Identification, Characteristics and Function of Phosphoglucomutase (PGM) in the Agar Biosynthesis and Carbon Flux in the Agarophyte Gracilariopsis lemaneiformis (Rhodophyta)
Abstract
:1. Introduction
2. Results
2.1. Correlation Analysis between Enzyme Activities and Agar Contents
2.2. Bioinformatic Characteristics of GlPGM
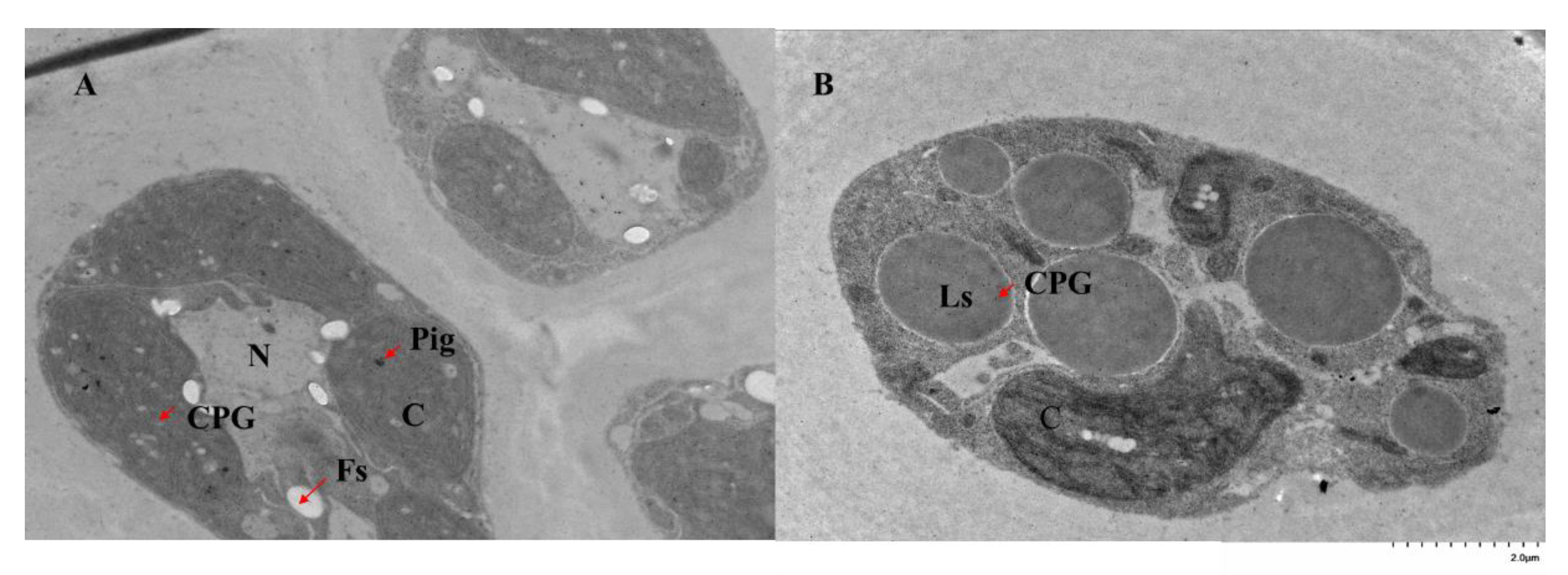
2.3. Subcellular Location of GlPGM1 and GlPGM3
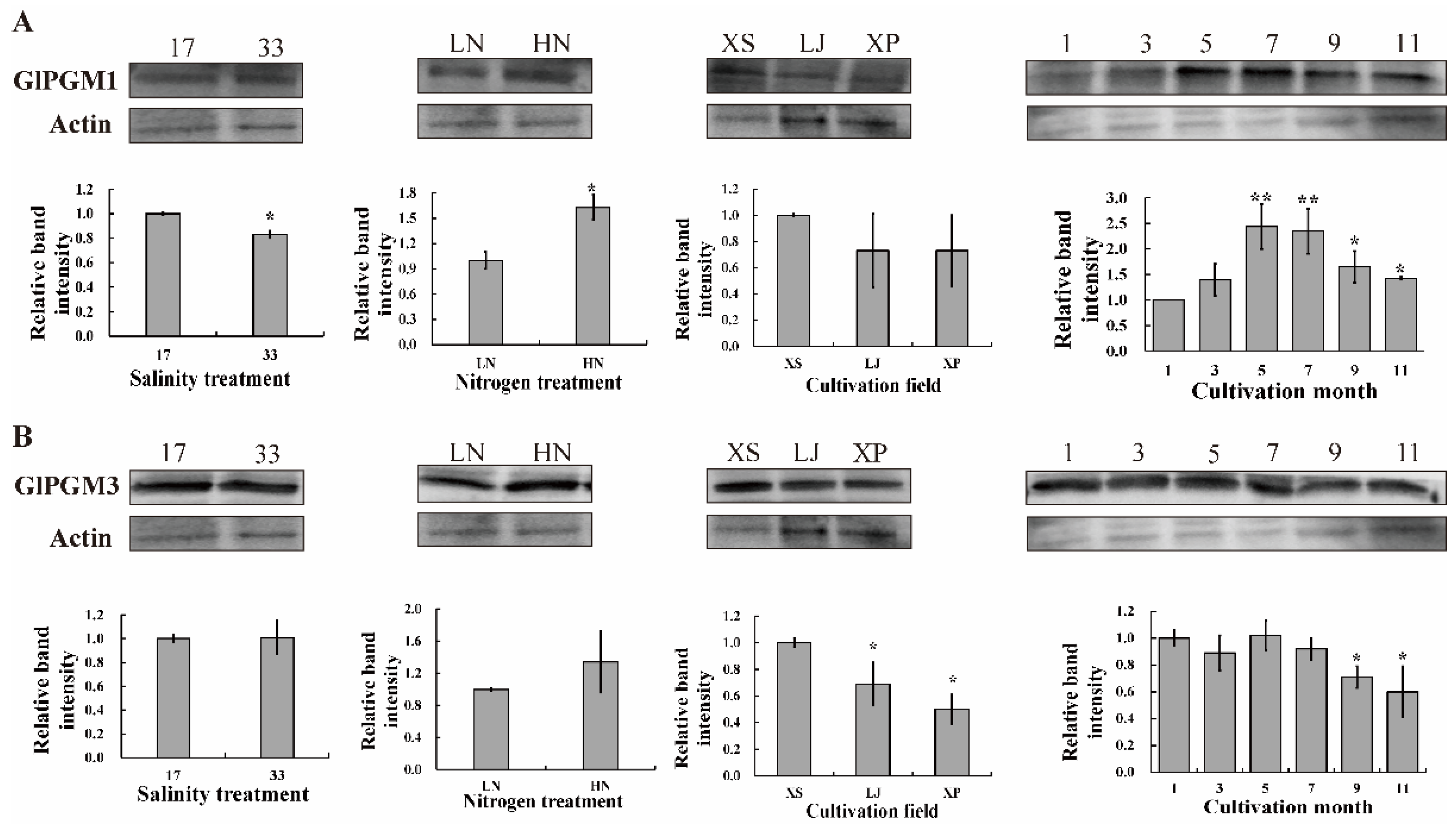
2.4. Immunoblotting Analysis of GlPGM1 and GlPGM3
2.5. Changes of Enzymic Activities under PGM Inhibited Condition
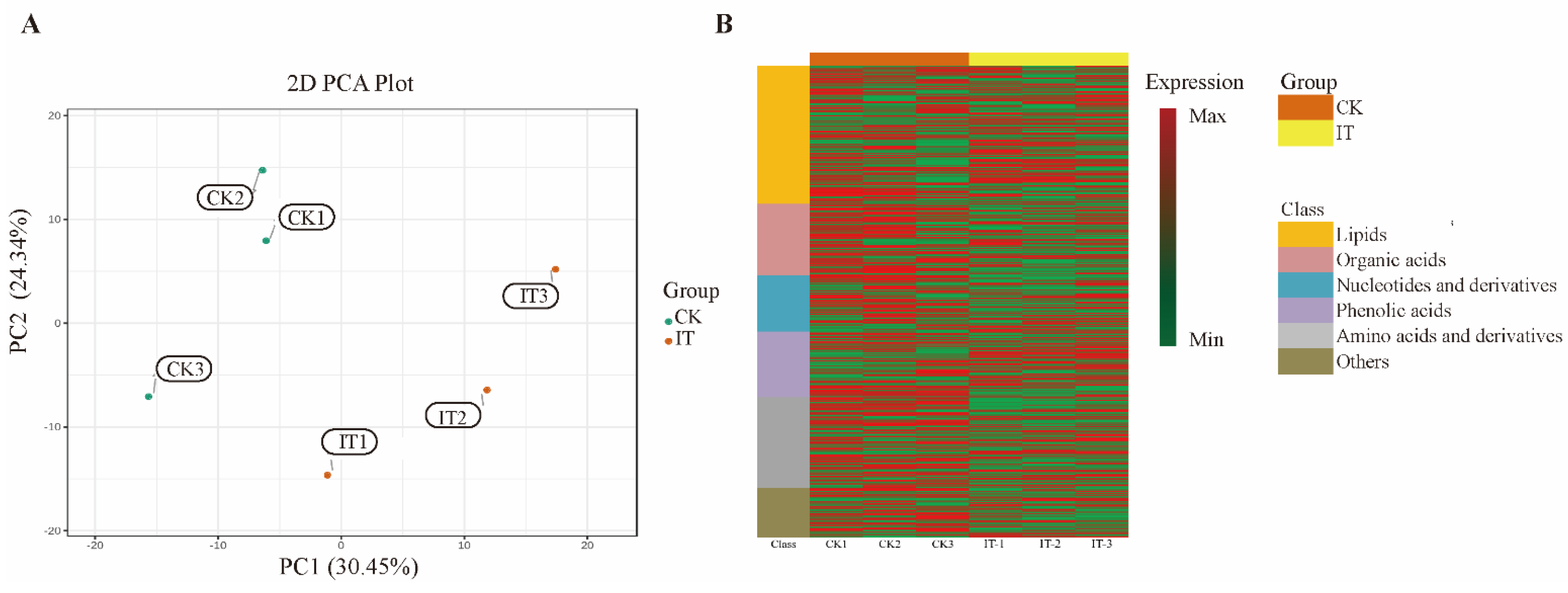
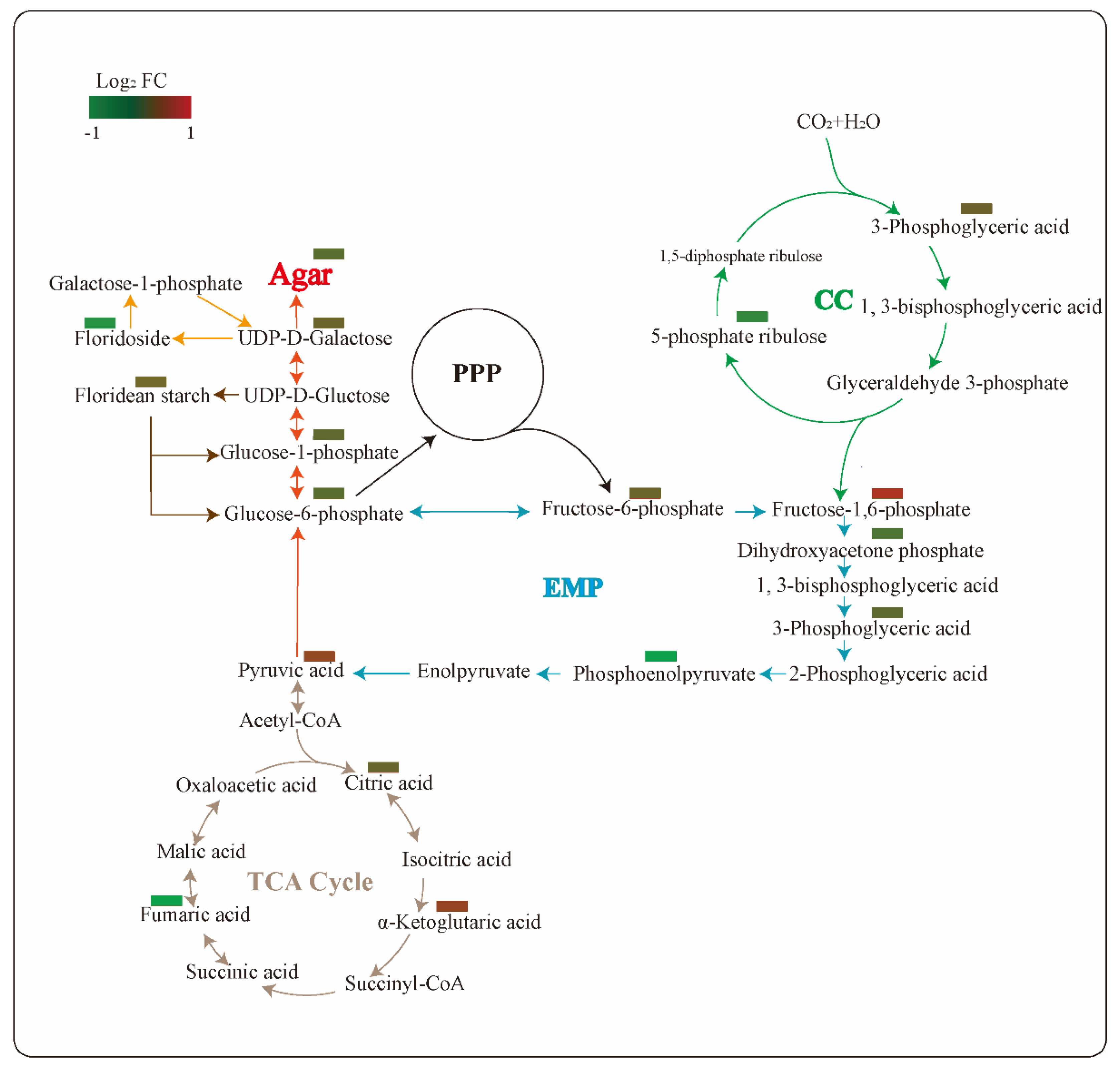
2.6. Changes of Primary Carbon Metabolism under PGM Inhibited Condition
3. Discussion
3.1. The Enzymes in Agar Biosynthesis Pathway
3.2. Characteristics and Functions of GlPGM
3.3. Agar and Carbon Flux Affected by PGM Inhibitor
4. Materials and Methods
4.1. Algal Materials and Culture Condition
4.2. Treatments for Correlation Analysis
4.3. Agar Extraction and Enzymic Activities Assays
4.4. GlPGM Identification
4.5. GlPGM Bioinformatic Analysis
4.6. GlPGM Recombinant and Polyclonal Antibody Preparation
4.7. Subcellular Localization
4.8. Western Blot Analysis
4.9. Analysis of Enzyme Activities under PGM Inhibited Condition
4.10. Carbon Metabolism Analysis under PGM Inhibited Condition
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, D.H.; Mahomoodally, M.F.; Sadeer, N.B.; Seok, P.G.; Zengin, G.; Palaniveloo, K.; Rengasamy, K.R.R. Nutritional and bioactive potential of seagrasses: A review. S. Afr. J. Bot. 2021, 137, 216–227. [Google Scholar] [CrossRef]
- Liu, Q.-M.; Yang, Y.; Maleki, S.J.; Alcocer, M.; Xu, S.S.; Shi, C.-L.; Cao, M.-J.; Liu, G.-M. Anti-food allergic activity of sulfated polysaccharide from Gracilaria lemaneiformis is dependent on immunosuppression and inhibition of p38 MAPK. J. Agric. Food Chem. 2016, 64, 4536–4544. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.K.; Lim, Y.Y.; Leow, A.T.; Namasivayam, P.; Ong Abdullah, J.; Ho, C.L. Biosynthesis of agar in red seaweeds: A review. Carbohydr. Polym. 2017, 164, 23–30. [Google Scholar] [CrossRef]
- Venugopal, V. Sulfated and non-sulfated polysaccharides from seaweeds and their uses: An overview. ECronicon. Nutr. 2018, 14, 126–141. [Google Scholar]
- Zhou, W.; Hu, Y.; Sui, Z.; Fu, F.; Wang, J.; Chang, L.; Li, B. Genome survey sequencing and genetic background characterization of Gracilariopsis lemaneiformis (Rhodophyta) based on next-generation sequencing. PLoS ONE 2013, 8, e69909. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Du, Q.; Mi, P.; Shang, E.; Sui, Z. Gene cloning and expression regulation in the pathway of agar and floridean starch synthesis of Gracilariopsis lemaneiformis (Rhodophyta). J. Appl. Phycol. 2018, 31, 1889–1896. [Google Scholar] [CrossRef]
- Li, M.; Sui, Z.; Kang, K.H.; Zhang, X.; Zhu, M.; Yan, B. Cloning and analysis of the galactose-1-phosphate uridylyltransferase (galt) gene of Gracilariopsis lemaneiformis (Rhodophyta) and correlation between gene expression and agar synthesis. J. Appl. Phycol. 2010, 22, 157–164. [Google Scholar] [CrossRef]
- Zhang, Y.; Sui, Z.H.; Ding, H.Y.; Zhong, J. Cloning of genes involved in agar synthesis and the analysis of its expressions and regulations in Gracilariopsis lemaneiformis (Rhodophyta). Period. Ocean. Univ. China 2011, 41, 7. (In Chinese) [Google Scholar] [CrossRef]
- Siow, R.S.; Teoh, S.; Teo, S.S.; Shukor, M.; Phang, S.M.; Ho, C.-L. Molecular cloning and characterization of GDP-mannose-3′,5′-epimerase from Gracilaria changii. J. Appl. Phycol. 2013, 25, 1309–1318. [Google Scholar] [CrossRef]
- Tang, X.B.; Xu, N.J.; Sun, X.; Li, Y.H.; Zhang, B. Effects of 24-epibrassinolide on the agar synthesis and expression of genes involved in marine alga Gracilariopsis lemaneiformis. J. Fish. China 2015, 39, 1788–1798. (In Chinese) [Google Scholar] [CrossRef]
- Pal, S.K.; Liput, M.; Piques, M.; Ishihara, H.; Obata, Y. Diurnal changes of polysome loading track sucrose content in the rosette of wild-type arabidopsis and the starchless pgm mutant. Plant Physiol. 2013, 162, 1246–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viola, R.; Nyvall, P.; Pedersen, M. The unique features of starch metabolism in red algae. Proc. R. Soc. Lond. Ser. B 2001, 268, 1417–1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, G.; Gao, L.; Jiang, M.; Jian, A.; He, L. The potential of seaweed cultivation to achieve carbon neutrality and mitigate deoxygenation and eutrophication. Environ. Res. Lett. 2022, 17, 014018. [Google Scholar] [CrossRef]
- Liu, H.Y.; Wang, Z.; Regni, C.; Zou, X.; Tipton, P.A. Detailed kinetic studies of an aggregating inhibitor; inhibition of phosphomannomutase/phosphoglucomutase by disperse blue 56. Biochemistry 2004, 43, 8662–8669. [Google Scholar] [CrossRef] [PubMed]
- Imbs, T.I.; Shevchenko, N.M.; Sukhoverkhov, S.V.; Semenova, T.L.; Skriptsova, A.V.; Zvyagintseva, T.N. Seasonal variations of the compostion and structural characteristics of polysaccharides from the brown alga Costaria costata. Chem. Nat. Compd. 2009, 45, 786–791. [Google Scholar] [CrossRef]
- Yoloxochitl, E.R.M.; Dora, L.A.H.; Gustavo, H.C.; Mauricio, M.O.; Jesús, I.M.l. Seasonal variation of the agar quality and chemical composition of Gracilaria veleroae and Gracilaria vermiculophylla (Rhodophyceae, Gracilariaceae) from Baja California Sur, Mexico. Phycol. Res. 2013, 61, 116–123. [Google Scholar] [CrossRef]
- Chang, L.; Sui, Z.; Fu, F.; Zhou, W.; Wang, J.; Kang, K.H.; Zhang, S.; Ma, J. Relationship between gene expression of UDP-glucose pyrophosphorylase and agar yield in Gracilariopsis lemaneiformis (Rhodophyta). J. Appl. Phycol. 2014, 26, 2435–2441. [Google Scholar] [CrossRef]
- Zhang, P.; Shao, Z.; Li, L.; Liu, S.; Yao, J.; Duan, D. Molecular characterisation and biochemical properties of phosphomannomutase/phosphoglucomutase (PMM/PGM) in the brown seaweed Saccharina japonica. J. Appl. Phycol. 2018, 30, 2687–2696. [Google Scholar] [CrossRef]
- Wang, B.; Gong, T.; Li, R.; Wang, Q.; Yang, J.; Liu, W.; Hu, G. Identification and analysis of PGM and SUS gene families in leymus Chinensis seed transcriptome. Mol. Plant Breed. 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Malinova, I.; Kunz, H.H.; Alseekh, S.; Herbst, K.; Fernie, A.R.; Gierth, M.; Fettke, J. Reduction of the cytosolic phosphoglu-comutase in Arabidopsis reveal impact on plant growth, seed and root development, and carbohydrate partitioning. PLoS ONE 2014, 9, e112468. [Google Scholar] [CrossRef]
- Hanson, K.R.; Mchale, N.A. A starchless mutant of Nicotiana sylvestris containing a modified plastid phosphoglucomutase. Plant Physiol. 1988, 88, 838–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tauberger, E.; Fernie, A.R.; Emmermann, M.; Renz, A.; Kossmann, J.; Willmitzer, L.; Trethewey, R.N. Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose-6-phosphate. Plant J. 2000, 23, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Lytovchenko, A.; Sweetlove, L.; Pauly, M.; Fernie, A.R. The influence of cytosolic phosphoglucomutase on photosynthetic carbohydrate metabolism. Planta 2002, 215, 1013–1021. [Google Scholar] [CrossRef]
- Nakamura-Gouvea, N.; Alves-Lima, C. Insights into agar and secondary metabolite pathways from the genome of the red alga Gracilaria domingensis (Rhodophyta, Gracilariales). J. Phycol. 2022, 58, 406–423. [Google Scholar] [CrossRef]
- Linka, M.; Jamai, A.; Weber, A.P.M. Functional characterization of the plastidic phosphate translocator gene family from the thermo-acidophilic red alga Galdieria sulphuraria reveals specific adaptations of primary carbon partitioning in green plants and red algae. Plant Physiol. 2008, 148, 1487–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Jia, X.; Wang, W.; Jin, Y.; Liu, W.; Wang, D.; Mao, Y.; Xie, C.; Liu, T. Floridean starch and floridoside metabolic pathways of Neoporphyra haitanensis and their regulatory mechanism under continuous darkness. Mar. Drugs 2021, 19, 664. [Google Scholar] [CrossRef]
- Macler, B.A. Regulation of carbon flow by nitrogen and light in the red alga, Gelidium coulteri. Plant Physiol. 1986, 82, 136–141. [Google Scholar] [CrossRef] [Green Version]
- Beither, R.; Haberman, S.; Livni, L. Complementarity in the regulation of phosphoglucomutase, phosphofructokinase and hexokinase; the role of glucose 1,6-bisphosphate. BBA-Enzymol. 1975, 2, 355–369. [Google Scholar] [CrossRef]
- Regni, C.; Shackelford, G.S.; Beamer, L.J. Complexes of the enzyme phosphomannomutase/phosphoglucomutase with a slow substrate and an inhibitor. Acta Crystallogr. Sect. F 2006, 62 Pt 8, 722–726. [Google Scholar] [CrossRef] [Green Version]
- Bartrons, R.; Carreras, M.; Climent, F.; Carreras, J. Inhibition of phosphoglucomutase by fructose 2,6-bisphosphate. BBA 1985, 842, 52–55. [Google Scholar] [CrossRef]
- Percival, M.D.; Doherty, K.; Gresser, M.J. Inhibition of phosphoglucomutase by vanadate. Biochemistry 1990, 29, 2764–2769. [Google Scholar] [CrossRef] [PubMed]
- Prathap, V.; Tyagi, A. Correlation between expression and activity of ADP- glucose pyrophosphorylase and starch synthase and their role in starch accumulation during grain filling under drought stress in rice. Plant Physiol. Biochem. 2020, 157, 239–243. [Google Scholar] [CrossRef]
- Siaut, M. Oil accumulation in the model green alga Chlamydomonas reinhardtii: Characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.S.; Koo, K.M.; Ryu, J.; Hong, M.J.; Kim, S.H.; Kwon, S.J.; Kim, J.B.; Choi, J.I.; Ahn, J.W. Overexpression of fructose-1,6-bisphosphate aldolase 1 enhances accumulation of fatty acids in Chlamydomonas reinhardtii. Algal Res. 2020, 47, 101825. [Google Scholar] [CrossRef]
- Lee, W.K.; Namasivayam, P.; Abdullah, J.O.; Ho, C.L. Transcriptome profiling of sulfate deprivation responses in two agarophytes Gracilaria changii and Gracilaria salicornia (Rhodophyta). Sci. Rep. 2017, 7, 46563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Zhao, X.; He, Y.; Yang, H. Cloning, purification and characterization of cytosolic fructose-1,6-bisphosphatase from mung bean (Vigna radiata). Food Chem. 2021, 347, 11. [Google Scholar] [CrossRef] [PubMed]
- Maruta, T.; Yonemitsu, M.; Yabuta, Y.; Tamoi, M.; Ishikawa, T.; Shigeoka, S. Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid biosynthesis. J. Biol. Chem. 2008, 283, 28842–28851. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Wu, J.; Wang, G.; Kang, Y.; Ooi, H.S.; Shen, T.; Zhao, X. Genomic analyses of unique carbohydrate and phytohormone metabolism in the macroalga Gracilariopsis lemaneiformis (Rhodophyta). BMC Plant Biol. 2018, 18, 94. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, L.; Wang, L.; Liu, L.; Li, L.; Sun, L.; Rao, Q.; Zhang, J.; Huang, S. JMJ704 positively regulates rice defense response against Xanthomonas oryzae pv. oryzae infection via reducing H3K4me2/3 associated with negative disease resistance regulators. BMC Plant Biol. 2015, 15, 286. [Google Scholar] [CrossRef] [Green Version]
- Hottiger, T.; Schmutz, P.; Wiemken, A. Heat-induced accumulation and futile cycling of trehalose in Saccharomyces cerevisiae. J. Bacteriol. 1987, 169, 1558–5522. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Sui, X.; Yu, Y.; Zhang, X. Effects of several environmental factors on the activity of α-galactosidase in Gracilaria lemaneiformis (Rhodophyta). Period. Ocean. Univ. China 2000, 30, 510–514. (In Chinese) [Google Scholar]
- Xiao, J.; Gu, C.; He, S.; Zhu, D.; Huang, Y.; Zhou, Q. Widely targeted metabolomics analysis reveals new biomarkers and mechanistic insights on chestnut (Castanea mollissima Bl.) calcification process. Food Res. Int. 2021, 141, 110–128. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Sun, P.; Zhang, Y.; Xuan, W.; Xu, N.; Sun, X. Response of trehalose, its degrading enzyme, sucrose, and floridoside/isofloridoside under abiotic stresses in Gracilariopsis lemaneiformis (Rhodophyta). J. Appl. Phycol. 2019, 31, 3861–3869. [Google Scholar] [CrossRef]



| Variables | PMI | PGM | PMM | UGPase | |
|---|---|---|---|---|---|
| Salinity treatment | |||||
| 7 d | Agar | −0.095 | 0.184 | −0.091 | 0.552 |
| 14 d | Agar | 0.033 | 0.476 | 0.572 | −0.017 |
| 21 d | Agar | −0.404 | 0.255 | −0.779 | 0.213 |
| Nitrogen treatment | |||||
| 7 d | Agar | 0.896 * | 0.955 * | 0.888 * | 0.787 |
| 14 d | Agar | 0.354 | 0.702 | 0.683 | 0.864 * |
| 21 d | Agar | 0.740 | 0.964 * | 0.612 | −0.980 * |
| Cultivation field | Agar | −0.043 | 0.999 * | 0.460 | 0.332 |
| Cultivation month | Agar | 0.763 | 0.973 * | 0.077 | 0.909 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Yu, X.; Liu, S.; Luo, S.; Chen, X.; Xu, N.; Sun, X. Identification, Characteristics and Function of Phosphoglucomutase (PGM) in the Agar Biosynthesis and Carbon Flux in the Agarophyte Gracilariopsis lemaneiformis (Rhodophyta). Mar. Drugs 2022, 20, 442. https://doi.org/10.3390/md20070442
Chen Q, Yu X, Liu S, Luo S, Chen X, Xu N, Sun X. Identification, Characteristics and Function of Phosphoglucomutase (PGM) in the Agar Biosynthesis and Carbon Flux in the Agarophyte Gracilariopsis lemaneiformis (Rhodophyta). Marine Drugs. 2022; 20(7):442. https://doi.org/10.3390/md20070442
Chicago/Turabian StyleChen, Qionglin, Xinlei Yu, Shixia Liu, Suya Luo, Xiaojiao Chen, Nianjun Xu, and Xue Sun. 2022. "Identification, Characteristics and Function of Phosphoglucomutase (PGM) in the Agar Biosynthesis and Carbon Flux in the Agarophyte Gracilariopsis lemaneiformis (Rhodophyta)" Marine Drugs 20, no. 7: 442. https://doi.org/10.3390/md20070442
APA StyleChen, Q., Yu, X., Liu, S., Luo, S., Chen, X., Xu, N., & Sun, X. (2022). Identification, Characteristics and Function of Phosphoglucomutase (PGM) in the Agar Biosynthesis and Carbon Flux in the Agarophyte Gracilariopsis lemaneiformis (Rhodophyta). Marine Drugs, 20(7), 442. https://doi.org/10.3390/md20070442






