Construction of Glucose-6-Phosphate Dehydrogenase Overexpression Strain of Schizochytrium sp. H016 to Improve Docosahexaenoic Acid Production
Abstract
1. Introduction
2. Results
2.1. Construction of G6PD Overexpression Strains in Schizochytrium sp. H016
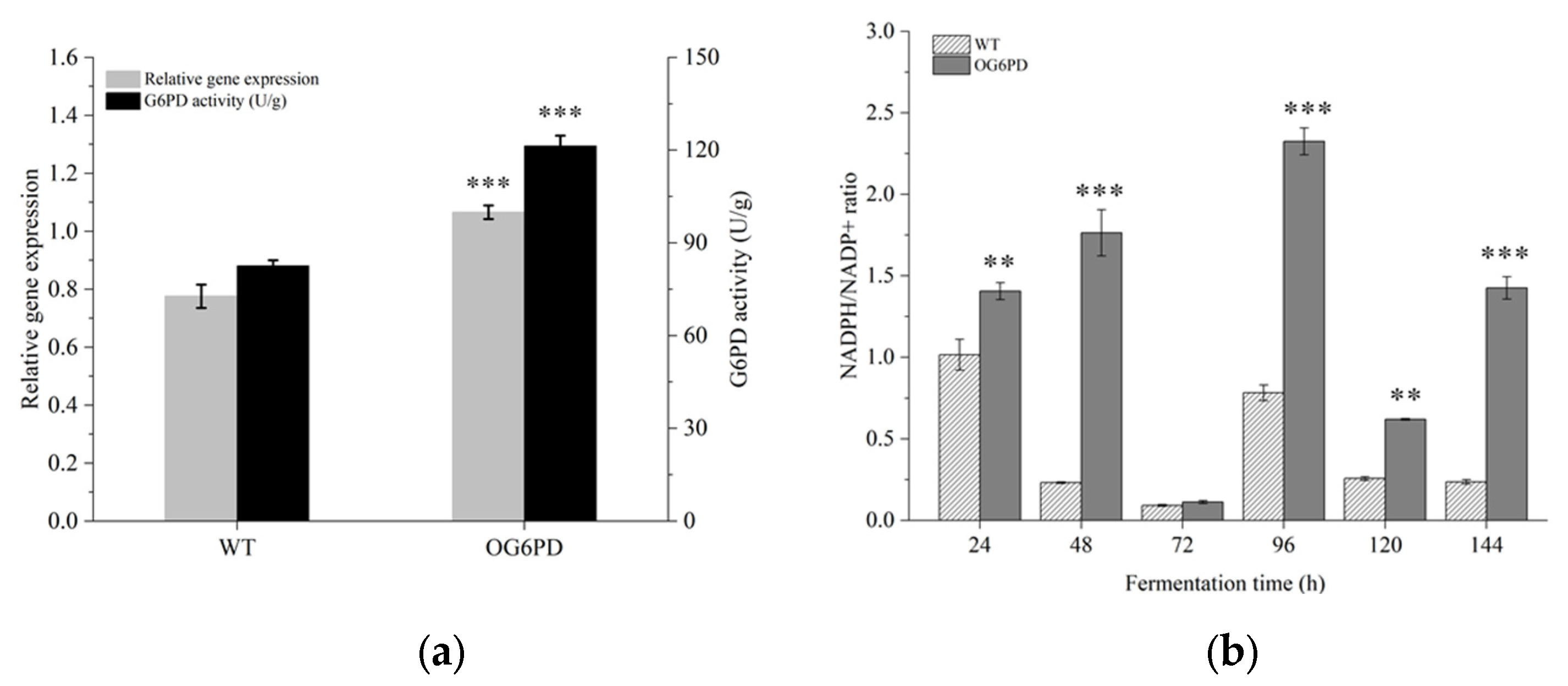
2.2. Validation of the Effects of G6PD Gene Overexpression in Schizochytrium sp. H016
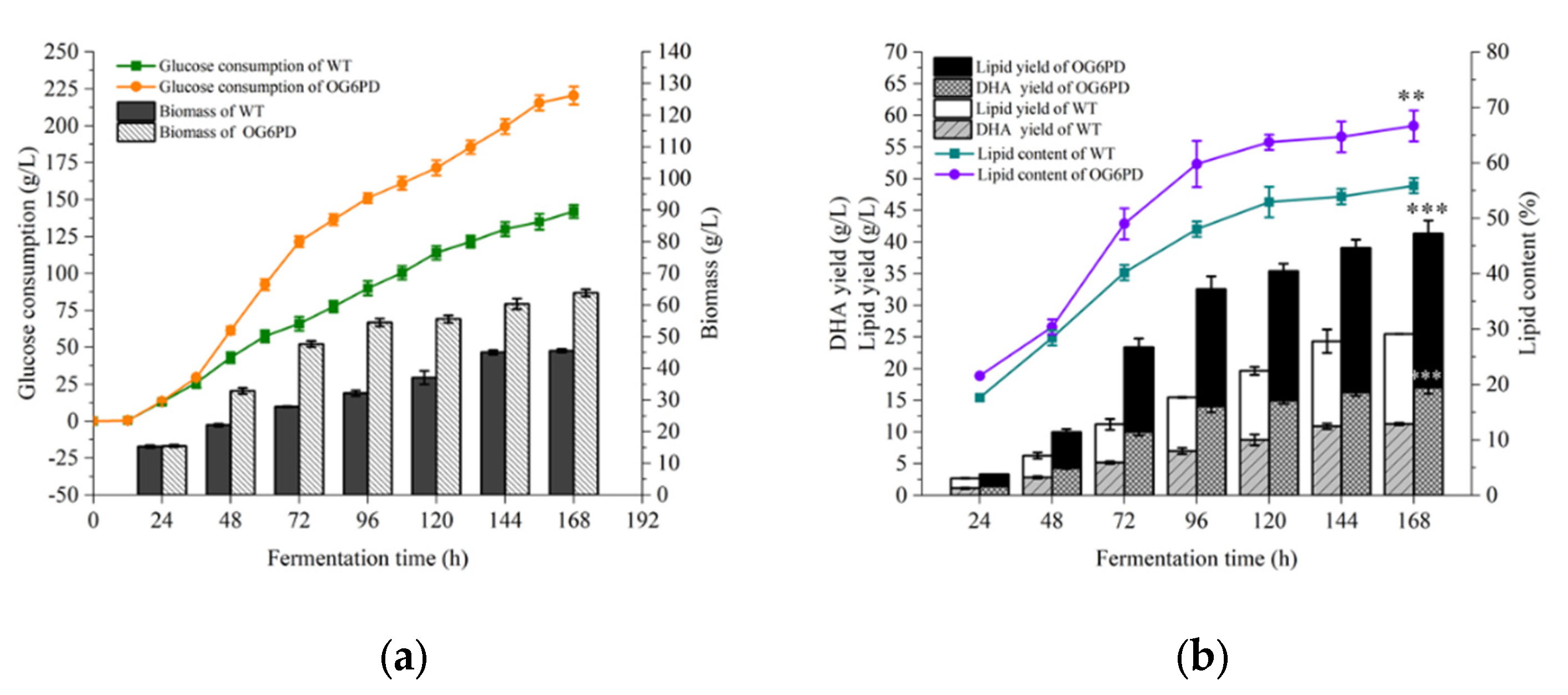
2.3. Influence of G6PD Gene Overexpression on the Fermentation Properties of Schizochytrium sp. H016
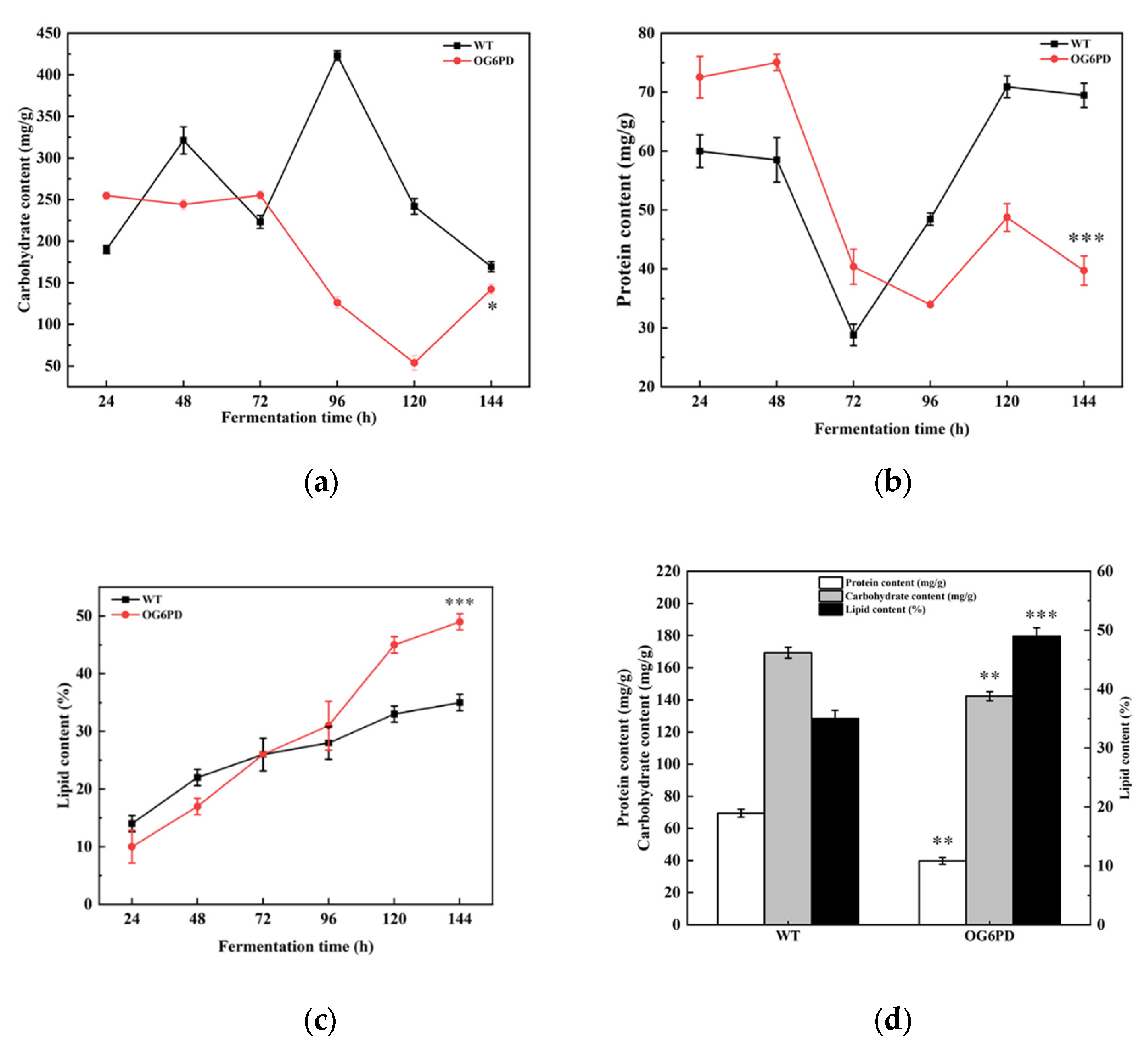
2.4. Overexpression of G6PD Gene can Promote the Accumulation of Lipid Content
2.5. Primary Metabolite Content Was Altered with G6PD Overexpression
2.6. Key Enzymes’ Expression Levels Were Verified by Quantitative Real-Time PCR
2.7. Fed-Batch Fermentation Characterization of the OG6PD Strain
3. Discussion
4. Materials and Methods
4.1. Microorganisms and Culture Conditions
4.2. G6PD Gene Cloning, Analysis, and Plasmid Construction
4.3. Validation Transformation of Schizochytrium sp. H016
4.4. Dry Cell Weight, Glucose Concentration, and Lipid Content Measurements
4.5. Analysis of Fatty Acid Component and Determination of Neutral Properties
4.6. Detection of Carbohydrate and Protein Content
4.7. Preparation of Schizochytrium sp. H016 Cell Extracts, Determination of Enzyme Activity and NADPH Content
4.8. Quantitative Real-Time PCR (RT-qPCR) Assay
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, S.; Chen, R. Docosahexaenoic acid-containing phospholipids and triglycerides based nutritional supplements. Recent Pat. Food Nutr. Agric. 2010, 2, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Docosahexaenoic Acid. Ann. Nutr. Metab. 2016, 69 (Suppl. 1), 7–21. [Google Scholar] [CrossRef] [PubMed]
- Jing, K.; Wu, T.; Lim, K. Omega-3 polyunsaturated fatty acids and cancer. Anticancer Agents Med. Chem. 2013, 13, 1162–1177. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Ambigaipalan, P. Omega-3 Polyunsaturated Fatty Acids and Their Health Benefits. Annu. Rev. Food Sci. Technol. 2018, 9, 345–381. [Google Scholar] [CrossRef] [PubMed]
- Weiser, M.J.; Butt, C.M.; Mohajeri, M.H. Docosahexaenoic Acid and Cognition throughout the Lifespan. Nutrients 2016, 8, 99. [Google Scholar] [CrossRef]
- Finco, A.M.O.; Mamani, L.D.G.; Carvalho, J.C.; de Melo Pereira, G.V.; Thomaz-Soccol, V.; Soccol, C.R. Technological trends and market perspectives for production of microbial oils rich in omega-3. Crit. Rev. Biotechnol. 2017, 37, 656–671. [Google Scholar] [CrossRef]
- Hathaway, D.; Pandav, K.; Patel, M.; Riva-Moscoso, A.; Singh, B.M.; Patel, A.; Min, Z.C.; Singh-Makkar, S.; Sana, M.K.; Sanchez-Dopazo, R.; et al. Omega 3 Fatty Acids and COVID-19: A Comprehensive Review. Infect. Chemother. 2020, 52, 478–495. [Google Scholar] [CrossRef]
- Rasool, M.S.; Siddiqui, F.; Hassan, M.A.; Hafiz, S. Novel Coronavirus-2019 (2019-nCoV): Perspectives of emergence, prophylaxis and predicted treatment approaches. Pak. J. Pharm. Sci. 2020, 33, 2199–2207. [Google Scholar]
- Rogero, M.M.; Leão, M.C.; Santana, T.M.; Pimentel, M.; Carlini, G.C.G.; da Silveira, T.F.F.; Gonçalves, R.C.; Castro, I.A. Potential benefits and risks of omega-3 fatty acids supplementation to patients with COVID-19. Free Radic. Biol. Med. 2020, 156, 190–199. [Google Scholar] [CrossRef]
- Martins, D.A.; Custodio, L.; Barreira, L.; Pereira, H.; Ben-Hamadou, R.; Varela, J.; Abu-Salah, K.M. Alternative Sources of n-3 Long-Chain Polyunsaturated Fatty Acids in Marine Microalgae. Mar. Drugs 2013, 11, 2259–2281. [Google Scholar] [CrossRef]
- Ward, O.P.; Singh, A. Omega-3/6 fatty acids: Alternative sources of production. Process Biochem. 2005, 40, 3627–3652. [Google Scholar] [CrossRef]
- Adarme-Vega, T.C.; Thomas-Hall, S.R.; Schenk, P.M. Towards sustainable sources for omega-3 fatty acids production. Curr. Opin. Biotechnol. 2014, 26, 14–18. [Google Scholar] [CrossRef]
- Dellero, Y.; Cagnac, O.; Rose, S.; Seddiki, K.; Cussac, M.; Morabito, C.; Lupette, J.; Aiese Cigliano, R.; Sanseverino, W.; Kuntz, M.; et al. Proposal of a new thraustochytrid genus Hondaea gen. nov. and comparison of its lipid dynamics with the closely related pseudo-cryptic genus Aurantiochytrium. Algal Res. 2018, 35, 125–141. [Google Scholar] [CrossRef]
- Metz, J.G.; Kuner, J.; Rosenzweig, B.; Lippmeier, J.C.; Roessler, P.; Zirkle, R. Biochemical characterization of polyunsaturated fatty acid synthesis in Schizochytrium: Release of the products as free fatty acids. Plant Physiol. Biochem. 2009, 47, 472–478. [Google Scholar] [CrossRef]
- Ling, X.; Guo, J.; Liu, X.; Zhang, X.; Wang, N.; Lu, Y.; Ng, I.S. Impact of carbon and nitrogen feeding strategy on high production of biomass and docosahexaenoic acid (DHA) by Schizochytrium sp. LU310. Bioresour. Technol. 2015, 184, 139–147. [Google Scholar] [CrossRef]
- Raghukumar, S.J.M.B. Thraustochytrid Marine Protists: Production of PUFAs and Other Emerging Technologies. Mar. Biotechnol. 2008, 10, 631–640. [Google Scholar] [CrossRef]
- Sun, X.M.; Xu, Y.S.; Huang, H. Thraustochytrid Cell Factories for Producing Lipid Compounds. Trends Biotechnol. 2021, 39, 648–650. [Google Scholar] [CrossRef]
- Cui, G.Z.; Ma, Z.X.; Liu, Y.J.; Feng, Y.G.; Sun, Z.J.; Cheng, Y.R.; Song, X.J.; Cui, Q. Overexpression of glucose-6-phosphate dehydrogenase enhanced the polyunsaturated fatty acid composition of Aurantiochytrium sp. SD116. Algal Res. 2016, 19, 138–145. [Google Scholar] [CrossRef]
- Ratledge, C. The role of malic enzyme as the provider of NADPH in oleaginous microorganisms: A reappraisal and unsolved problems. Biotechnol. Lett. 2014, 36, 1557–1568. [Google Scholar] [CrossRef]
- McKenna, M.C.; Stevenson, J.H.; Huang, X.L.; Tildon, J.T.; Zielke, C.L.; Hopkins, I.B. Mitochondrial malic enzyme activity is much higher in mitochondria from cortical synaptic terminals compared with mitochondria from primary cultures of cortical neurons or cerebellar granule cells. Neurochem. Int. 2000, 36, 451–459. [Google Scholar] [CrossRef]
- Li, Z.; Sun, H.X.; Mo, X.M.; Li, X.Y.; Xu, B.; Tian, P. Overexpression of malic enzyme (ME) of Mucor circinelloides improved lipid accumulation in engineered Rhodotorula glutinis. Appl. Microbiol. Biot. 2013, 97, 4927–4936. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.H.; Kim, M.D.; Jin, Y.S.; Seo, J.H. Engineering of NADPH regenerators in Escherichia coli for enhanced biotransformation. Appl. Microbiol. Biot. 2013, 97, 2761–2772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Adams, I.P.; Ratledge, C. Malic enzyme: The controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiol. Read. 2007, 153, 2013–2025. [Google Scholar] [CrossRef]
- Xue, J.; Chen, T.T.; Zheng, J.W.; Balamurugan, S.; Liu, Y.H.; Yang, W.D.; Liu, J.S.; Li, H.Y. Glucose-6-Phosphate Dehydrogenase from the Oleaginous Microalga Nannochloropsis Uncovers Its Potential Role in Promoting Lipogenesis. Biotechnol. J. 2020, 15, 1900135. [Google Scholar] [CrossRef] [PubMed]
- Kruger, N.J.; von Schaewen, A. The oxidative pentose phosphate pathway: Structure and organisation. Curr. Opin. Plant Biol. 2003, 6, 236–246. [Google Scholar] [CrossRef]
- Ren, L.J.; Huang, H.; Xiao, A.H.; Lian, M.; Jin, L.J.; Ji, X.J. Enhanced docosahexaenoic acid production by reinforcing acetyl-CoA and NADPH supply in Schizochytrium sp. HX-308. Bioproc. Biosyst. Eng. 2009, 32, 837–843. [Google Scholar] [CrossRef]
- Li, Z.; Ling, X.; Zhou, H.; Meng, T.; Zeng, J.; Hang, W.; Shi, Y.; He, N. Screening chemical modulators of benzoic acid derivatives to improve lipid accumulation in Schizochytrium limacinum SR21 with metabolomics analysis. Biotechnol. Biofuels 2019, 12, 209. [Google Scholar] [CrossRef]
- Bao, Z.; Zhu, Y.; Feng, Y.; Zhang, K.; Zhang, M.; Wang, Z.; Yu, L. Enhancement of lipid accumulation and docosahexaenoic acid synthesis in Schizochytrium sp. H016 by exogenous supplementation of sesamol. Bioresour. Technol. 2022, 345, 126527. [Google Scholar] [CrossRef]
- Yuzbasheva, E.Y.; Mostova, E.B.; Andreeva, N.I.; Yuzbashev, T.V.; Laptev, I.A.; Sobolevskaya, T.I.; Sineoky, S.P. Co-expression of glucose-6-phosphate dehydrogenase and acyl-CoA binding protein enhances lipid accumulation in the yeast Yarrowia lipolytica. New Biotechnol. 2017, 39, 18–21. [Google Scholar] [CrossRef]
- Reisz, J.A.; Tzounakas, V.L.; Nemkov, T.; Voulgaridou, A.I.; Papassideri, I.S.; Kriebardis, A.G.; D’Alessandro, A.; Antonelou, M.H. Metabolic Linkage and Correlations to Storage Capacity in Erythrocytes from Glucose 6-Phosphate Dehydrogenase-Deficient Donors. Front. Med. 2017, 4, 248. [Google Scholar] [CrossRef]
- Xue, J.; Balamurugan, S.; Li, D.-W.; Liu, Y.-H.; Zeng, H.; Wang, L.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Glucose-6-phosphate dehydrogenase as a target for highly efficient fatty acid biosynthesis in microalgae by enhancing NADPH supply. Metab. Eng. 2017, 41, 212–221. [Google Scholar] [CrossRef]
- Hao, G.; Chen, H.; Gu, Z.; Zhang, H.; Chen, W.; Chen, Y.Q. Metabolic Engineering of Mortierella alpina for Enhanced Arachidonic Acid Production through the NADPH-Supplying Strategy. Appl. Environ. Microbiol. 2016, 82, 3280–3288. [Google Scholar] [CrossRef]
- Hurbain, J.; Thommen, Q.; Anquez, F.; Pfeuty, B. Quantitative modeling of pentose phosphate pathway response to oxidative stress reveals a cooperative regulatory strategy. Iscience 2022, 25, 104681. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, X.; Balamurugan, S.; Yang, W.D.; Liu, J.S.; Dong, H.P.; Li, H.Y. Identification of a putative seipin ortholog involved in lipid accumulation in marine microalga Phaeodactylum tricornutum. J. Appl. Phycol. 2017, 29, 2821–2829. [Google Scholar] [CrossRef]
- Aleman-Nava, G.S.; Cuellar-Bermudez, S.P.; Cuaresma, M.; Bosma, R.; Muylaert, K.; Ritmann, B.E.; Parra, R. How to use Nile Red, a selective fluorescent stain for microalgal neutral lipids. J. Microbiol. Methods 2016, 128, 74–79. [Google Scholar] [CrossRef]
- Escorcia, W.; Ruter, D.L.; Nhan, J.; Curran, S.P. Quantification of Lipid Abundance and Evaluation of Lipid Distribution in Caenorhabditis elegans by Nile Red and Oil Red O Staining. Jove-J. Vis. Exp. 2018, 133, e57352. [Google Scholar] [CrossRef]
- Bianchi, G.; Marzocchi, R.; Lorusso, C.; Ridolfi, V.; Marchesini, G. Nutritional treatment of chronic liver failure. Hepatol. Res. 2008, 38 (Suppl. 1), S93–S101. [Google Scholar] [CrossRef]
- Chen, Y.; Vaidyanathan, S. Simultaneous assay of pigments, carbohydrates, proteins and lipids in microalgae. Anal. Chim. Acta 2013, 776, 31–40. [Google Scholar] [CrossRef]
- Lv, J.-M.; Cheng, L.-H.; Xu, X.-H.; Zhang, L.; Chen, H.-L. Enhanced lipid production of Chlorella vulgaris by adjustment of cultivation conditions. Bioresour. Technol. 2010, 101, 6797–6804. [Google Scholar] [CrossRef]
- Hasan, H.; Abd Rahim, M.H.; Campbell, L.; Carter, D.; Abbas, A.; Montoya, A. Overexpression of acetyl-CoA carboxylase in Aspergillus terreus to increase lovastatin production. New Biotechnol. 2018, 44, 64–71. [Google Scholar] [CrossRef]
- Han, X.; Zhao, Z.N.; Wen, Y.; Chen, Z. Enhancement of docosahexaenoic acid production by overexpression of ATP-citrate lyase and acetyl-CoA carboxylase in Schizochytrium sp. Biotechnol. Biofuels 2020, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zamboni, N.; Fischer, E.; Laudert, D.; Aymerich, S.; Hohmann, H.P.; Sauer, U. The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway. J. Bacteriol. 2004, 186, 4528–4534. [Google Scholar] [CrossRef] [PubMed]
- Metz, J.G.; Roessler, P.; Facciotti, D.; Levering, C.; Dittrich, F.; Lassner, M.; Valentine, R.; Lardizabal, K.; Domergue, F.; Yamada, A.; et al. Production of Polyunsaturated Fatty Acids by Polyketide Synthases in Both Prokaryotes and Eukaryotes. Science 2001, 293, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Kiegerl, B.; Tavassoli, M.; Smart, H.; Shabits, B.N.; Zaremberg, V.; Athenstaedt, K. Phosphorylation of the lipid droplet localized glycerol-3-phosphate acyltransferase Gpt2 prevents a futile triacylglycerol cycle in yeast. Biochim. Biophys. Acta BBA–Mol. Cell Biol. Lipids 2019, 1864, 158509. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Hirasawa, E.; Takemura, T.; Takahashi, S.; Chokshi, K.; Pancha, I.; Tanaka, K.; Imamura, S. Accelerated triacylglycerol production without growth inhibition by overexpression of a glycerol-3-phosphate acyltransferase in the unicellular red alga Cyanidioschyzon merolae. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Pietrocola, F.; Galluzzi, L.; Bravo-San Pedro, J.M.; Madeo, F.; Kroemer, G. Acetyl Coenzyme A: A Central Metabolite and Second Messenger. Cell Metab. 2015, 21, 805–821. [Google Scholar] [CrossRef]
- Liu, Y.-Z.; Xu, M.-Y.; Dai, X.-Y.; Yan, L.; Li, L.; Zhu, R.-Z.; Ren, L.-J.; Zhang, J.-Q.-Z.; Zhang, X.-F.; Li, J.-F.; et al. Pyruvate Kinase M2 Mediates Glycolysis Contributes to Psoriasis by Promoting Keratinocyte Proliferation. Front. Pharmacol. 2021, 12, 765790. [Google Scholar] [CrossRef]
- Jia, J.; Han, D.X.; Gerken, H.G.; Li, Y.T.; Sommerfeld, M.; Hu, Q.; Xu, J. Molecular mechanisms for photosynthetic carbon partitioning into storage neutral lipids in Nannochloropsis oceanica under nitrogen-depletion conditions. Algal Res. 2015, 7, 66–77. [Google Scholar] [CrossRef]
- Michel, G.; Tonon, T.; Scornet, D.; Cock, J.M.; Kloareg, B. Central and storage carbon metabolism of the brown alga Ectocarpus siliculosus: Insights into the origin and evolution of storage carbohydrates in Eukaryotes. New Phytol. 2010, 188, 67–81. [Google Scholar] [CrossRef]
- Andre, C.; Froehlich, J.E.; Moll, M.R.; Benning, C. A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2006–2022. [Google Scholar] [CrossRef]
- Baud, S.; Wuilleme, S.; Dubreucq, B.; de Almeida, A.; Vuagnat, C.; Lepiniec, L.; Miquel, M.; Rochat, C. Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J. 2007, 52, 405–419. [Google Scholar] [CrossRef]
- Tian, W.-N.; Braunstein, L.D.; Pang, J.; Stuhlmeier, K.M.; Xi, Q.-C.; Tian, X.; Stanton, R.C. Importance of Glucose-6-phosphate Dehydrogenase Activity for Cell Growth*. J. Biol. Chem. 1998, 273, 10609–10617. [Google Scholar] [CrossRef]
- Ho, H.-y.; Cheng, M.-l.; Lu, F.-j.; Chou, Y.-h.; Stern, A.; Liang, C.-m.; Chiu, D.T.-y. Enhanced oxidative stress and accelerated cellular senescence in glucose-6-phosphate dehydrogenase (G6PD)-deficient human fibroblasts. Free Radic. Biol. Med. 2000, 29, 156–169. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Z.; Hoshino, A.; Zheng, H.D.; Morley, M.; Arany, Z.; Rabinowitz, J.D. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat. Metab. 2019, 1, 404–415. [Google Scholar] [CrossRef]
- Urbano, S.B.; Di Capua, C.; Cortez, N.; Farias, M.E.; Alvarez, H.M. Triacylglycerol accumulation and oxidative stress in Rhodococcus species: Differential effects of pro-oxidants on lipid metabolism. Extremophiles 2014, 18, 375–384. [Google Scholar] [CrossRef]
- Zhang, Z.; Liew, C.W.; Handy, D.E.; Zhang, Y.; Leopold, J.A.; Hu, J.; Guo, L.; Kulkarni, R.N.; Loscalzo, J.; Stanton, R.C. High glucose inhibits glucose-6-phosphate dehydrogenase, leading to increased oxidative stress and beta-cell apoptosis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 1497–1505. [Google Scholar]
- Chen, M.; Tang, H.; Ma, H.; Holland, T.C.; Ng, K.Y.; Salley, S.O. Effect of nutrients on growth and lipid accumulation in the green algae Dunaliella tertiolecta. Bioresour. Technol. 2011, 102, 1649–1655. [Google Scholar] [CrossRef]
- Converti, A.; Perego, P.; Sordi, A.; Torre, P. Effect of starting xylose concentration on the microaerobic metabolism of Debaryomyces hansenii—The use of carbon material balances. Appl. Biochem. Biotech. 2002, 101, 15–29. [Google Scholar] [CrossRef]
- Patra, K.C.; Hay, N. The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014, 39, 347–354. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J.; Sun, P.P.; Ma, X.N.; Jiang, Y.; Chen, F. Sesamol Enhances Cell Growth and the Biosynthesis and Accumulation of Docosahexaenoic Acid in the Microalga Crypthecodinium cohnii. J. Agric. Food Chem. 2015, 63, 5640–5645. [Google Scholar] [CrossRef]
- Song, X.J.; Tan, Y.Z.; Liu, Y.J.; Zhang, J.T.; Liu, G.L.; Feng, Y.G.; Cui, Q. Different Impacts of Short-Chain Fatty Acids on Saturated and Polyunsaturated Fatty Acid Biosynthesis in Aurantiochytrium sp. SD116. J. Agric. Food Chem. 2013, 61, 9876–9881. [Google Scholar] [CrossRef] [PubMed]
- Indarti, E.; Majid, M.I.A.; Hashim, R.; Chong, A. Direct FAME synthesis for rapid total lipid analysis from fish oil and cod liver oil. J. Food Compos. Anal. 2005, 18, 161–170. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, C.; Song, L.; Sommerfeld, M.; Hu, Q. A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J. Microbiol. Methods 2009, 77, 41–47. [Google Scholar] [CrossRef] [PubMed]




| Fatty Acid Composition | Fatty Acid Content (% of Total FA) | Fatty Acid Yield (g/L) | ||
|---|---|---|---|---|
| WT | OG6PD | WT | OG6PD | |
| C14:0 | 1.65 ± 0.22 | 1.35 ± 0.01 | 0.21 ± 0.03 | 0.25 ± 0.01 |
| C15:0 | 7.68 ± 1.46 | 3.32 ± 0.54 | 0.97 ± 0.18 | 0.61 ± 0.10 |
| C16:0 | 36.33 ± 0.78 | 28.22 ± 0.16 | 4.57 ± 0.10 | 5.16 ± 0.03 |
| C17:0 | 2.73 ± 0.43 | 1.20 ± 0.18 | 0.34 ± 0.05 | 0.22 ± 0.04 |
| C18:0 | 1.39 ± 0.04 | 1.12 ± 0.08 | 0.17 ± 0.01 | 0.20 ± 0.01 |
| EPA | 0.50 ± 0.03 | 0.51 ± 0.07 | 0.06 ± 0.02 | 0.09 ± 0.01 |
| DPA | 8.79 ± 0.30 | 11.07 ± 0.01 | 1.10 ± 0.04 | 2.02 ± 0.02 |
| DHA | 36.64 ± 0.23 | 48.17 ± 1.10 | 4.60 ± 0.03 | 8.81 ± 0.20 |
| SFA | 49.77 ± 1.29 | 35.20 ± 0.64 | 6.25 ± 0.16 | 6.43 ± 0.12 |
| PUFA | 45.93 ± 0.49 | 59.75 ± 1.04 | 5.78 ± 0.07 | 10.92 ± 0.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Y.; Zhu, Y.; Bao, Z.; Wang, B.; Liu, T.; Wang, H.; Yu, T.; Yang, Y.; Yu, L. Construction of Glucose-6-Phosphate Dehydrogenase Overexpression Strain of Schizochytrium sp. H016 to Improve Docosahexaenoic Acid Production. Mar. Drugs 2023, 21, 17. https://doi.org/10.3390/md21010017
Feng Y, Zhu Y, Bao Z, Wang B, Liu T, Wang H, Yu T, Yang Y, Yu L. Construction of Glucose-6-Phosphate Dehydrogenase Overexpression Strain of Schizochytrium sp. H016 to Improve Docosahexaenoic Acid Production. Marine Drugs. 2023; 21(1):17. https://doi.org/10.3390/md21010017
Chicago/Turabian StyleFeng, Yumei, Yuanmin Zhu, Zhendong Bao, Bohan Wang, Tingting Liu, Huihui Wang, Tianyi Yu, Ying Yang, and Longjiang Yu. 2023. "Construction of Glucose-6-Phosphate Dehydrogenase Overexpression Strain of Schizochytrium sp. H016 to Improve Docosahexaenoic Acid Production" Marine Drugs 21, no. 1: 17. https://doi.org/10.3390/md21010017
APA StyleFeng, Y., Zhu, Y., Bao, Z., Wang, B., Liu, T., Wang, H., Yu, T., Yang, Y., & Yu, L. (2023). Construction of Glucose-6-Phosphate Dehydrogenase Overexpression Strain of Schizochytrium sp. H016 to Improve Docosahexaenoic Acid Production. Marine Drugs, 21(1), 17. https://doi.org/10.3390/md21010017