolide, and -heptolide, respectively, from (R)- or (S)-3-hydroxybutanoic acid under Yamaguchi’s macrolactonization conditions. Helv. Chim. Acta 1988, 71, 155–167. [Google Scholar] [CrossRef]
- de Carvalho, M.P.; Abraham, W.R. Antimicrobial and biofilm inhibiting diketopiperazines. Curr. Med. Chem. 2012, 19, 3564–3577. [Google Scholar] [CrossRef] [PubMed]
- Maeda, J.; Nishida, M.; Takikawa, H.; Yoshida, H.; Azuma, T.; Yoshida, M.; Mizushina, Y. Inhibitory effects of sulfobacin B on DNA polymerase and inflammation. Int. J. Mol. Med. 2010, 26, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, H.W.; Christian, M.J.D.; Hay, S.; Nicolson, J.; Sutherland, D.; Crumlish, M. Jellyfish as vectors of bacterial disease for farmed salmon (Salmo Salar). J. Vet. Diagn. Investig. 2010, 22, 376–382. [Google Scholar] [CrossRef]
- Delannoy, C.M.J.; Houghton, J.D.R.; Fleming, N.E.C.; Ferguson, H.W. Mauve stingers (Pelagia noctiluca) as carriers of the bacterial fish pathogen Tenacibaculum maritimum. Aquaculture 2011, 311, 255–257. [Google Scholar] [CrossRef]
- Småge, S.B.; Brevik, Ø.J.; Frisch, K.; Watanabe, K.; Duesund, H.; Nylund, A. Concurrent jellyfish blooms and tenacibaculosis outbreaks in Northern Norwegian Atlantic salmon (Salmo salar) farms. PLoS ONE 2017, 12, e0187476. [Google Scholar] [CrossRef]
- Rodrigues, C.J.; de Carvalho, C.C. Cultivating marine bacteria under laboratory conditions: Overcoming the “unculturable” dogma. Front. Bioeng. Biotechnol. 2022, 10, 1476. [Google Scholar] [CrossRef]
- Kos Kramar, M.; Tinta, T.; Lučić, D.; Malej, A.; Turk, V. Bacteria associated with moon jellyfish during bloom and post-bloom periods in the Gulf of Trieste (northern Adriatic). PLoS ONE 2019, 14, e0198056. [Google Scholar] [CrossRef]
- Dobretsov, S.; Qian, P.-Y. The role of epibotic bacteria from the surface of the soft coral Dendronephthya sp. in the inhibition of larval settlement. J. Exp. Mar. Biol. Ecol. 2004, 299, 35–50. [Google Scholar] [CrossRef]
- Shnit-Orland, M.; Kushmaro, A. Coral mucus-associated bacteria: A possible first line of defense. FEMS Microbiol. Ecol. 2009, 67, 371–380. [Google Scholar] [CrossRef]
- Hao, W.; Wang, L.; Li, F.; Sun, T.; Peng, S.; Li, Y.; Zhao, J.; Dong, Z. Bacterial communities associated with hydromedusa Gonionemus vertens in different regions in Chinese coastal waters. J. Oceanol. Limnol. 2022, 40, 1530–1543. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N.; Pinnow, N.; Langfeldt, D.; Roik, A.; Güllert, S.; Chibani, C.M.; Reusch, T.B.H.; Schmitz, R.A. The native microbiome is crucial for offspring generation and fitness of Aurelia aurita. mBio 2020, 11, e02336-20. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N.; Neulinger, S.C.; Pinnow, N.; Künzel, S.; Baines, J.F.; Schmitz, R.A. Composition of bacterial communities associated with Aurelia aurita changes with compartment, life stage, and population. Appl. Environ. Microbiol. 2015, 81, 6038–6052. [Google Scholar] [CrossRef]
- Dong, C.; Bai, X.; Sheng, H.; Jiao, L.; Zhou, H.; Shao, Z. Distribution of PAHs and the PAH-degrading bacteria in the deep-sea sediments of the high-latitude Arctic Ocean. Biogeosci. Disc. 2014, 11, 13985–14021. [Google Scholar] [CrossRef]
- Gerdes, B.; Brinkmeyer, R.; Dieckmann, G.; Helmke, E. Influence of crude oil on changes of bacterial communities in Arctic sea-ice. FEMS Microbiol. Ecol. 2005, 53, 129–139. [Google Scholar] [CrossRef]
- Urbanczyk, H.; Ast, J.C.; Higgins, M.J.; Carson, J.; Dunlap, P.V. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 2823–2829. [Google Scholar] [CrossRef] [PubMed]
- Nyholm, S.V.; McFall-Ngai, M. The winnowing: Establishing the squid–vibrio symbiosis. Nat. Rev. Microbiol. 2004, 2, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Whistler, C.A.; Ruby, E.G. GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association withan animal host. J. Bacteriol. 2003, 185, 7202–7212. [Google Scholar] [CrossRef] [PubMed]
- Widder, E.A. Bioluminescence in the ocean: Origins of biological, chemical, and ecological diversity. Science 2010, 328, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.W.; Nishiguchi, M.K. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar. Biol. 2004, 144, 1151–1155. [Google Scholar] [CrossRef]
- Ruby, E.G. Symbiotic conversations are revealed under genetic interrogation. Nat. Rev. Microbiol. 2008, 6, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Yu, H.; Li, R.; Xing, R.; Liu, S.; Li, P. Exploring the antibacterial and antifungal potential of jellyfish-associated marine fungi by cultivation-dependent approaches. PLoS ONE 2015, 10, e0144394. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.D.; Osterhage, C.; König, G.M. Epicoccamide, a novel secondary metabolite from a jellyfish-derived culture of Epicoccum purpurascens. Org. Biomol. Chem. 2003, 1, 507–510. [Google Scholar] [CrossRef]
- Cleary, D.F.R.; Becking, L.E.; Polónia, A.R.M.; Freitas, R.M.; Gomes, N.C.M. Jellyfish-associated bacterial communities and bacterioplankton in Indonesian marine lakes. FEMS Microbiol. Ecol. 2016, 92, fiw064. [Google Scholar] [CrossRef]
- Ghotbi, M.; Kelting, O.; Blümel, M.; Tasdemir, D. Gut and gill-associated microbiota of the flatfish European plaice (Pleuronectes platessa): Diversity, metabolome and bioactivity against human and aquaculture pathogens. Mar. Drugs 2022, 20, 573. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef]
- Rohwer, F.; Seguritan, V.; Azam, F.; Knowlton, N. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 2002, 243, 1–10. [Google Scholar] [CrossRef]
- Iacuaniello, C.M. An Examination of Intestinal Microbiota of Mesopelagic Fish Reveals Microbial Community Diversity across Fish Families. Master’s Thesis, University of California San Diego, La Jolla, CA, USA, 2019. [Google Scholar]
- Ovchinnikova, T.V.; Balandin, S.V.; Aleshina, G.M.; Tagaev, A.A.; Leonova, Y.F.; Krasnodembsky, E.D.; Men’shenin, A.V.; Kokryakov, V.N. Aurelin, a novel antimicrobial peptide from jellyfish Aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem. Biophys. Res. Commun. 2006, 348, 514–523. [Google Scholar] [CrossRef]
- Masschelein, J.; Jenner, M.; Challis, G.L. Antibiotics from Gram-negative bacteria: A comprehensive overview and selected biosynthetic highlights. Nat. Prod. Rep. 2017, 34, 712–783. [Google Scholar] [CrossRef] [PubMed]
- Elshafie, H.S.; Viggiani, L.; Mostafa, M.S.; El-Hashash, M.A.; Camele, I.; Bufo, S.A. Biological activity and chemical identification of ornithine lipid produced by Burkholderia gladioli pv. agaricicola ICMP 11096 using LC-MS and NMR analyses. J. Biol. Res. 2017, 90, 1–8. [Google Scholar] [CrossRef]
- Tahara, Y.; Yamada, Y.; Kondo, K. Antimicrobial activity of the ornithine-containing lipid isolated from Gluconobacter cerinus. Agric. Biol. Chem. 1977, 41, 417–418. [Google Scholar] [CrossRef]
- Kristoffersen, V.; Jenssen, M.; Jawad, H.R.; Isaksson, J.; Hansen, E.H.; Rämä, T.; Hansen, K.Ø.; Andersen, J.H. Two novel lyso-ornithine lipids isolated from an Arctic marine Lacinutrix sp. bacterium. Molecules 2021, 26, 5295. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, Z.; Li, J.; Yang, X.; Fei, B.; Leung, P.H.; Tao, X. A new antimicrobial agent: Poly (3-hydroxybutyric acid) oligomer. Macromol. Biosci. 2019, 19, 1800432. [Google Scholar] [CrossRef]
- Dalton, B.; Bhagabati, P.; De Micco, J.; Padamati, R.B.; O’Connor, K. A review on biological synthesis of the biodegradable polymers polyhydroxyalkanoates and the development of multiple applications. Catalysts 2022, 12, 319. [Google Scholar] [CrossRef]
- Abdelwahab, M.A.; Ahmed, A.; El-Said, K.S.; El Naggar, S.A.; ElKholy, H.M. Evaluation of antibacterial and anticancer properties of poly (3-hydroxybutyrate) functionalized with different amino compounds. Int. J. Biol. Macromol. 2019, 122, 793–805. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Kumar, K.; Arora, A.; Katti, D.S. Fabrication and characterization of Pluronic modified poly (hydroxybutyrate) fibers for potential wound dressing applications. Mater. Sci. Eng. C 2016, 63, 266–273. [Google Scholar] [CrossRef]
- Karahaliloğlu, Z.; Ercan, B.; Taylor, E.N.; Chung, S.; Denkbaş, E.B.; Webster, T.J. Antibacterial polyhydroxybutyrate (PHB) membranes for guided bone regeneration. In Proceedings of the Society for Biomaterials Annual Meeting and Exposition, Denver, CO, USA, 16–19 April 2014. [Google Scholar]
- Xavier, J.R.; Babusha, S.T.; George, J.; Ramana, K.V. Material properties and antimicrobial activity of polyhydroxybutyrate (PHB) films incorporated with vanillin. Appl. Biochem. Biotechnol. 2015, 176, 1498–1510. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, X.; Xu, T.; Yang, X.; Liu, Y. Diketopiperazines from marine organisms. Chem. Biodivers. 2010, 7, 2809–2829. [Google Scholar] [CrossRef]
- Huang, R.-M.; Yi, X.-X.; Zhou, Y.; Su, X.; Peng, Y.; Gao, C.-H. An update on 2,5-Diketopiperazines from marine organisms. Mar. Drugs 2014, 12, 6213–6235. [Google Scholar] [CrossRef] [PubMed]
- Utermann, C.; Parrot, D.; Breusing, C.; Stuckas, H.; Staufenberger, T.; Blümel, M.; Labes, A.; Tasdemir, D. Combined genotyping, microbial diversity and metabolite profiling studies on farmed Mytilus spp. from Kiel Fjord. Sci. Rep. 2018, 8, 7983. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Katajamaa, M.; Miettinen, J.; Orešič, M. MZmine: Toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 2006, 22, 634–636. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Allen, F.; Pon, A.; Wilson, M.; Greiner, R.; Wishart, D. CFM-ID: A web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res. 2014, 42, 94–99. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Fan, B.; Parrot, D.; Blümel, M.; Labes, A.; Tasdemir, D. Influence of OSMAC-based cultivation in metabolome and anticancer activity of fungi associated with the brown Alga Fucus vesiculosus. Mar. Drugs 2019, 17, 67. [Google Scholar] [CrossRef]
- Petersen, L.-E.; Marner, M.; Labes, A.; Tasdemir, D. Rapid metabolome and bioactivity profiling of fungi associated with the leaf and rhizosphere of the Baltic seagrass Zostera marina. Mar. Drugs 2019, 17, 419. [Google Scholar] [CrossRef] [PubMed]



| Source | Isolate Number | Isolation Medium | Genus | Class | Phylum |
|---|---|---|---|---|---|
| Outer umbrella | SU122 | MA | Bizionia sp. | Flavobacteriia | Bacteroidota |
| SU123 | MA | Psychrobacter sp. | Gammaproteobacteria | Pseudomonadota | |
| SU124 | MA | Polaribacter sp. | Flavobacteriia | Bacteroidota | |
| SU125 | MA | Salinibacterium sp. | Actinomycetes | Actinomycetota | |
| SU126 | MA | Shewanella sp. | Gammaproteobacteria | Pseudomonadota | |
| SU127 | MA | Pseudoalteromonas sp. | Gammaproteobacteria | Pseudomonadota | |
| SU128 | MA | Psychrobacter sp. | Gammaproteobacteria | Pseudomonadota | |
| SU129 | HS | Vibrio sp. | Gammaproteobacteria | Pseudomonadota | |
| Inner umbrella | SU134 | MA | Polaribacter sp. | Flavobacteriia | Bacteroidota |
| SU135 | MA | Pseudoalteromonas sp. | Gammaproteobacteria | Pseudomonadota | |
| SU136 | MA | Vibrio sp. | Gammaproteobacteria | Pseudomonadota | |
| SU137 | MA | Psychrobacter sp. | Gammaproteobacteria | Pseudomonadota | |
| SU139 | HS | Aliivibrio sp. | Gammaproteobacteria | Pseudomonadota | |
| SU140 | HS | Pseudoalteromonas sp. | Gammaproteobacteria | Pseudomonadota | |
| SU143 | WSP | Psychrobacter sp. | Gammaproteobacteria | Pseudomonadota | |
| SU147 | WSP | Shewanella sp. | Gammaproteobacteria | Pseudomonadota |
| Isolate | Medium-Regime | Efm | MRSA | Lg |
|---|---|---|---|---|
| Polaribacter sp. SU124 | MB-liquid | 67.3 | 7.3 | >100 |
| MB-solid | >100 | 20.9 | >100 | |
| GYM-liquid | >100 | 80.8 | >100 | |
| GYM-solid | >100 | 23.2 | >100 | |
| Shewanella sp. SU126 | MB-liquid | 59.9 | 20.7 | >100 |
| MB-solid | 18.7 | 8.5 | 53.4 | |
| GYM-liquid | >100 | >100 | >100 | |
| GYM-solid | n.t | n.t | n.t | |
| Psychrobacter sp. SU137 | MB-liquid | 37.8 | 21.3 | 69.6 |
| MB-solid | 19.4 | 18.7 | 43.3 | |
| GYM-liquid | 18.7 | 18.5 | 56.2 | |
| GYM-solid | 7.3 | 8.1 | 20.1 | |
| Psychrobacter sp. SU143 | MB-liquid | >100 | 20.9 | >100 |
| MB-solid | >100 | 25.7 | >100 | |
| GYM-liquid | 20.5 | 20.2 | >100 | |
| GYM-solid | 39.6 | 9.9 | >100 | |
| Positive control | 1.6 | 3.1 | 0.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oppong-Danquah, E.; Miranda, M.; Blümel, M.; Tasdemir, D. Bioactivity Profiling and Untargeted Metabolomics of Microbiota Associated with Mesopelagic Jellyfish Periphylla periphylla. Mar. Drugs 2023, 21, 129. https://doi.org/10.3390/md21020129
Oppong-Danquah E, Miranda M, Blümel M, Tasdemir D. Bioactivity Profiling and Untargeted Metabolomics of Microbiota Associated with Mesopelagic Jellyfish Periphylla periphylla. Marine Drugs. 2023; 21(2):129. https://doi.org/10.3390/md21020129
Chicago/Turabian StyleOppong-Danquah, Ernest, Martina Miranda, Martina Blümel, and Deniz Tasdemir. 2023. "Bioactivity Profiling and Untargeted Metabolomics of Microbiota Associated with Mesopelagic Jellyfish Periphylla periphylla" Marine Drugs 21, no. 2: 129. https://doi.org/10.3390/md21020129
APA StyleOppong-Danquah, E., Miranda, M., Blümel, M., & Tasdemir, D. (2023). Bioactivity Profiling and Untargeted Metabolomics of Microbiota Associated with Mesopelagic Jellyfish Periphylla periphylla. Marine Drugs, 21(2), 129. https://doi.org/10.3390/md21020129







