Overexpressing CrePAPS Polyadenylate Activity Enhances Protein Translation and Accumulation in Chlamydomonas reinhardtii
Abstract
1. Introduction
2. Results
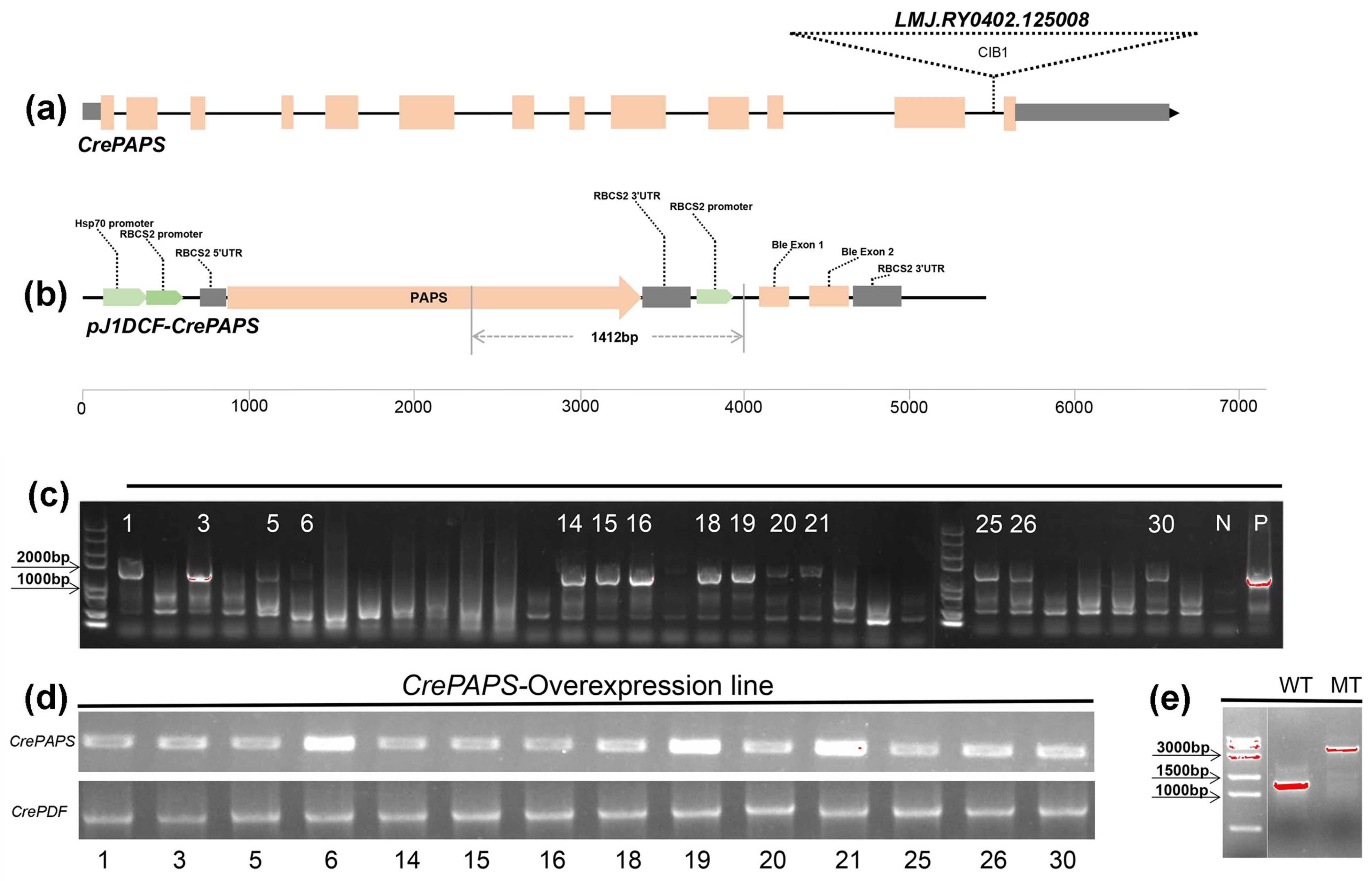
2.1. Generation of CrePAPS-Overexpressing Lines and Identification of Insertion Mutants
2.2. Analysis of CrePAPS Polyadenylate Activity
2.3. Label-Free Quantitative Proteomic Analysis of C. reinhardtii Strains Exhibiting Higher Polyadenylate Activity
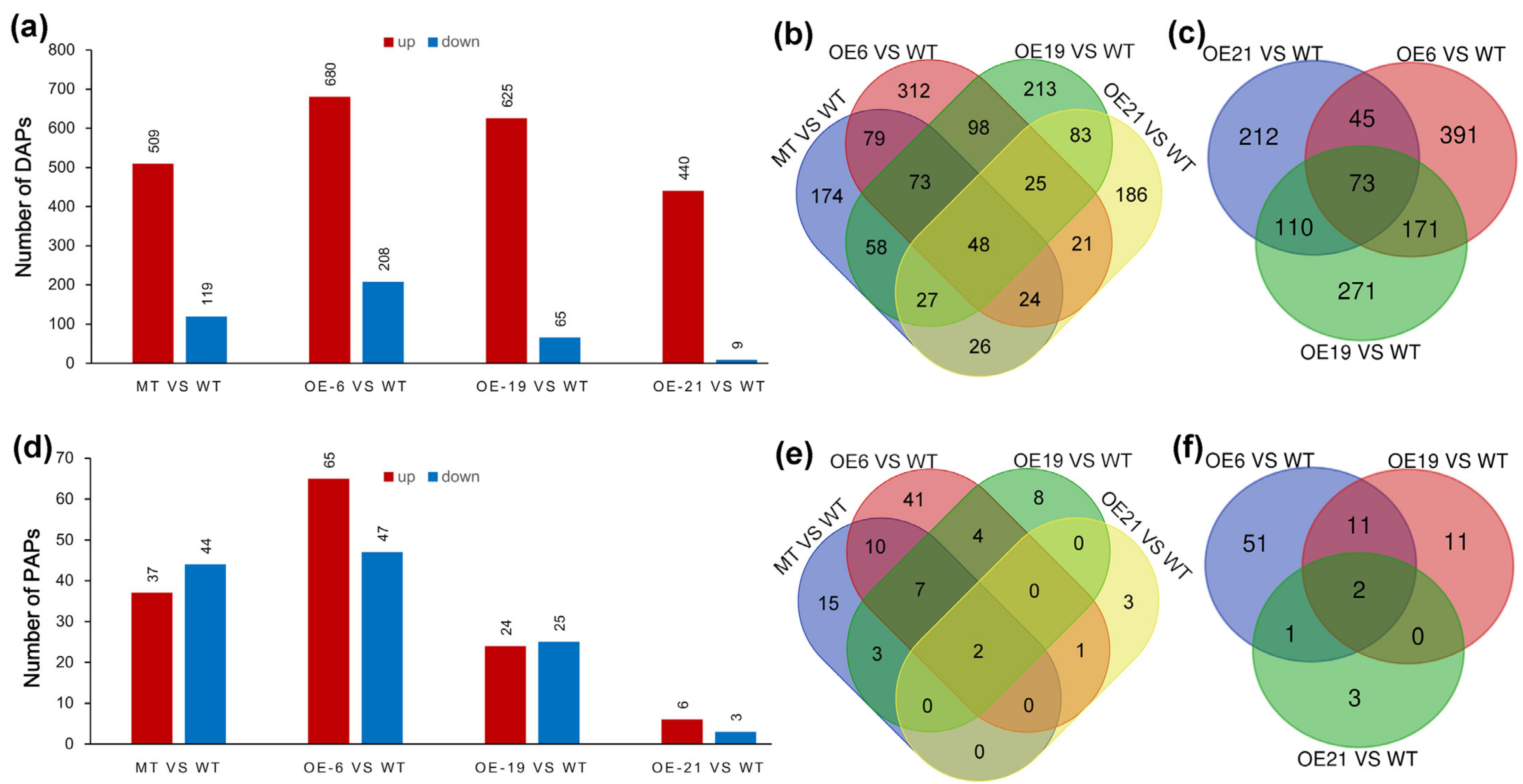
2.4. Screening Differentially Accumulated Proteins (DAPs) and Proportionally Altered Proteins (PAPs)
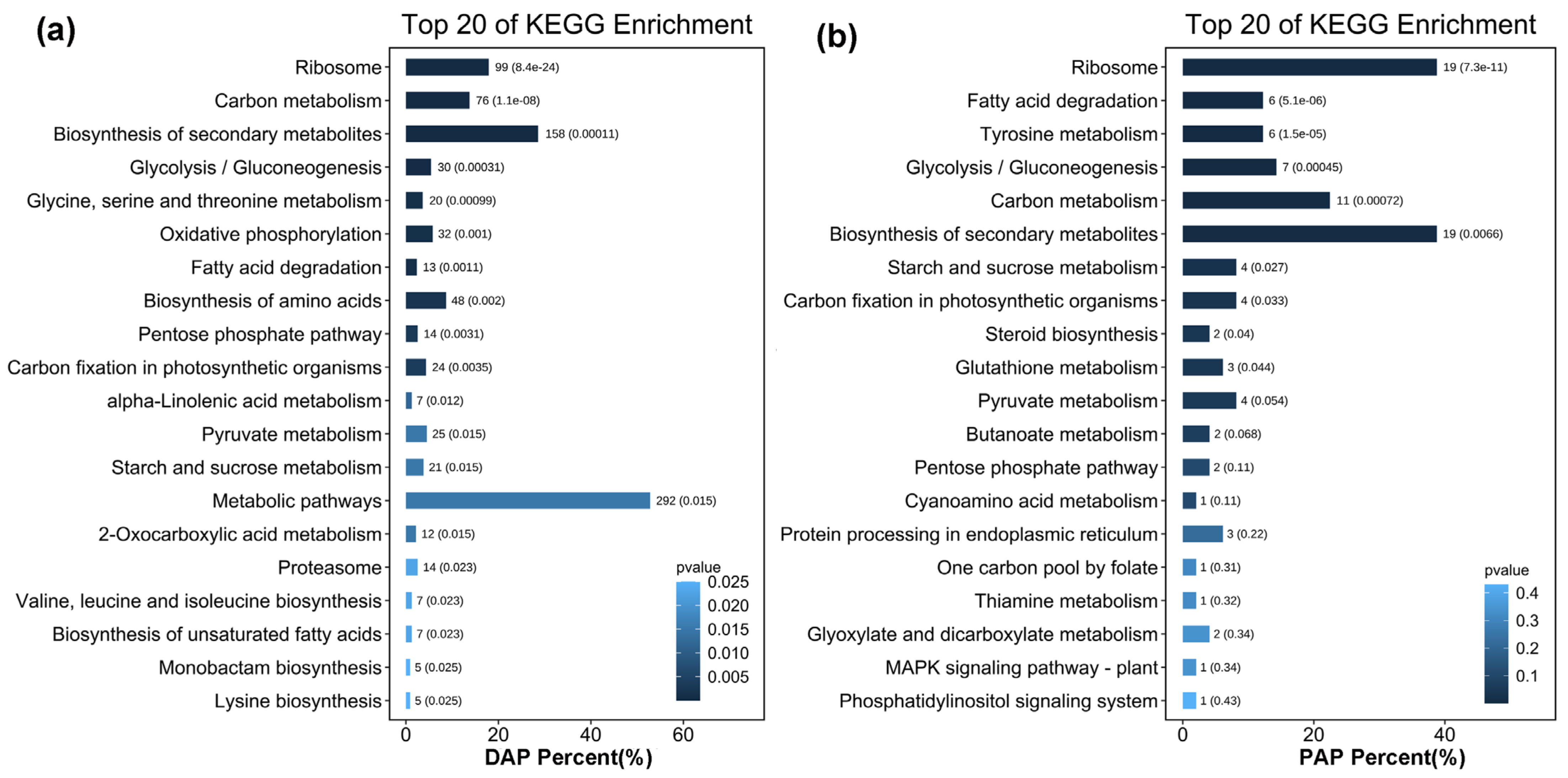
2.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Functional Annotation Analyses of Upregulated DAPs and PAPs
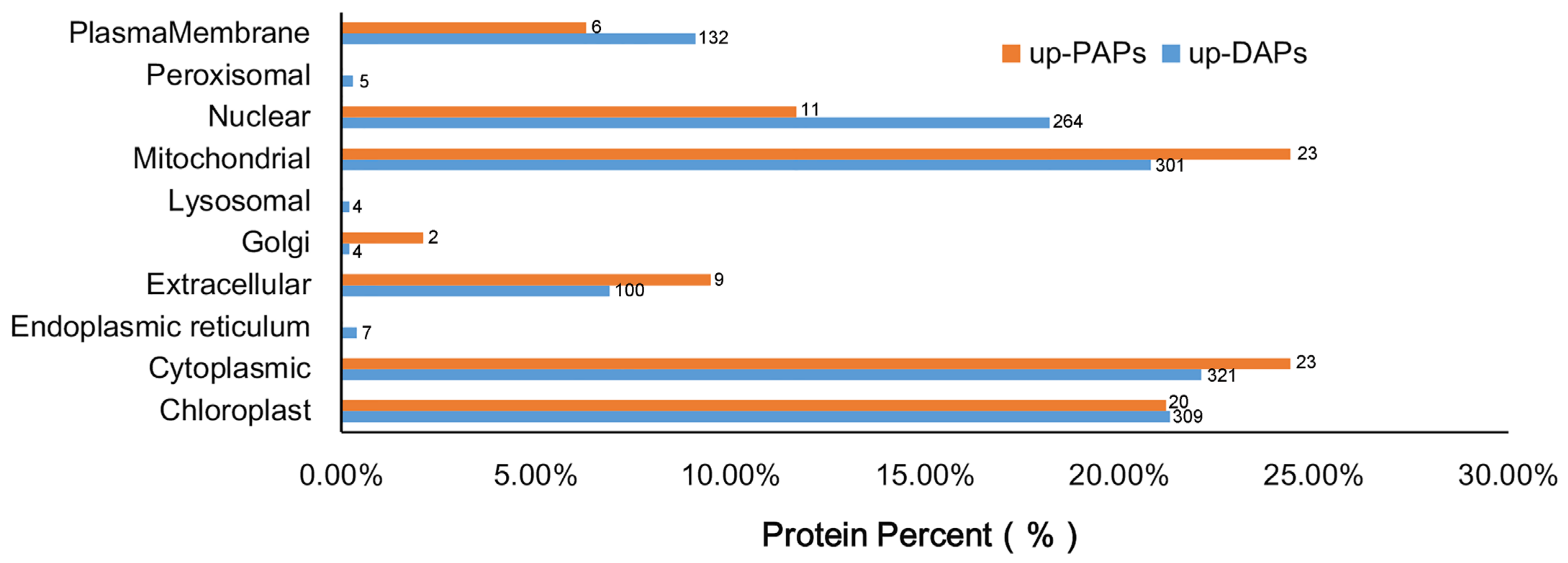
2.6. Subcellular Localisation of Up-DAPs and Up-PAPs
2.7. Analysis of Crude total Protein Content and Dissolved Protein Content
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. RNA Isolation and qRT-PCR
4.3. Overexpression Plasmid Construction and Transformation
4.4. Expression and Purification of CrePAPS Proteins in E. coli
4.5. In Vitro Polyadenylation Assay
4.6. Protein Extraction from Algal Cells
4.7. Protein Digestion and LC- MS/MS Analysis
4.8. Proteomic Data Statistics and Analysis
4.9. The Kjeldahl Method and BCA Assays
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pavlou, A.K.; Reichert, J.M. Recombinant protein therapeutics—success rates, market trends and values to 2010. Nat. Biotechnol. 2004, 22, 1513–1519. [Google Scholar] [CrossRef] [PubMed]
- Kesik-Brodacka, M. Progress in biopharmaceutical development. Biotechnol. Appl. Biochem. 2018, 65, 306–322. [Google Scholar] [CrossRef] [PubMed]
- Ansari, R.; Saghir, S.; Torisky, R.; Husain, K. Recombinant Protein Production: From Bench to Biopharming. Front. Drug Des. Discov. 2021, 10, 1–31. [Google Scholar] [CrossRef]
- Rech, E.; Vianna, G.; Melro Murad, A.; Cunha, N.; Lacorte, C.; Araujo, A.; Brigido, M.; Waters, M.; Barry, O.K.; Andrew, S.; et al. Recombinant proteins in plants. BMC Proc. 2014, 8, O1. [Google Scholar] [CrossRef]
- Gifre-Renom, L.; Aris, A.; Bach, A.; Garcia-Fruitos, E. Trends in recombinant protein use in animal production. Microb. Cell Factories 2017, 16, 40. [Google Scholar] [CrossRef] [PubMed]
- Gasser, B.; Mattanovich, D. Recombinant Protein Production in Yeast; Springer: New York, NY, USA, 2019. [Google Scholar] [CrossRef]
- Nosaki, S.; Miura, K. Transient Expression of Recombinant Proteins in Plants; Academic Press: Amsterdam, The Netherlands, 2021; Volume 660, pp. 193–203. [Google Scholar]
- Zahurancik, W.; Szkoda, B.; Lai, L.; Gopalan, V. Ramping Recombinant Protein Expression in Bacteria. Biochemistry 2020, 59, 2122–2124. [Google Scholar] [CrossRef]
- Grahl, I.; Reumann, S. Stramenopile microalgae as “green biofactories” for recombinant protein production. World J. Microbiol. Biotechnol. 2021, 37, 163. [Google Scholar] [CrossRef]
- Kindle, K.; Sodeinde, O. Nuclear and chloroplast transformation in Chlamydomonas reinhardtii: Strategies for genetic manipulation and gene expression. J. Appl. Phycol. 1994, 6, 231–238. [Google Scholar] [CrossRef]
- Rochaix, J.D. Chlamydomonas Reinhardtii as the Photosynthetic Yeast. Annu. Rev. Genet. 1995, 29, 209–230. [Google Scholar] [CrossRef]
- Rochaix, J.D. The three genomes of Chlamydomonas. Photosynth. Res. 2002, 73, 285–293. [Google Scholar] [CrossRef]
- Remacle, C.; Cardol, P.; Coosemans, N.; Gaisne, M.; Bonnefoy, N. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl. Acad. Sci. USA 2006, 103, 4771–4776. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhao, Z.; Wu, Z.; Fan, Z.; Chen, J.; Wu, J.; Li, J. Successful expression of heterologous egfp gene in the mitochondria of a photosynthetic eukaryote Chlamydomonas reinhardtii. Mitochondrion 2011, 11, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Fan, Z.; Zhao, Z.; Chen, J.; Li, J. Stable expression of antibiotic-resistant gene ble from Streptoalloteichus hindustanus in the mitochondria of Chlamydomonas reinhardtii. PLoS ONE 2012, 7, e35542. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, S.; Franklin, S.; Lerner, R. Expression and assembly of a fully active antibody in algae. Proc. Natl. Acad. Sci. USA 2003, 100, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Rasala, B.; Mayfield, S. The microalga Chlamydomonas reinhardtii as a platform for the production of human protein therapeutics. Bioeng. Bugs 2011, 2, 50–54. [Google Scholar] [CrossRef]
- Tran, M.; Van, C.; Barrera, D.; Pettersson, P.; Peinado, C.; Bui, J.; Mayfield, S. Production of unique immunotoxin cancer therapeutics in algal chloroplasts. Proc. Natl. Acad. Sci. USA 2012, 110, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Demurtas, O.; Massa, S.; Ferrante, P.; Venuti, A.; Franconi, R.; Giuliano, G. A Chlamydomonas-Derived Human Papillomavirus 16 E7 Vaccine Induces Specific Tumor Protection. PLoS ONE 2013, 8, e61473. [Google Scholar] [CrossRef]
- Zedler, J.; Gangl, D.; Hamberger, B.; Purton, S.; Robinson, C. Stable expression of a bifunctional diterpene synthase in the chloroplast of Chlamydomonas reinhardtii. J. Appl. Phycol. 2014, 27, 2271–2277. [Google Scholar] [CrossRef]
- Passmore, L.; Coller, J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell Biol. 2021, 23, 93–106. [Google Scholar] [CrossRef]
- Wu, C.; Roy, B.; He, F.; Yan, K.; Jacobson, A. Poly(A)-Binding Protein Regulates the Efficiency of Translation Termination. Cell Rep. 2020, 33, 108399. [Google Scholar] [CrossRef]
- Eckmann, C.; Rammelt, C.; Wahle, E. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA 2011, 2, 348–361. [Google Scholar] [CrossRef] [PubMed]
- Eliseeva, I.A.; Lyabin, D.N.; Ovchinnikov, L.P. Poly(A)-binding proteins: Structure, domain organization, and activity regulation. Biochem. Biokhimiia 2013, 78, 1377–1391. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.; Much, C.; DiGiacomo, M.; Azzi, C.; Ivanova, I.; Vitsios, D.M.; Pistolic, J.; Collier, P.; Moreira, P.N.; Benes, V.; et al. mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 2017, 548, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Alles, J.; Legnini, I.; Pacelli, M.; Rajewsky, N. Rapid Nuclear Deadenylation of Mammalian Messenger RNA. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kühn, U.; Wahle, E. Structure and function of poly(A) binding protein. Biochim. Biophys. Acta 2004, 1678, 67–84. [Google Scholar] [CrossRef]
- Machida, K.; Shigeta, T.; Yamamoto, Y.; Ito, T.; Svitkin, Y.; Sonenberg, N.; Imataka, H. Dynamic interaction of poly(A)-binding protein with the ribosome. Sci. Rep. 2018, 8, 17435. [Google Scholar] [CrossRef]
- Dunn, E.F.; Hammell, C.M.; Hodge, C.A.; Cole, C.N. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 2005, 19, 90. [Google Scholar] [CrossRef]
- Sato, H.; Maquat, L. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin. Genes Dev. 2009, 23, 2537–2550. [Google Scholar] [CrossRef]
- Nicholson, A.; Pasquinelli, A. Tales of Detailed Poly(A) Tails. Trends Cell Biol. 2018, 29, 191–200. [Google Scholar] [CrossRef]
- Rutledge, C.; Lau, H.-T.; Mangan, H.; Hardy, L.; Sunnotel, O.; Guo, F.; MacNicol, A.; Walsh, C.; Lees-Murdock, D. Efficient Translation of Dnmt1 Requires Cytoplasmic Polyadenylation and Musashi Binding Elements. PLoS ONE 2014, 9, e88385. [Google Scholar] [CrossRef]
- Preiss, T.; Muckenthaler, M.; Hentze, M.W. Poly(A)-tail-promoted translation in yeast: Implications for translational control. RNA 1998, 4, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- Beilharz, T.; Preiss, T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 2007, 13, 982–997. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Green, C. Analysis of Circadian Regulation of Poly(A)-Tail Length. Methods Enzymol. 2015, 551, 387–403. [Google Scholar] [CrossRef] [PubMed]
- Safaee, N.; Kozlov, G.; Noronha, A.; Xie, J.; Wilds, C.; Gehring, K. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol. Cell 2012, 48, 375–386. [Google Scholar] [CrossRef]
- Lingner, J.; Kellermann, J.; Keller, W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature 1991, 354, 496–498. [Google Scholar] [CrossRef]
- Hunt, A.; Xu, R.; Addepalli, B.; Rao, S.; Forbes, K.; Meeks, L.; Xing, D.; Mo, M.; Zhao, H.; Majumder, A.; et al. Arabidopsis mRNA polyadenylation machinery: Comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genom. 2008, 9, 220. [Google Scholar] [CrossRef]
- Meeks, L.R.; Addepalli, B.; Hunt, A.G. Characterization of genes encoding poly(A) polymerases in plants: Evidence for duplication and functional specialization. PLoS ONE 2009, 4, e8082. [Google Scholar] [CrossRef]
- Hafe, L.; Keller, E. The polyadenylate polymerases from yeast. J. Biol. Chem. 1975, 250, 1838–1846. [Google Scholar] [CrossRef]
- Kioka, T.; Yamagami, S.; Mui, K.; Onishi, H.; Kawakita, Y. Polyadenylate polymerase of the E1 mouse brain. Neurosci. Res. Suppl. 1991, 14, S93. [Google Scholar] [CrossRef]
- Thuresson, A.C.; Astrom, J.; Astrom, A.; Gronvik, K.O.; Virtanen, A. Multiple forms of poly(A) polymerases in human cells. Proc. Natl. Acad. Sci. USA 1994, 91, 979–983. [Google Scholar] [CrossRef]
- Czesnick, H.; Lenhard, M. Antagonistic control of flowering time by functionally specialized poly(A) polymerases in Arabidopsis thaliana. Plant J. 2016, 88, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Addepalli, B.; Meeks, L.R.; Forbes, K.P.; Hunt, A.G. Novel alternative splicing of mRNAs encoding poly(A) polymerases in Arabidopsis. Biochim. Biophys. Acta 2004, 1679, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Vi, S.L.; Trost, G.; Lange, P.; Czesnick, H.; Rao, N.; Lieber, D.; Laux, T.; Gray, W.M.; Manley, J.L.; Groth, D.; et al. Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. Proc. Natl. Acad. Sci. USA 2013, 110, 13994–13999. [Google Scholar] [CrossRef] [PubMed]
- Kappel, C.; Trost, G.; Czesnick, H.; Ramming, A.; Kolbe, B.; Vi, S.L.; Bispo, C.; Becker, J.D.; de Moor, C.; Lenhard, M. Genome-Wide Analysis of PAPS1-Dependent Polyadenylation Identifies Novel Roles for Functionally Specialized Poly(A) Polymerases in Arabidopsis thaliana. PLoS Genet. 2015, 11, e1005474. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mohamad, W.H.H.; Raus, R.; Zainuddin, Z.; Mat Amin, N.; Basherudin, N. Chlamydomonas reinhardtii as host in production of recombinant proteins for medical uses. Asia-Pac. J. Mol. Biol. Biotechnol. 2017, 25, 82–90. [Google Scholar]
- Shamriz, S.; Ofoghi, H. Outlook in the application of Chlamydomonas reinhardtii chloroplast as a platform for recombinant protein production. Biotechnol. Genet. Eng. Rev. 2017, 32, 92–106. [Google Scholar] [CrossRef]
- Mathieu-Rivet, E.; Marie-Christine, K.-M.; Vanier, G.; Ovide, C.; Burel, C.; Lerouge, P.; Bardor, M. Protein N-glycosylation in eukaryotic microalgae and its impact on the production of nuclear expressed biopharmaceuticals. Front. Plant Sci. 2014, 5, 359. [Google Scholar] [CrossRef]
- Ramos-Martinez, E.; Fimognari, L.; Sakuragi, Y. High yield secretion of recombinant proteins from the microalga Chlamydomonas reinhardtii. Plant Biotechnol. J. 2017, 15, 1214–1224. [Google Scholar] [CrossRef]
- Smejkal, G.; Robinson, M.; Lawrence, N.; Tao, F.; Saravis, C.; Schumacher, R. Increased Protein Yields from Escherichia coli Using Pressure-Cycling Technology. J. Biomol. Tech. 2006, 17, 173–175. [Google Scholar]
- Bora, N.; Bawa, Z.; Bill, R.; Wilks, M. The Implementation of a Design of Experiments Strategy to Increase Recombinant Protein Yields in Yeast (Review). Methods Mol. Biol. 2012, 866, 115–127. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Liu, H. Enhanced Recombinant Protein Production Under Special Environmental Stress. Front. Microbiol. 2021, 12, 630814. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, N.; Sun, R.; Wang, J.; Lu, F. Enhancement of synthetic mRNA translation efficiency through engineered poly(A) tails. bioRxiv 2021. [Google Scholar] [CrossRef]
- Gallie, D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991, 5, 2108–2116. [Google Scholar] [CrossRef]
- Xiang, K.; Bartel, D.P. The molecular basis of coupling between poly(A)-tail length and translational efficiency. eLife 2021, 10, e66493. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.; Weixlbaumer, A. The intricate relationship between transcription and translation. Proc. Natl. Acad. Sci. USA 2021, 118, e2106284118. [Google Scholar] [CrossRef] [PubMed]
- Piqué, M.; Lopez, J.; Méndez, R. Cytoplasmic mRNA Polyadenylation and Translation Assays. Methods Mol. Biol. 2006, 322, 183–198. [Google Scholar] [CrossRef]
- Richter, J. Breaking the Code of Polyadenylation-Induced Translation. Cell 2008, 132, 335–337. [Google Scholar] [CrossRef]
- Villalba, A.; Coll, O.; Gebauer, F. Cytoplasmic polyadenylation and translational control. Curr. Opin. Genet. Dev. 2011, 21, 452–457. [Google Scholar] [CrossRef]
- Conklin-Brittain, N.; Dierenfeld, E.; Wrangham, R.; Norconk, M.; Silver, S. Chemical Protein Analysis: A Comparison of Kjeldahl Crude Protein and Total Ninhydrin Protein from Wild, Tropical Vegetation. J. Chem. Ecol. 1999, 25, 2601–2622. [Google Scholar] [CrossRef]
- Gray, G.; Gray, N. A tail of translational regulation. eLife 2017, 6, e29104. [Google Scholar] [CrossRef]
- Zhu, W.; Smith, J.; Huang, C.-M. Mass Spectrometry-Based Label-Free Quantitative Proteomics. J. Biomed. Biotechnol. 2010, 2010, 840518. [Google Scholar] [CrossRef] [PubMed]
- Campion, E.; Loughran, S.; Walls, D. Protein Quantitation and Analysis of Purity; Springer: New York, NY, USA, 2017; Volume 1485, pp. 225–255. [Google Scholar]




| Strains | No. of MS/MS Fragments | No. of Unique Peptides | No. of Unique Proteins |
|---|---|---|---|
| MT-1 | 24,711 | 7131 | 2262 |
| MT-2 | 24,745 | 7185 | 2187 |
| MT-3 | 24,760 | 7013 | 2188 |
| WT-1 | 24,687 | 6814 | 2452 |
| WT-2 | 24,711 | 6931 | 2376 |
| WT-3 | 24,732 | 6982 | 2399 |
| OE-6-1 | 24,766 | 7038 | 2197 |
| OE-6-2 | 24,777 | 7347 | 2181 |
| OE-6-3 | 24,762 | 6818 | 2203 |
| OE-19-1 | 24,787 | 6836 | 2178 |
| OE-19-2 | 24,802 | 6858 | 2168 |
| OE-19-3 | 24,849 | 7513 | 2199 |
| OE-21-1 | 24,775 | 7014 | 2494 |
| OE-21-2 | 24,739 | 6939 | 2492 |
| OE-21-3 | 24,790 | 8293 | 2553 |
| Total | 371,393 | 15,265 | 3613 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhuang, J.; Ni, S.; Luo, H.; Zheng, K.; Li, X.; Lan, C.; Zhao, D.; Bai, Y.; Jia, B.; et al. Overexpressing CrePAPS Polyadenylate Activity Enhances Protein Translation and Accumulation in Chlamydomonas reinhardtii. Mar. Drugs 2022, 20, 276. https://doi.org/10.3390/md20050276
Wang Q, Zhuang J, Ni S, Luo H, Zheng K, Li X, Lan C, Zhao D, Bai Y, Jia B, et al. Overexpressing CrePAPS Polyadenylate Activity Enhances Protein Translation and Accumulation in Chlamydomonas reinhardtii. Marine Drugs. 2022; 20(5):276. https://doi.org/10.3390/md20050276
Chicago/Turabian StyleWang, Quan, Jieyi Zhuang, Shuai Ni, Haolin Luo, Kaijie Zheng, Xinyi Li, Chengxiang Lan, Di Zhao, Yongsheng Bai, Bin Jia, and et al. 2022. "Overexpressing CrePAPS Polyadenylate Activity Enhances Protein Translation and Accumulation in Chlamydomonas reinhardtii" Marine Drugs 20, no. 5: 276. https://doi.org/10.3390/md20050276
APA StyleWang, Q., Zhuang, J., Ni, S., Luo, H., Zheng, K., Li, X., Lan, C., Zhao, D., Bai, Y., Jia, B., & Hu, Z. (2022). Overexpressing CrePAPS Polyadenylate Activity Enhances Protein Translation and Accumulation in Chlamydomonas reinhardtii. Marine Drugs, 20(5), 276. https://doi.org/10.3390/md20050276








