Antibacterial and Anticancer Activities of Pleurocidin-Amide, a Potent Marine Antimicrobial Peptide Derived from Winter Flounder, Pleuronectes americanus
Abstract
:1. Introduction
2. Results
2.1. Characteristics of Pleurocidin (Ple) and Its C-terminal Amidation Derivative Ple-a
2.2. Antibacterial Activity of Ple and Ple-a against Gram-Positive and Gram-Negative Bacteria
2.3. Synergistic Eeffect of Ple-a Used in Combination with Antibiotics against MDR E. coli
2.4. Selective Cytotoxicity of Ple and Ple-a against A549 and NIH-3T3 Cell Lines
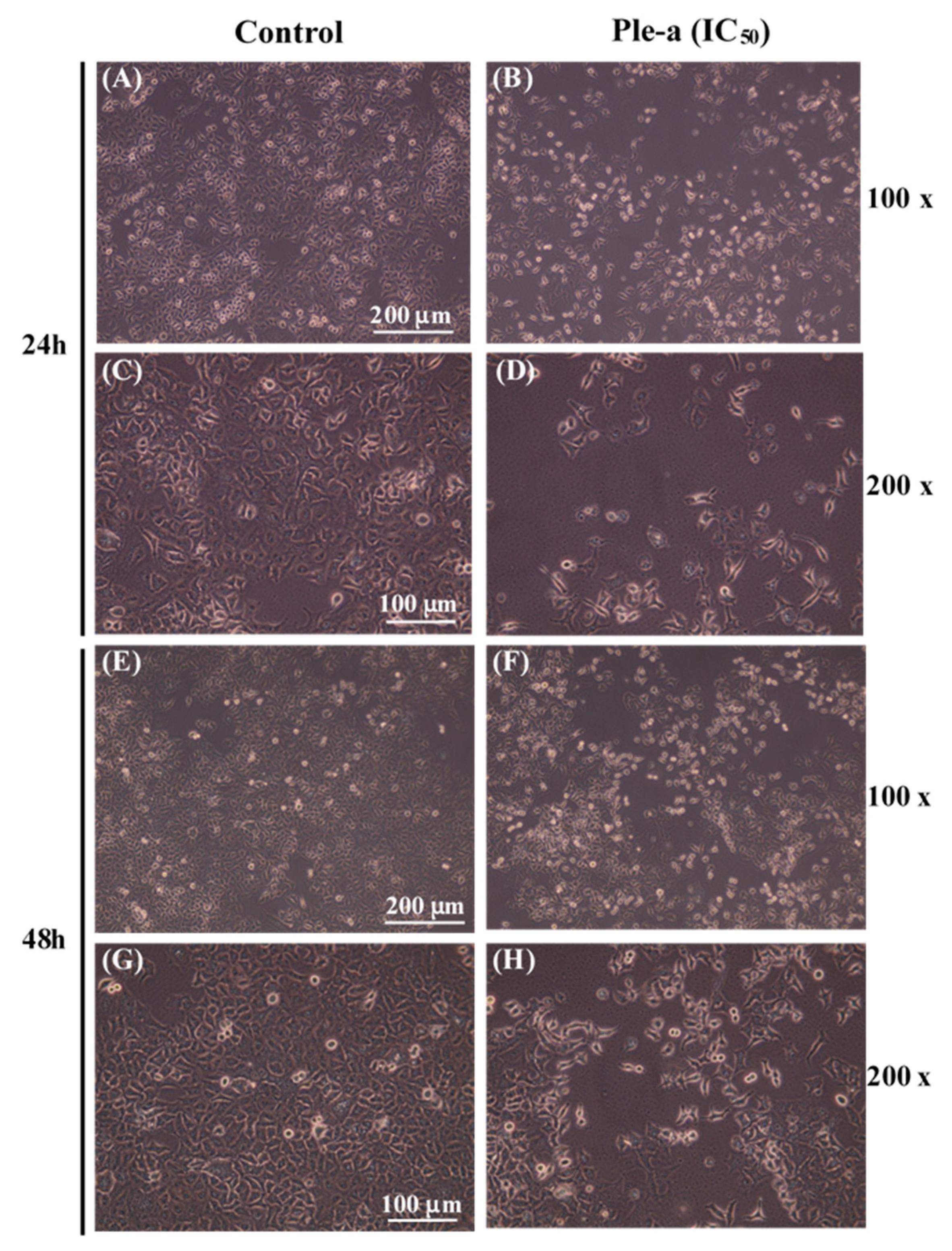
2.5. Ple-a Alters A549 Cell Morphology
2.6. Ple-a Increased the Number of Cells in the Sub-G1 Phase as Revealed by Flow Cytometry
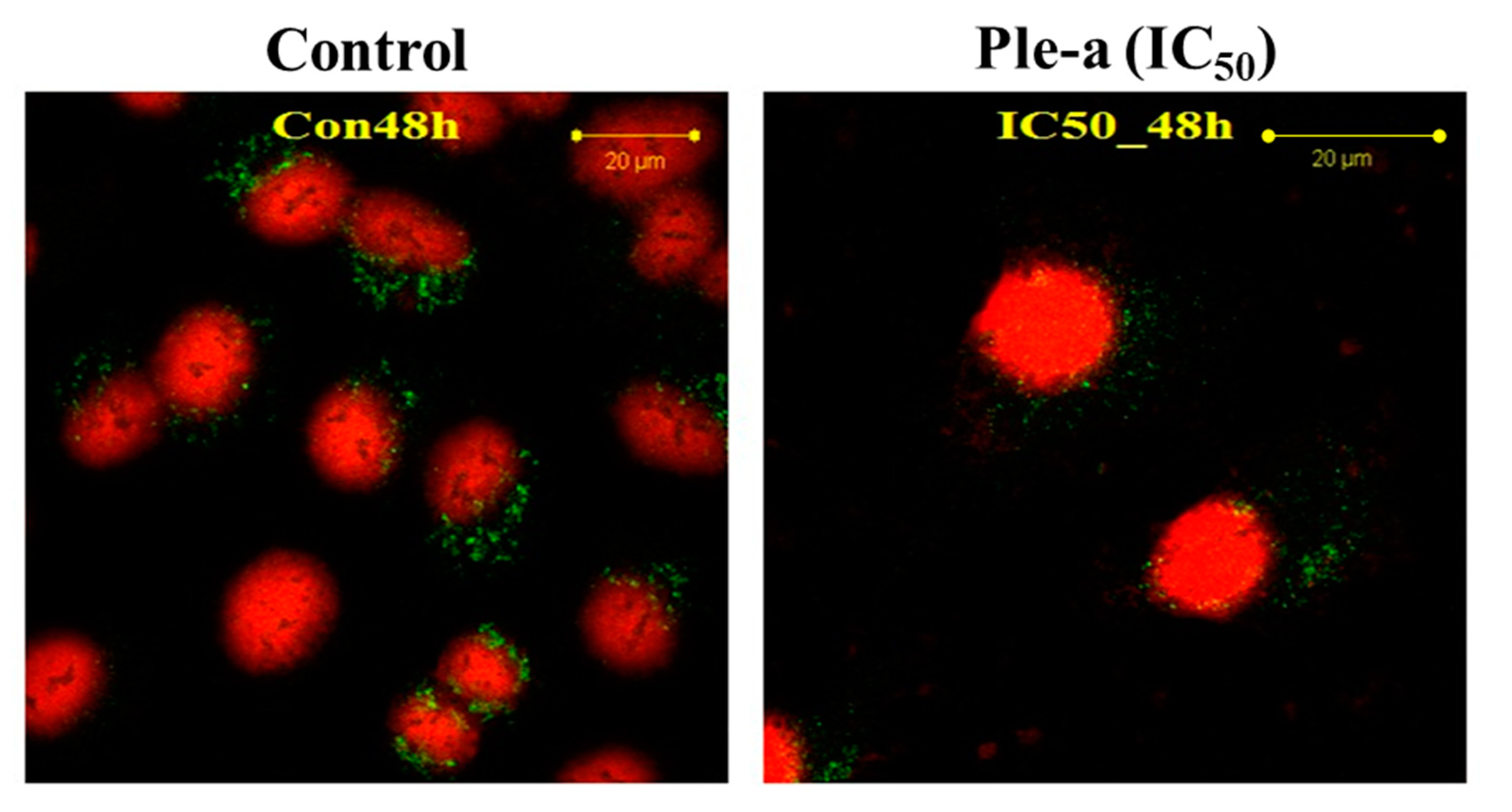
2.7. Ple-a Induced Both Apoptosis and Autophagy of A549 Cells at the Early Stage
2.8. Ple-a-Induced Apoptosis of A549 Cells Was Enhanced by Inhibition of Autophagy at a Late Stage
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. AMPs and Their Aantibacterial Activity
4.3. Checkerboard Assay for Drug Combination Effect
4.4. Cell Lines and Culture Conditions
4.5. Cell Morphology Monitoring
4.6. Cell Viability Assay
4.7. Flow Cytometric Analysis of Cell Cycle
4.8. Western Blot Analysis
4.9. Fluorescent Staining and Confocal Microscopy
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davies, J. Inactivation of antibiotics and the dissemination of resistance genes. Science 1994, 264, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.M.; Bonomo, R.A. The threat of antibiotic resistance in Gram-negative pathogenic bacteria: Beta-lactams in peril. Curr. Opin. Microbiol. 2005, 8, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Patrzykat, A. Clinical development of cationic antimicrobial peptides: From natural to novel antibiotics. Curr. Drug. Targets. Infect. Disord. 2002, 2, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef]
- Scott, M.G.; Hancock, R.E.W. Cationic antimicrobial peptides and their multifunctional role in the immune system. Crit. Rev. Immunol. 2000, 20, 407–431. [Google Scholar] [CrossRef]
- Hancock, R.E.W. Cationic peptides: Effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 2001, 1, 156–164. [Google Scholar] [CrossRef]
- Dathe, M.; Wieprecht, T. Structural features of helical antimicrobial peptides: Their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta 1999, 1462, 71–87. [Google Scholar] [CrossRef]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Rational design of α-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Lehrer, R. Cationic peptides: A newsource of antibiotics. Trends Biotechnol. 1998, 16, 82–88. [Google Scholar] [CrossRef]
- Cole, A.M.; Weis, P.; Diamond, G. Isolation and characterization of pleurocidin, an antimicrobial petpide in the skin secretions of winter flounder. J. Biol. Chem. 1997, 272, 12008–12013. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.E.; Patrzykat, A.; Pytyck, J.; Gallant, J.W. Identification, structure and differential expression of novel pleurocidins clustered on the genome of the winter flounder, Pseudopleuronectes americanus (Walbaum). Eur. J. Biochem. 2003, 270, 3720–3730. [Google Scholar] [CrossRef] [PubMed]
- McMillan, K.A.M.; Coombs, M.R.P. Investigating potential applications of the fish anti-microbial peptide pleurocidin: A systematic review. Pharmaceuticals 2021, 14, 687. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.J.; Chotimah, I.N.; Bertani, P.; Bechinger, B. A spectroscopic study of the membrane interaction of the antimicrobial peptide Pleurocidin. Mol. Membr. Biol. 2006, 23, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.J.; Marquette, A.; Bechinger, B. Zwitterionic phospholipids and sterols modulate antimicrobial peptide-induced membrane destabilization. Biophys. J. 2007, 93, 4289–4299. [Google Scholar] [CrossRef]
- Syvitski, R.T.; Burton, I.; Mattatall, N.R.; Douglas, S.E.; Jakeman, D.L. Structural characterization of the antimicrobial peptide pleurocidin from winter flounder. Biochemistry 2005, 44, 7282–7293. [Google Scholar] [CrossRef]
- Talandashti, R.; Mahdiuni, H.; Jafari, M.; Mehrnejad, F. Molecular basis for membrane selectivity of antimicrobial peptide Pleurocidin in the presence of different eukaryotic and prokaryotic model membranes. J. Chem. Inf. Model. 2019, 59, 3262–3276. [Google Scholar] [CrossRef]
- Ko, S.J.; Kang, N.H.; Kim, M.K.; Park, J.; Park, E.; Park, G.H.; Kang, T.W.; Na, D.E.; Park, J.B.; Yi, Y.E.; et al. Antibacterial and anti-biofilm activity, and mechanism of action of pleurocidin against drug resistant Staphylococcus aureus. Microb. Pathog. 2019, 127, 70–78. [Google Scholar] [CrossRef]
- Tao, R.; Tong, Z.; Lin, Y.; Xue, Y.; Wang, W.; Kuang, R.; Wang, P.; Tian, Y.; Ni, L. Antimicrobial and antibiofilm activity of pleurocidin against cariogenic microorganisms. Peptides 2011, 32, 1748–1754. [Google Scholar] [CrossRef]
- Patrzykat, A.; Friedrich, C.L.; Zhang, L.; Mendoza, V.; Hancock, R.E.W. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 2002, 46, 605–614. [Google Scholar] [CrossRef]
- Lan, Y.; Ye, Y.; Kozlowska, J.; Lam, J.K.; Drake, A.F.; Mason, A.J. Structural contribution to the intracellular targeting strategies of antimicrobial peptides. Biochim. Biophys. Acta 2010, 1798, 1934–1943. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Lee, D.G. Oxidative stress by antimicrobial peptide pleurocidin triggers apoptosis in Candida albicans. Biochimie 2011, 93, 1873–1879. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Choi, H.; Lee, D.G. Influence of the N- and C-terminal regions of antimicrobial peptide pleurocidin on antibacterial activity. J. Microbiol. Biotechnol. 2012, 22, 1367–1374. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Lee, D.G. The influence of the N-terminal region of antimicrobial peptide pleurocidin on fungal apoptosis. J. Microbiol. Biotechnol. 2013, 23, 1386–1394. [Google Scholar] [CrossRef]
- Yoshida, K.; Mukai, Y.; Niidome, T.; Takashi, C.; Tokunaga, Y.; Hatakeyama, T.; Aoyagi, H. Interaction of pleurocidin and its analogs with phospholipid membrane and their antibacterial activity. J. Pept. Res. 2001, 57, 119–126. [Google Scholar] [CrossRef]
- Sung, W.S.; Lee, D.G. Pleurocidin-derived antifungal peptides with selective membrane-disruption effect. Biochem. Biophys. Res. Commun. 2008, 369, 858–861. [Google Scholar] [CrossRef]
- Hilchie, A.L.; Haney, E.F.; Pinto, D.M.; Hancock, R.E.W.; Hoskin, D.W. Enhanced killing of breast cancer cells by a d-amino acid analog of the winter flounder-derived pleurocidin NRC-03. Exp. Mol. Pathol. 2015, 99, 426–434. [Google Scholar] [CrossRef]
- Hilchie, A.L.; Conrad, D.M.; Coombs, M.R.P.; Zemlak, T.; Doucette, C.D.; Liwski, R.S.; Hoskin, D.W. Pleurocidin-family cationic antimicrobial peptides mediate lysis of multiple myeloma cells and impair the growth of multiple myeloma xenografts. Leuk. Lymphoma 2013, 54, 2255–2262. [Google Scholar] [CrossRef]
- Hilchie, A.L.; Doucette, C.D.; Pinto, D.M.; Patrzykat, A.; Douglas, S.; Hoskin, D.W. Pleurocidin-family cationic antimicrobial peptides are cytolytic for breast carcinoma cells and prevent growth of tumor xenografts. Breast Cancer Res. 2011, 13, R102. [Google Scholar] [CrossRef]
- Chou, H.T.; Kuo, T.Y.; Chiang, J.C.; Pei, M.J.; Yang, W.T.; Yu, H.C.; Lin, S.B.; Chen, W.J. Design and synthesis of cationic antimicrobial peptides with improved activity and selectivity against Vibrio spp. Int. J. Antimicrob. Agents 2008, 32, 130–138. [Google Scholar] [CrossRef]
- Chou, H.T.; Wen, H.W.; Kuo, T.Y.; Lin, C.C.; Chen, W.J. Interaction of cationic antimicrobial peptides with phospholipid vesicles and their antibacterial activity. Peptides 2010, 31, 1811–1820. [Google Scholar] [CrossRef] [PubMed]
- Shalev, D.E.; Mor, A.; Kustanovich, I. Structural consequences of carboxyamidation of dermaseptin S3. Biochemistry 2002, 41, 7312–7317. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Weiss, R.M.; Terwilliger, T.C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. USA 1984, 81, 140–144. [Google Scholar] [CrossRef]
- Zhou, Y.; Peng, Y. Synergistic effect of clinically used antibiotics and peptide antibiotics against Gram-positive and Gram-negative bacteria. Exp. Ther. Med. 2013, 6, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M.C.; Bolton, W.E. Comparison of cell viability probes compatible with fixation and permeabilization for combined surface and intracellular staining in flow cytometry. Cytometry 1995, 19, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Boya, P.; Reggiori, F.; Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013, 15, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.Z.; Shi, Y.Y.; Wei, L.L.; Sun, G.C. Autophagy induction and antiproliferative effect of a novel curcumin derivative MOMI-1 on the human lung cancer cells A549. J. Biochem. Mol. Toxicol. 2019, 33, e22280. [Google Scholar] [CrossRef]
- Talandashti, R.; Mehrnejad, F.; Rostamipour, K.; Doustdar, F.; Lavasanifar, A. Molecular insights into pore formation mechanism, membrane perturbation, and water permeation by the antimicrobial peptide Pleurocidin: A combined all-atom and coarse-grained molecular dynamics simulation study. J. Phys. Chem. B 2021, 125, 7163–7176. [Google Scholar] [CrossRef]
- O’Rourke, A.; Beyhan, S.; Choi, Y.; Morales, P.; Chan, A.P.; Espinoza, J.L.; Dupont, C.L.; Meyer, K.J.; Spoering, A.; Lewis, K.; et al. Mechanism-of-action classification of antibiotics by global transcriptome profiling. Antimicrob. Agents Chemother. 2020, 64, e01207-19. [Google Scholar] [CrossRef]
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef]
- Koval, O.A.; Tkachenko, A.V.; Fomin, A.S.; Semenov, D.V.; Nushtaeva, A.A.; Kuligina, E.V.; Zavjalov, E.L.; Richter, V.A. Lactaptin induces p53-independent cell death associated with features of apoptosis and autophagy and delays growth of breast cancer cells in mouse xenografts. PLoS ONE 2014, 9, e93921. [Google Scholar] [CrossRef]
- Pan, W.R.; Chen, P.W.; Chen, Y.L.; Hsu, H.C.; Lin, C.C.; Chen, W.J. Bovine lactoferricin B induces apoptosis of human gastric cancer cell line AGS by inhibition of autophagy at a late stage. J. Dairy Sci. 2013, 96, 7511–7520. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.R.; Chen, Y.L.; Hsu, H.C.; Chen, W.J. Antimicrobial peptide GW-H1-induced apoptosis of human gastric cancer AGS cell lin is enhanced by suppression of autophagy. Mol. Cell. Biochem. 2015, 400, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Li, J.H.; Yu, C.Y.; Lin, C.J.; Chiu, P.H.; Chen, P.W.; Lin, C.C.; Chen, W.J. Novel cationic antimicrobial peptide GW-H1 induced caspase-dependent apoptosis of hepatocellular carcinoma cell lines. Peptides 2012, 36, 257–265. [Google Scholar] [CrossRef] [PubMed]
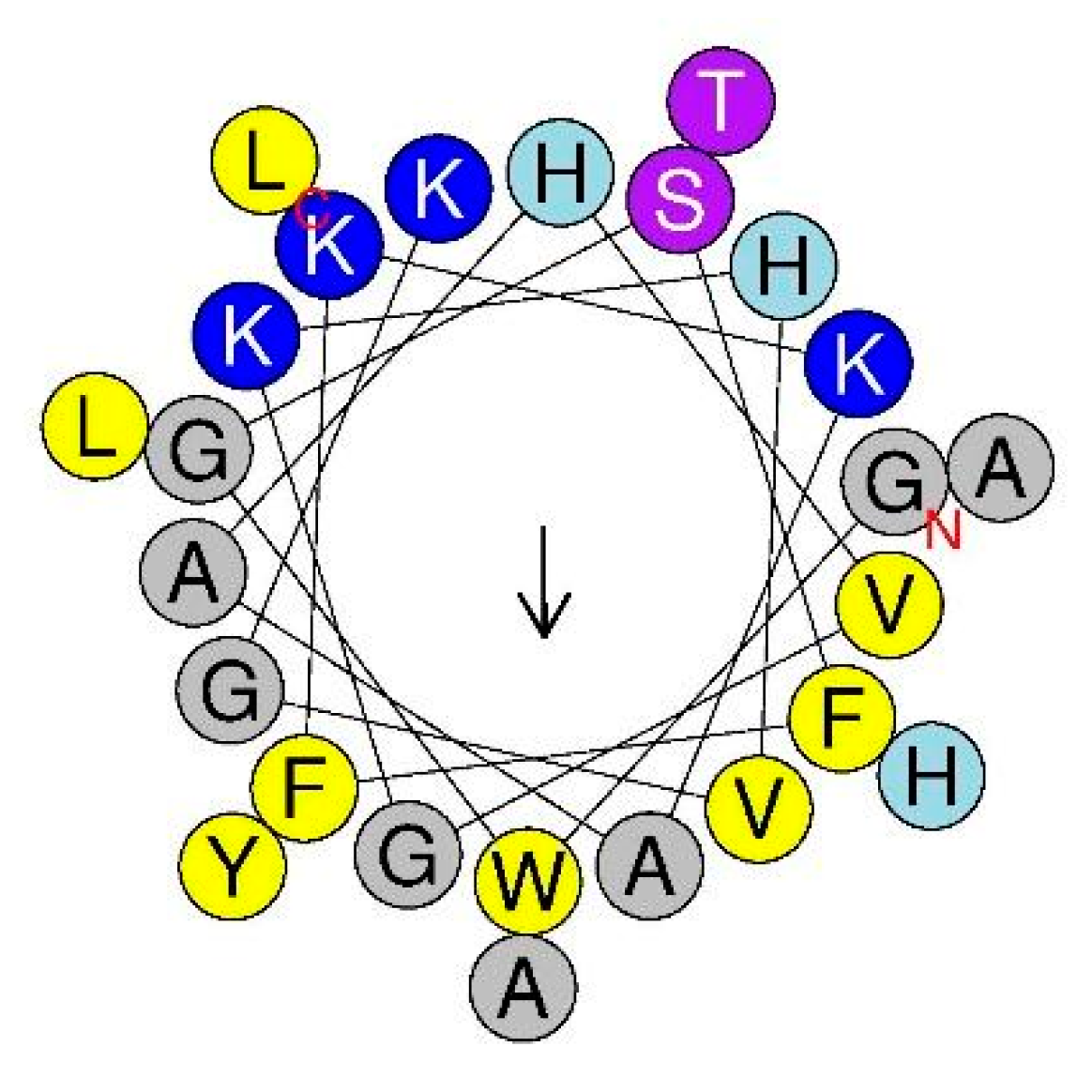





| AMP | Amino Acid Sequence | Charge | MW | Hyrophobicity a | Hydrophobic Moment a |
|---|---|---|---|---|---|
| Ple | GWGSFFKKAAHVGKHVGKAALTHYL | +7 | 2711.17 | −0.026 | 0.287 |
| Ple-a | GWGSFFKKAAHVGKHVGKAALTHYL-NH2 | +8 | 2710.18 | −0.026 | 0.287 |
| Bacterial Strain | G(+)/G(−) | MIC (μg/mL) a | ||
|---|---|---|---|---|
| Ple | Ple-a | |||
| Typical G(+) and G(−) bacterial strains | Staphylococcus aureus | G(+) | 16 | 4 |
| Staphylococcus xylosus | G(+) | 16 | 8 | |
| Listeria monocytogenes | G(+) | 32 | 16 | |
| Streptococcu bovis | G(+) | 128 | 32 | |
| Escherichia coli | G(−) | 16 | 2 | |
| Enterobacter aerogenes | G(−) | 2 | 1 | |
| Enterobacter cloacae | G(−) | 4 | 1 | |
| Yersinia enterocolitica | G(−) | 32 | 8 | |
| Pseudomonas aeruginosa | G(−) | 8 | 2 | |
| Salmonella enterica | G(−) | 64 | 8 | |
| Klebsiella oxytoca | G(−) | 8 | 2 | |
| Multidrug-resistant G(+) and G(−) bacterial strains | Enterococcus faecium-4R | G(+) | 256 | 32 |
| Escherichia coli-7R | G(−) | 16 | 2 | |
| Pseudomonas aeruginosa-5R | G(−) | 64 | 16 | |
| Klebsiella pneumoniae-7R | G(−) | 64 | 8 | |
| Klebsiella pneumoniae-10R | G(−) | 128 | 8 | |
| Salmonella enterica serovar Choleraesuis-13R | G(−) | 32 | 8 | |
| Acinetobacter baumannii-8R | G(−) | 8 | 4 | |
| Clinically isolated marine bacterial pathogens | Streptococcus iniae | G(+) | 128 | 32 |
| Lactococcus garvieae | G(+) | 128 | 32 | |
| Vibrio alginolyticus | G(−) | 8 | 1 | |
| Vibrio harveyi | G(−) | 8 | 2 | |
| Vibrio parahaemolyticus | G(−) | 8 | 1 | |
| Vibrio anguillarum | G(−) | 128 | 32 | |
| Photobacterium damselae subsp. piscicida | G(−) | 1 | 0.5 | |
| Ple-a:Antibiotic (1:1) | ||||
|---|---|---|---|---|
| MICo | MICc (1:1) | FICI | Type of Interaction | |
| Ple-a | 2 | |||
| Ampicillin | 256 | |||
| Ple-a/Ampicillin | 1/1 | 0.50 | Synergistic | |
| Imipenem | 16 | |||
| Ple-a/Imipenem | 1/1 | 0.56 | Additive | |
| Meropenem | 0.5 | |||
| Ple-a/Meropenem | 0.5/0.5 | 1.25 | Indifference | |
| Ceftazidime | 128 | |||
| Ple-a/Ceftazidime | 1/1 | 0.51 | Additive | |
| Cefotaxime | 256 | |||
| Ple-a/Cefotaxime | 1/1 | 0.50 | Synergistic | |
| Amikacin | 16 | |||
| Ple-a/Amikacin | 1/1 | 0.56 | Additive | |
| Gentamycin | 256 | |||
| Ple-a/Gentamycin | 1/1 | 0.50 | Synergistic | |
| Levofloxacin | 2 | |||
| Ple-a/Levofloxacin | 1/1 | 1.0 | Additive | |
| Ampicillin-sulbactam | 128 | |||
| Ple-a/Ampicillin-sulbactam | 1/1 | 0.51 | Additive | |
| Ple-a:Antibiotic (2:1) | ||||
| MICo | MICc (2:1) | FICI | Type of interaction | |
| Ple-a | 2 | |||
| Ampicillin | 256 | |||
| Ple-a/Ampicillin | 0.5/0.5 | 0.25 | Synergistic | |
| Imipenem | 16 | |||
| Ple-a/Imipenem | 0.5/0.5 | 0.28 | Synergistic | |
| Meropenem | 0.5 | |||
| Ple-a/Meropenem | 0.5/0.5 | 1.25 | Indifference | |
| Ceftazidime | 128 | |||
| Ple-a/Ceftazidime | 0.5/0.5 | 0.25 | Synergistic | |
| Cefotaxime | 256 | |||
| Ple-a/Cefotaxime | 0.5/0.5 | 0.25 | Synergistic | |
| Amikacin | 16 | |||
| Ple-a/Amikacin | 1/1 | 0.56 | Additive | |
| Gentamycin | 256 | |||
| Ple-a/Gentamycin | 1/1 | 0.50 | Synergistic | |
| Levofloxacin | 2 | |||
| Ple-a/Levofloxacin | 1/1 | 1.0 | Additive | |
| Ampicillin-sulbactam | 128 | |||
| Ple-a/Ampicillin-sulbactam | 1/1 | 0.51 | Additive | |
| Ple-a:Antibiotic (1:2) | ||||
| MICo | MICc (1:2) | FICI | Type of interaction | |
| Ple-a | 2 | |||
| Ampicillin | 256 | |||
| Ple-a/Ampicillin | 2/2 | 1.01 | Indifference | |
| Imipenem | 16 | |||
| Ple-a/Imipenem | 1/1 | 0.56 | Additive | |
| Meropenem | 0.5 | |||
| Ple-a/Meropenem | 0.5/0.5 | 1.25 | Indifference | |
| Ceftazidime | 128 | |||
| Ple-a/Ceftazidime | 2/2 | 1.02 | Indifference | |
| Cefotaxime | 256 | |||
| Ple-a/Cefotaxime | 1/1 | 0.50 | Synergistic | |
| Amikacin | 16 | |||
| Ple-a/Amikacin | 2/2 | 1.13 | Indifference | |
| Gentamycin | 256 | |||
| Ple-a/Gentamycin | 2/2 | 1.01 | Indifference | |
| Levofloxacin | 2 | |||
| Ple-a/Levofloxacin | 1/1 | 1.0 | Additive | |
| Ampicillin-sulbactam | 128 | |||
| Ple-a/Ampicillin-sulbactam | 2/2 | 1.02 | Indifference | |
| Cell line | Description | IC50 (μM) a | |
|---|---|---|---|
| Ple | Ple-a | ||
| J5 | Hepatocellular carcinoma | 54.9 | 11.0 |
| Huh7 | Hepatocellular carcinoma | n.d. b | 60.0 |
| Hep3B | Hepatocellular carcinoma | 340.9 | 77.5 |
| A549 | Non-small cell lung adenocarcinoma | 300.8 | 42.1 |
| AGS | Stomach adenocarcinoma | 186.5 | 29.8 |
| WiDr | Colon adenocarcinoma | n.d. | 197.3 |
| NIH-3T3 | Mouse fibroblast | n.d. | 313 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, H.-C.; Chen, M.-H.; Yeh, M.-L.; Chen, W.-J. Antibacterial and Anticancer Activities of Pleurocidin-Amide, a Potent Marine Antimicrobial Peptide Derived from Winter Flounder, Pleuronectes americanus. Mar. Drugs 2022, 20, 519. https://doi.org/10.3390/md20080519
Hsu H-C, Chen M-H, Yeh M-L, Chen W-J. Antibacterial and Anticancer Activities of Pleurocidin-Amide, a Potent Marine Antimicrobial Peptide Derived from Winter Flounder, Pleuronectes americanus. Marine Drugs. 2022; 20(8):519. https://doi.org/10.3390/md20080519
Chicago/Turabian StyleHsu, Hui-Chen, Ming-Hsin Chen, Ming-Lung Yeh, and Wei-Jung Chen. 2022. "Antibacterial and Anticancer Activities of Pleurocidin-Amide, a Potent Marine Antimicrobial Peptide Derived from Winter Flounder, Pleuronectes americanus" Marine Drugs 20, no. 8: 519. https://doi.org/10.3390/md20080519
APA StyleHsu, H.-C., Chen, M.-H., Yeh, M.-L., & Chen, W.-J. (2022). Antibacterial and Anticancer Activities of Pleurocidin-Amide, a Potent Marine Antimicrobial Peptide Derived from Winter Flounder, Pleuronectes americanus. Marine Drugs, 20(8), 519. https://doi.org/10.3390/md20080519






