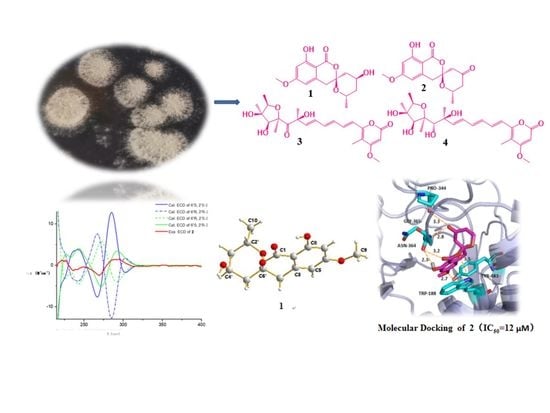
New Polyketides from Mangrove Endophytic Fungus Penicillium sp. BJR-P2 and Their Anti-Inflammatory Activity
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification and Purification
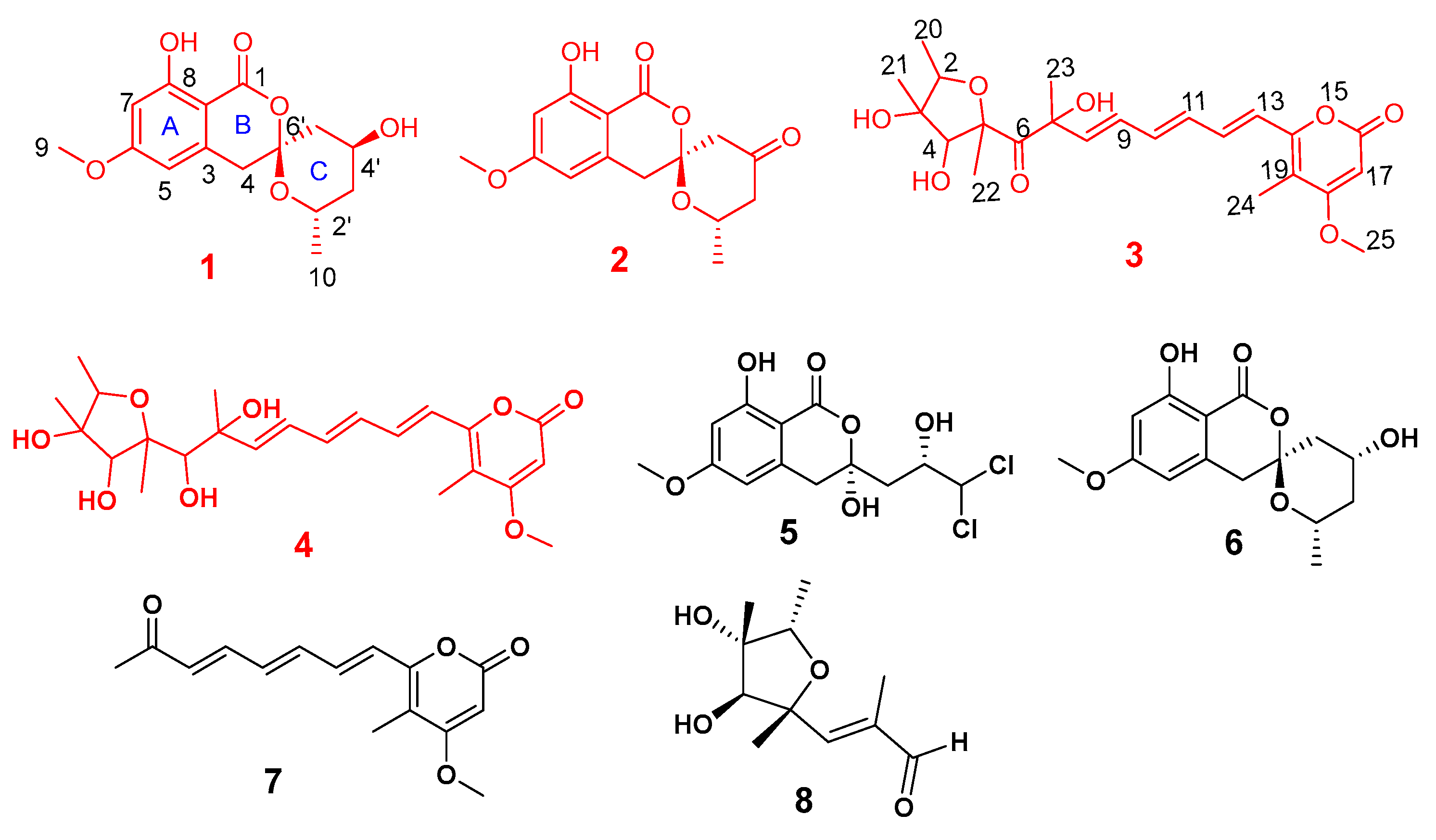
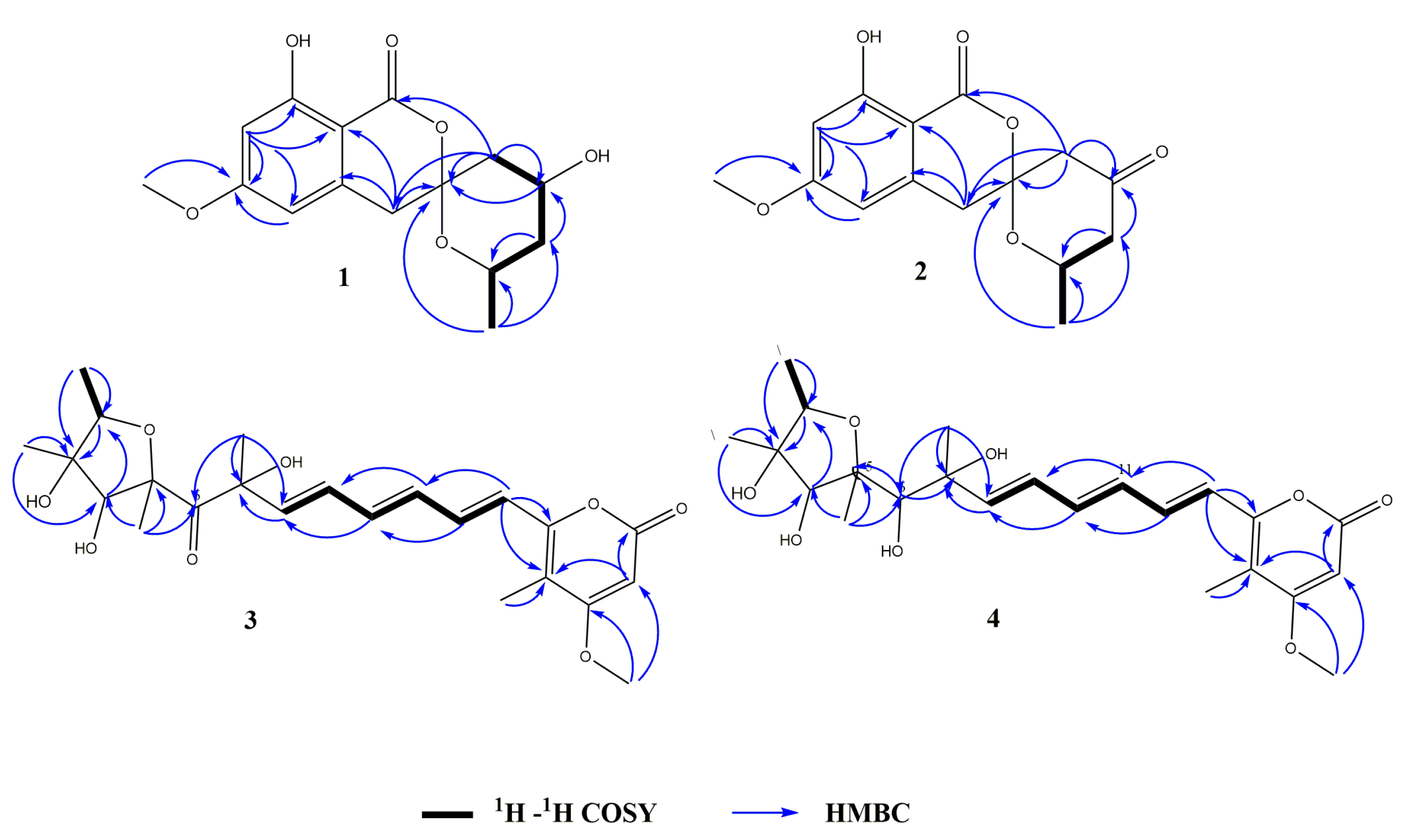
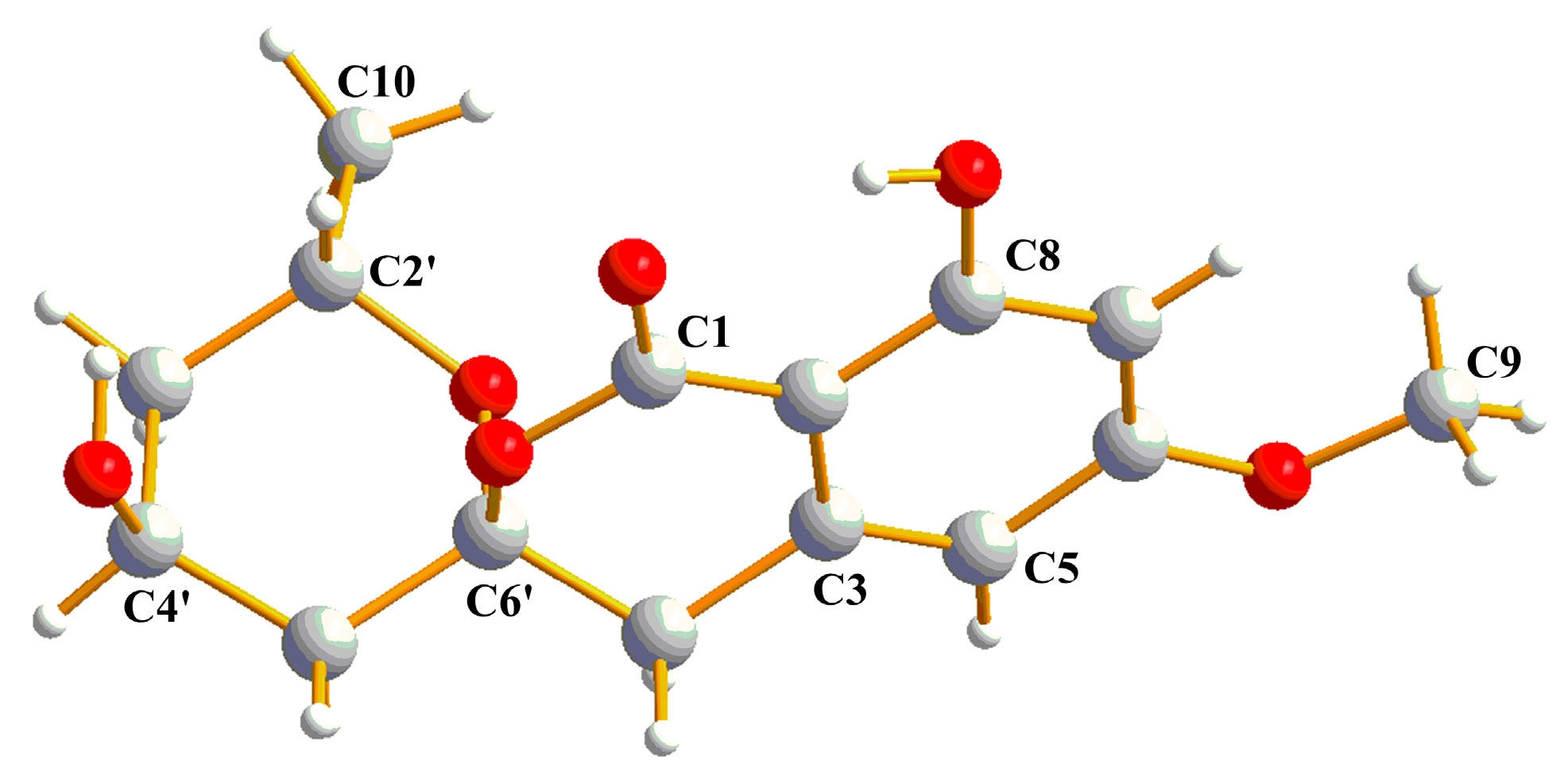
2.2. Structural Elucidation
2.3. Anti-Inflammatory Activity
2.4. Molecular Docking Studies
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Extraction and Isolation
3.4. ECD and 13C NMR Calculation
3.5. Anti-Inflammatory Assays
3.6. Molecular Docking Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Savill, J. Resolution of inflammation: The beginning programs the end. Nat. Immunol. 2005, 6, 1191–1197. [Google Scholar] [CrossRef]
- Pierce, G.F. Macrophages: Important physiologic and pathologic sources of polypeptide growth factors. Am. J. Resp. Cell Mol. 1990, 2, 233–234. [Google Scholar] [CrossRef]
- Moghaddam, A.S.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, M.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef] [PubMed]
- Carl, N. Points of control in inflammation. Nature 2002, 420, 846–852. [Google Scholar]
- Glancy, R.M.; Amin, A.R.; Abramson, S.B. The role of nitric oxide in inflammation and immunity. Arthritis Rheum. 1998, 41, 1111–1151. [Google Scholar]
- Yang, Y.Z.; Wei, Z.; Teichmann, A.T.; Wieland, F.H.; Wang, A.; Lei, X.G.; Zhu, Y.; Yin, J.X.; Fan, T.T.; Zhou, L.; et al. Development of a novel nitric Oxide (NO) production inhibitor with potential therapeutic effect on chronic inflammation. Eur. J. Med. Chem. 2020, 193, 112216. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.N.; Mejia, E.G.D.; Wu, J.S. Inhibitory effect of a glycoprotein isolated from golden oyster mushroom (pleurotus citrinopileatus) on the lipopolysaccharide-induced inflammatory reaction in RAW 264.7 macrophage. J. Agric. Food Chem. 2011, 59, 7092–7097. [Google Scholar] [CrossRef]
- Dalsgaard, P.W.; Blunt, J.W.; Munro, M.H.; Frisvad, J.C.; Christophersen, C. Communesins G and H, new alkaloids from the psychrotolerant fungus Penicillium rivulum. J. Nat. Prod. 2005, 68, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Orfali, R.; Perveen, S.; Al-Taweel, A.; Ahmed, A.F.; Majrashi, N.; Alluhay, K.; Khan, A.; Luciano, P.; Taglialatela-Scafati, O. Penipyranicins A-C: Antibacterial methylpyran polyketides from a hydrothermal spring sediment Penicillium sp. J. Nat. Prod. 2020, 83, 3591–3597. [Google Scholar] [CrossRef]
- Dalsgaard, P.W.; Larsen, T.O.; Frydenvang, K.; Christophersen, C. Psychrophilin A and cycloaspeptide D, novel cyclic peptides from the psychrotolerant fungus Penicillium ribeum. J. Nat. Prod. 2004, 67, 878–881. [Google Scholar] [CrossRef] [PubMed]
- Zhuravleva, O.I.; Sobolevskaya, M.P.; Leshchenko, E.V.; Kirichuk, N.N.; Denisenko, V.A.; Dmitrenok, P.S.; Dyshlovoy, S.A.; Zakharenko, A.M.; Kim, N.Y.; Afiyatullov, S. Meroterpenoids from the alga-derived fungi Penicillium thomii Maire and Penicillium lividum Westling. J. Nat. Prod. 2014, 77, 1390–1395. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Wang, W.; Li, X.; Zhang, Y.; Wang, L.; Yuan, C. A novel isocoumarin with anti-influenza virus activity from strobilanthes cusia. Fitoterapia 2015, 107, 60–62. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Lu, C.H.; Zheng, Z.H. A new 3,4-dihydroisocoumarin isolated from Botryosphaeria sp. F00741. Chem. Nat. Comp. 2012, 48, 205–207. [Google Scholar] [CrossRef][Green Version]
- Zhao, Y.; Liu, D.; Proksch, P.; Yu, S.; Lin, W.H. Isocoumarin derivatives from the sponge-associated fungus Peyronellaea glomerata with antioxidant activities. Chem. Biodivers. 2016, 3, 1186–1193. [Google Scholar] [CrossRef] [PubMed]
- Girich, E.V.; Yurchenko, A.N.; Smetanina, O.F.; Trinh, P.T.H.; Ngoc, M.T.D.; Pivkin, M.V.; Popov, R.S.; Pislyagin, E.A.; Menchinskaya, E.S.; Chingizova, E.A.; et al. Neuroprotective metabolites from vietnamese marine derived fungi of Aspergillus and Penicillium genera. Mar. Drugs 2020, 18, 608. [Google Scholar] [CrossRef]
- Gu, B.B.; Wu, Y.; Tang, J.; Jiao, W.H.; Li, L.; Sun, F.; Wang, S.P.; Yang, F.; Lin, H.W. Azaphilone and isocoumarin derivatives from the sponge-derived fungus Eupenicillium sp. 6A-9. Tetrahedron Lett. 2018, 59, 3345–3348. [Google Scholar] [CrossRef]
- Shabir, G.; Saeed, A.; El-Seedi, H.R. Natural isocoumarins: Structural styles and biological activities, the revelations carry on. Phytochemistry 2021, 181, 112568. [Google Scholar] [CrossRef] [PubMed]
- Noor, A.O.; Almasri, D.M.; Bagalagel, A.A.; Abdallah, H.M.; Mohamed, S.G.A.; Mohamed, G.A.; Ibrahim, S.R.M. Naturally Occurring Isocoumarins Derivatives from Endophytic Fungi: Sources, Isolation, Structural Characterization, Biosynthesis, and Biological Activities. Molecules 2020, 25, 395. [Google Scholar] [CrossRef] [PubMed]
- Shizuri, Y.; Shigemori, H.; Sato, R.; Yamamura, S.; Kawai, K.; Furukawa, H. Four new metabolites produced by Penicillium citreo-viride B. on addition of NaBr. Chem. Lett. 2006, 17, 1419–1422. [Google Scholar] [CrossRef]
- Nagel, D.W.; Steyn, P.S.; Scott, D.B. Production of citreoviridin by Penicillium pulvillorum. Phytochemistry 1972, 11, 627–630. [Google Scholar] [CrossRef]
- Yang, M.H.; Li, T.X.; Wang, Y.; Liu, R.H.; Luo, J.; Kong, L.Y. Antimicrobial metabolites from the plant endophytic fungus Penicillium sp. Fitoterapia 2017, 116, 72–76. [Google Scholar] [CrossRef]
- Kosemura, S.; Kojima, S.I.; Yamamura, S. Citreopyrones, new metabolites of two hybrid strains, KO 0092 and KO 0141, derived from the Penicillium species. Chem. Lett. 1997, 26, 33–34. [Google Scholar] [CrossRef]
- Shizuri, Y.; Nishiyama, S.; Imai, D.; Yamamura, S. Isolation and stereostructures of citreoviral, citreodiol, and epicitreodiol. Tetrahedron Lett. 1984, 25, 4771–4774. [Google Scholar] [CrossRef]
- Oikawa, H. Biosynthesis of structurally unique fungal metabolite GKK1032A2: Indication of novel carbocyclic formation mechanism in polyketide biosynthesis. J. Org. Chem. 2003, 68, 3552–3557. [Google Scholar] [CrossRef]
- Li, H.; Jiang, J.; Liu, Z.; Lin, S.; Xia, G.; Xia, X.; Ding, B.; He, L.; Lu, Y.; She, Z. Peniphenones A−D from the mangrove fungus Penicillium dipodomyicola HN4-3A as inhibitors of mycobacterium tuberculosis phosphatase MptpB. J. Nat. Prod. 2014, 77, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Vallance, P.; Leiper, J. Blocking NO synthesis: How, where and why? Nat. Rev. Drug Discov. 2002, 1, 939–950. [Google Scholar] [CrossRef]
- Eiserich, J.P.; Hristova, M.; Cross, C.E.; Jones, A.D.; Freeman, B.A.; Halliwell, B.; Vliet, A. Formation of nitric oxide-derived inflammatory oxidants by myeloperoxidase in neutrophils. Nature 1998, 391, 393–397. [Google Scholar] [CrossRef]
- Garcin, E.D.; Arvai, A.S.; Rosenfeld, R.J.; Kroeger, M.D.; Crane, B.R.; Andersson, G.; Andrews, G.; Hamley, P.J.; Mallinder, P.R.; Nicholls, D.J.; et al. Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nat. Chem. Biol. 2008, 4, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Liu, Y.N.; Li, J.; Huang, X.S.; Yan, T.; Cao, W.H.; Liu, H.J.; Long, Y.H.; She, Z.G. Diaporindenes A–D: Four unusual 2, 3-dihydro-1H -indene analogues with anti-inflammatory activities from the mangrove endophytic fungus Diaporthe sp. SYSU-HQ3. J. Org. Chem. 2018, 83, 11804–11813. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, T.; Matsuda, H.; Toguchida, I.; Ueda, K.; Yoshikawa, M. Absolute stereostructures of three new sesquiterpenes from the fruit of alpiniaoxyphylla with inhibitory effects on nitric oxide production and degranulation in RBL-2H3 Cells. J. Nat. Prod. 2002, 65, 1468–1474. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Z.M.; Liu, H.J.; Pan, Y.H.; Li, J.; Liu, L. Dichloroisocoumarins with potential anti-inflammatory activity from the mangrove endophytic fungus Ascomycota sp. CYSK-4. Mar. Drugs 2018, 16, 54. [Google Scholar] [CrossRef] [PubMed]
- Garrett, M.M.; Huey, R.; Lingstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar]
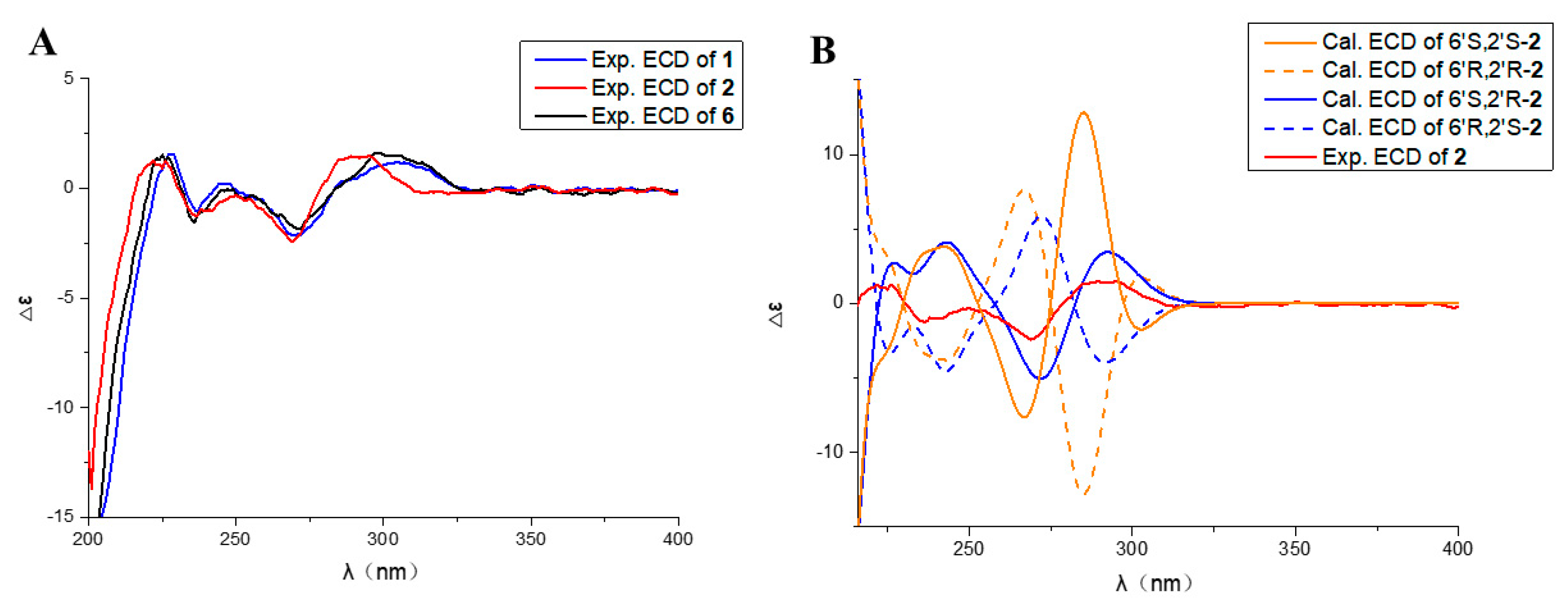
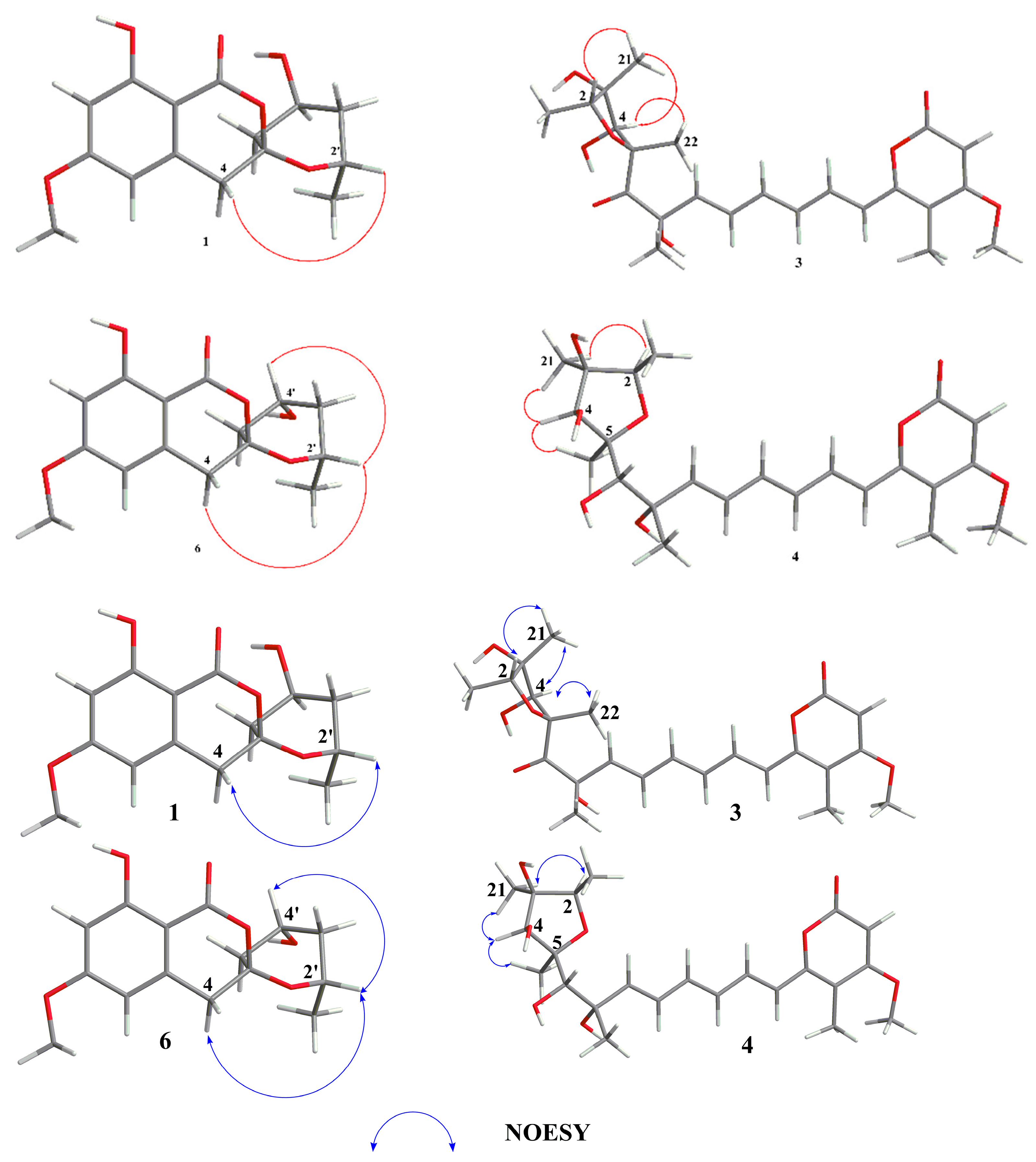
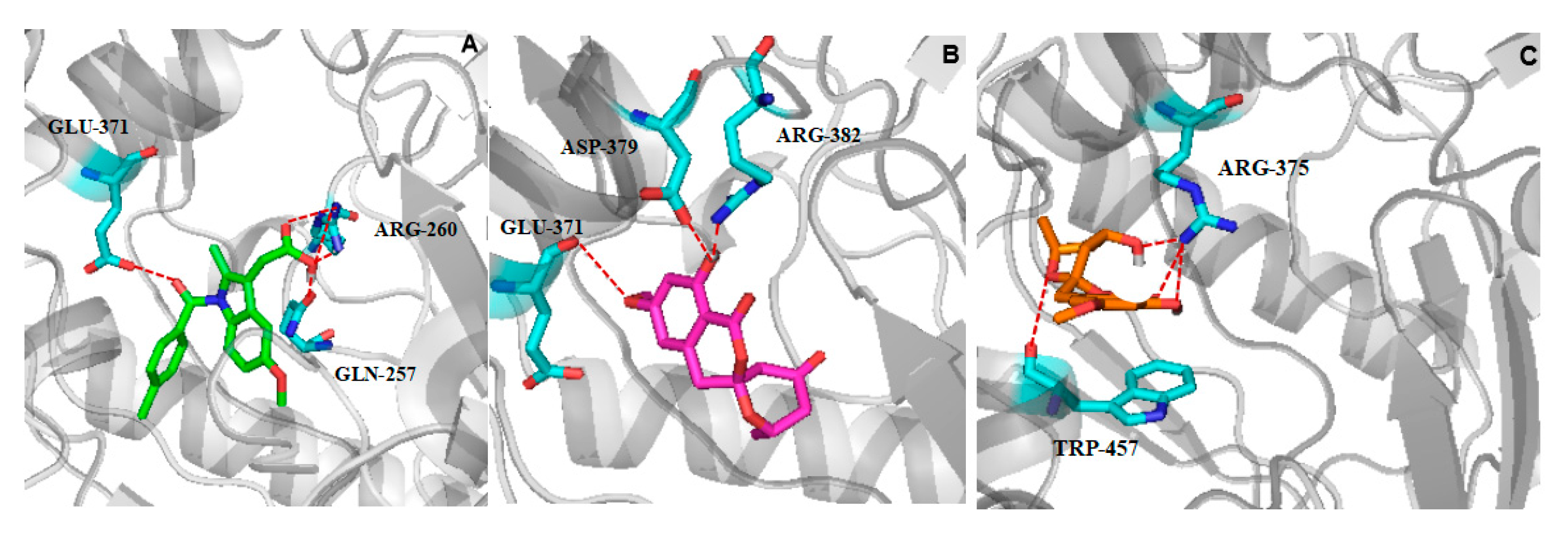
| Position | 1 a (δC, Type) | 1 a (δH, Type) | 2 b (δC, Type) | 2 b (δH, Type) |
|---|---|---|---|---|
| 1 | 169.2, C | 167.5, C | ||
| 2 | 101.0, C | 100.6, C | ||
| 3 | 140.2, C | 138.3, C | ||
| 4 | 39.1, CH2 | 2.95, d (16.5) | 38.6, CH2 | 3.14, d (16.4) |
| 3.17, d (16.5) | 3.20, d (16.4) | |||
| 5 | 106.8, CH | 6.34, s | 107.4, CH | 6.29, s |
| 6 | 166.7, C | 166.4, C | ||
| 7 | 99.3, CH | 6.35, d (2.1) | 99.7, CH | 6.38, d (2.3) |
| 8 | 164.4, C | 164.7, C | ||
| 9 | 55.2, CH3 | 3.82, s | 55.8, CH3 | 3.84, s |
| 10 | 20.5, CH3 | 1.07, d (6.3) | 21.6, CH3 | 1.24, d (6.2) |
| 2′ | 63.0, CH | 4.39, m | 67.9, CH | 4.41, m |
| 3′ | 38.5, CH2 | 1.53, m | 47.9, CH2 | 2.29, dd (11.5, 15.1) |
| 1.78, m | 2.53, m | |||
| 4′ | 63.4, CH | 4.17, m | 202.5, C | |
| 5′ | 38.9, CH2 | 1.87, dd (4.1, 15.0) | 49.5, CH2 | 2.59, d (15.4) |
| 2.21, dt (2.2, 15.0) | 2.85, dd (1.56, 15.3) | |||
| 6′ | 104.6, C | 104.7, C | ||
| 8-OH | 11.0, s |
| Position | 3 b (δC, Type) | 3 b (δH, Type) | 4 a (δC, Type) | 4 a (δH, Type) |
|---|---|---|---|---|
| 2 | 80.2, CH | 4.24, q (6.4) | 78.8, CH | 4.08, q (6.4) |
| 3 | 86.3, C | 83.9, C | ||
| 4 | 78.1, CH | 3.94, s | 78.1, CH | 3.79, s |
| 5 | 84.9, C | 86.3, C | ||
| 6 | 204.7, C | 91.2, CH | 3.69, s | |
| 7 | 82.3, C | 73.3, C | ||
| 8 | 139.1, CH | 6.05, d (15.5) | 140.9, CH | 6.01, d (15.4) |
| 9 | 129.1, CH | 6.27, dd (15.5, 10.5) | 128.5, CH | 6.43, dd (11.0, 15.4) |
| 10 | 137.3, CH | 6.43, dd (15.0, 10.5) | 138.1, CH | 6.65, dd (11.0, 15.0) |
| 11 | 132.5, CH | 6.36, dd (15.0, 10.5) | 131.5, CH | 6.50, dd (11.0, 15.0) |
| 12 | 135.8, CH | 7.16, dd (15.0, 10.9) | 136.1, CH | 7.16, dd (11.0, 15.0) |
| 13 | 119.6, CH | 6.34, d (15.0) | 119.4, CH | 6.59, d (15.0) |
| 14 | 154.5, C | 154.9, C | ||
| 16 | 163.9, C | 165.4, C | ||
| 17 | 88.9, CH | 5.49, s | 88.1, CH | 5.64, s |
| 18 | 170.8, C | 172.1, C | ||
| 19 | 108.2, C | 108.8, C | ||
| 20 | 13.1, CH3 | 1.23, d (6.5) | 12.8, CH3 | 1.20, d (6.4) |
| 21 | 16.9, CH3 | 1.35, s | 12.8, CH3 | 1.22, s |
| 22 | 12.5, CH3 | 1.39, s | 26.3, CH3 | 1.30, s |
| 23 | 31.4, CH3 | 1.46, s | 12.2, CH3 | 1.32, s |
| 24 | 9.0, CH3 | 1.96, s | 7.9, CH3 | 2.02, s |
| 25 | 56.3, CH3 | 3.82, s | 56.3, CH3 | 3.92, s |
| Compound | IC50 (μM) | Inhibition Ratio at 50 μM |
|---|---|---|
| 1 | - | <50% |
| 2 | 12 | 97% |
| 3 | - | <50% |
| 4 | - | <50% |
| Indometacin a | 35.8 ± 5.7 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Ye, G.; Tang, J.; Li, J.; Liu, W.; Wu, L.; Long, Y. New Polyketides from Mangrove Endophytic Fungus Penicillium sp. BJR-P2 and Their Anti-Inflammatory Activity. Mar. Drugs 2022, 20, 583. https://doi.org/10.3390/md20090583
Chen C, Ye G, Tang J, Li J, Liu W, Wu L, Long Y. New Polyketides from Mangrove Endophytic Fungus Penicillium sp. BJR-P2 and Their Anti-Inflammatory Activity. Marine Drugs. 2022; 20(9):583. https://doi.org/10.3390/md20090583
Chicago/Turabian StyleChen, Chen, Geting Ye, Jing Tang, Jialin Li, Wenbin Liu, Li Wu, and Yuhua Long. 2022. "New Polyketides from Mangrove Endophytic Fungus Penicillium sp. BJR-P2 and Their Anti-Inflammatory Activity" Marine Drugs 20, no. 9: 583. https://doi.org/10.3390/md20090583
APA StyleChen, C., Ye, G., Tang, J., Li, J., Liu, W., Wu, L., & Long, Y. (2022). New Polyketides from Mangrove Endophytic Fungus Penicillium sp. BJR-P2 and Their Anti-Inflammatory Activity. Marine Drugs, 20(9), 583. https://doi.org/10.3390/md20090583