Comparative Venomics of C. flavidus and C. frigidus and Closely Related Vermivorous Cone Snails
Abstract
:1. Introduction
2. Results
2.1. Comparative Conotoxin Profiles of C. flavidus and C. frigidus
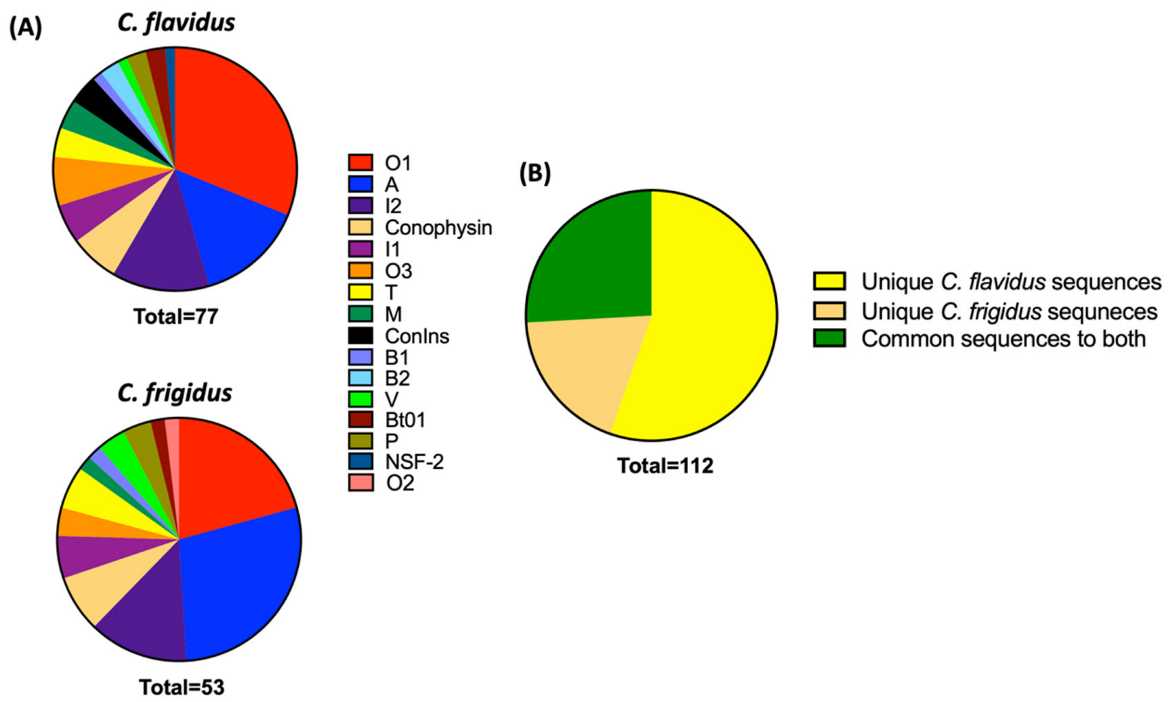
Comparative Proteomic Search of the Trasncriptomic Sequences in Respective Venom Duct Proteomes
2.2. Conotoxin Diversity in O1, I2, A and Contryphan Superfamilies of C. flavidus and C. frigidus at the Transcriptomic Level
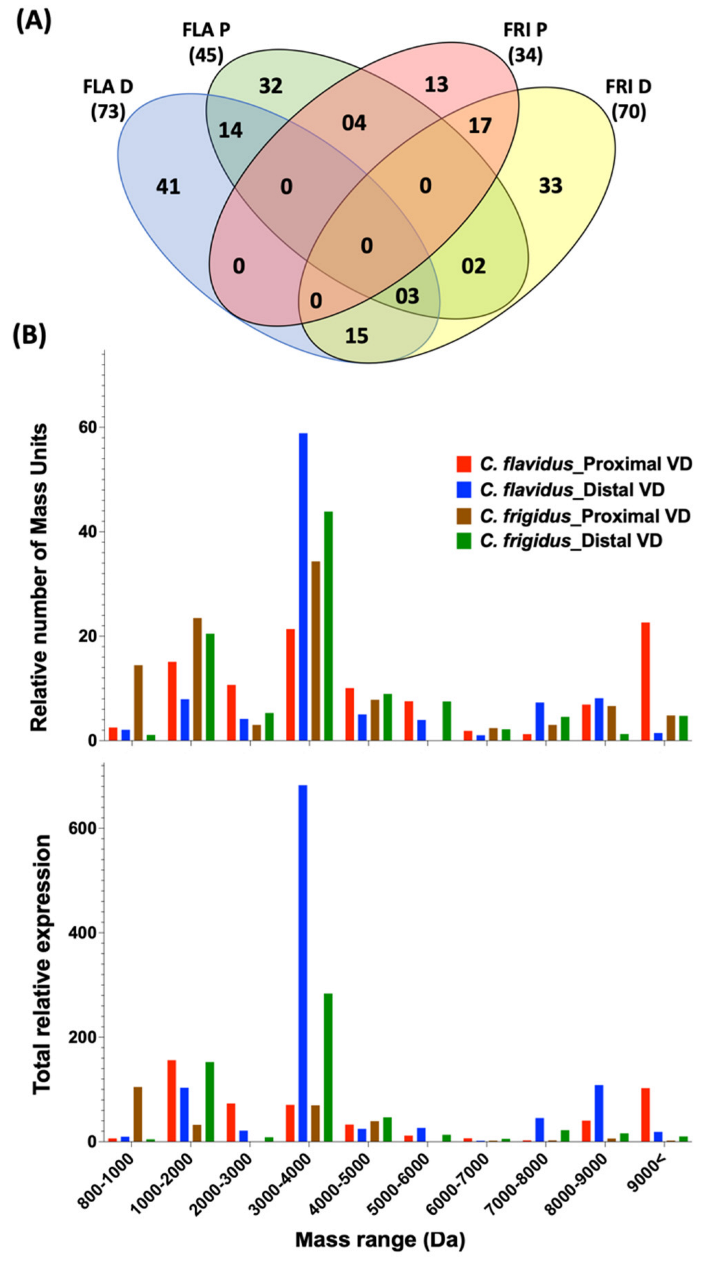
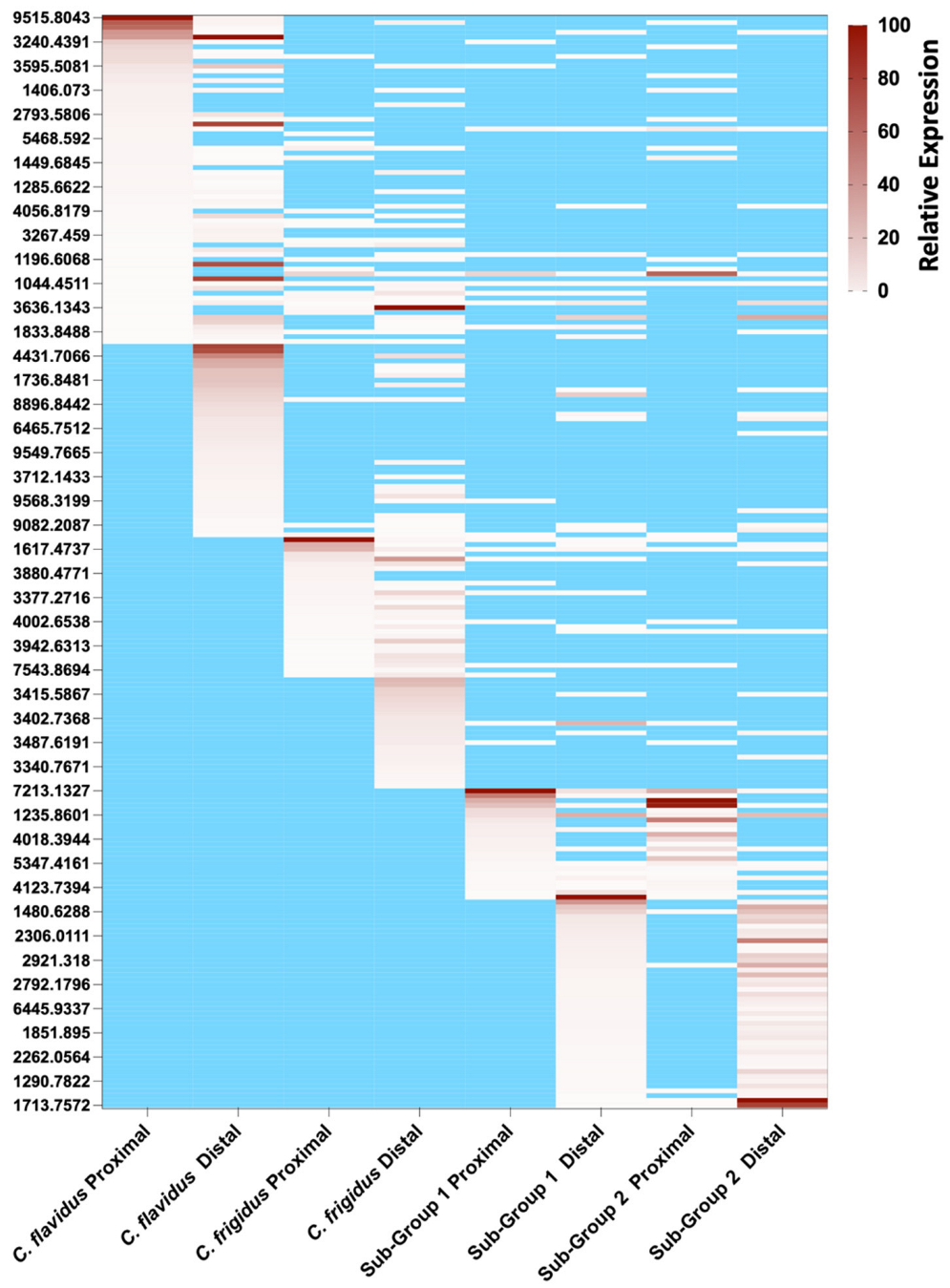
2.3. Peptide Mass Expression Patterns in the C. flavidus and C. frigidus Venom Duct Sections
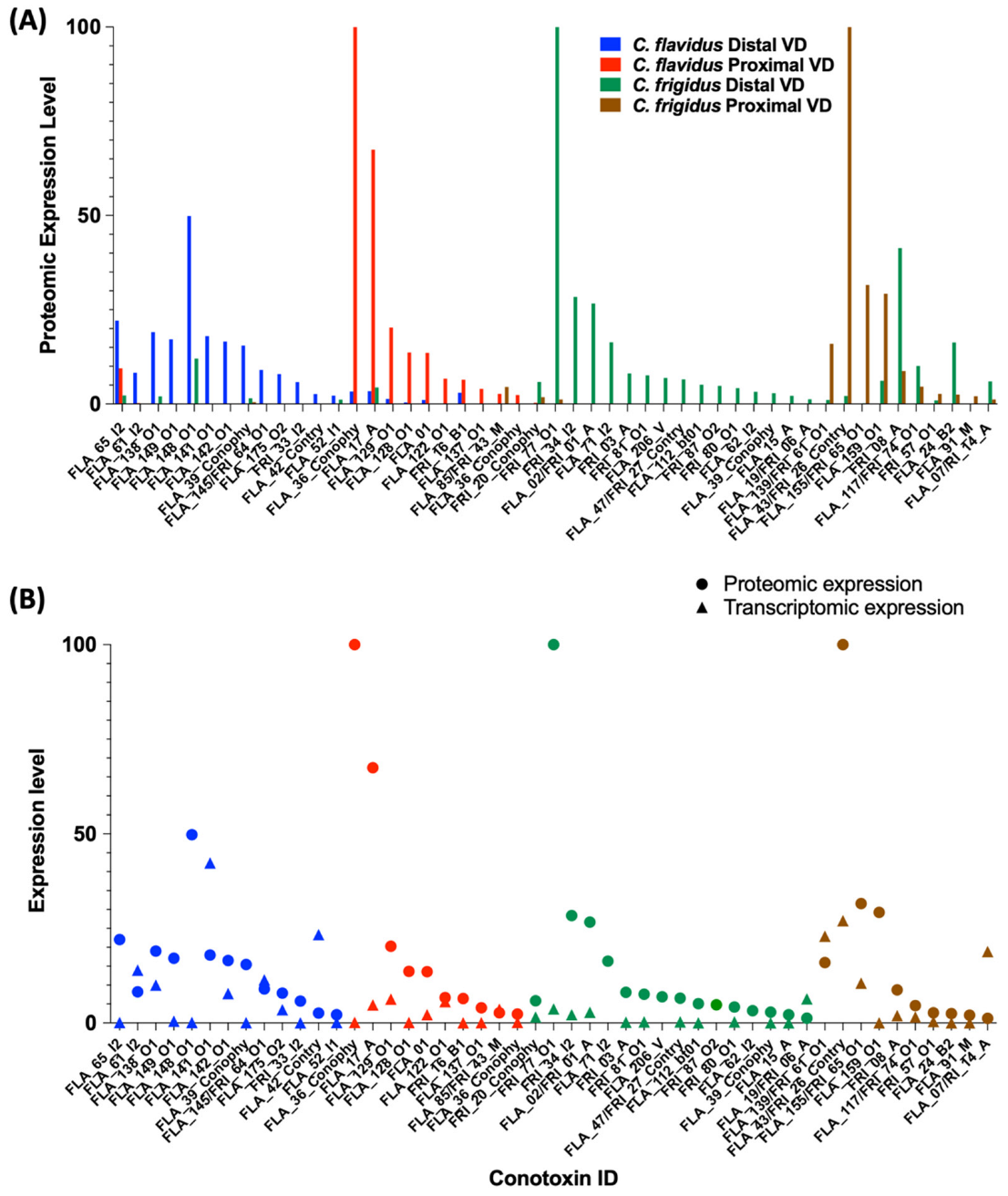
Venom Duct Localisation Patterns of Dominant Peptide Masses in C. flavidus and C. frigidus and Sequence Predictions for Dominant Peptide Masses
2.4. Comparison of C. frigidus and C. flavidus Toxin Expression Patterns to Two Morphologically Similar Unidentified Sub-Groups
3. Discussion
4. Materials and Methods
4.1. RNA Extraction, cDNA Library, 454 Sequencing, and Assembly
4.2. Conopeptide Sequence Analysis
4.3. Specimen Collection for Proteomics
4.4. Cone Snail Venom Peptide Extraction
4.5. Reduction, Alkylation, and Trypsin Digestion of Extracted Venoms
4.6. LC-ESI-MS Analysis
4.7. Proteomic Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MolluscaBase. Conus Linnaeus, 1758. World Register of Marine Species. Available online: http://www.marinespecies.org/aphia.php?p=taxdetails&id=137813 (accessed on 10 January 2022).
- Uribe, J.E.; Puillandre, N.; Zardoya, R. Beyond conus: Phylogenetic relationships of Conidae based on complete mitochondrial genomes. Mol. Phylogenet. Evol. 2017, 107, 142–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohn, A.J. Maximal species richness in Conus: Diversity, diet and habitat on reefs of northeast Papua New Guinea. Coral Reefs 2001, 20, 25–38. [Google Scholar]
- Vallejo, B. Inferring the mode of speciation in Indo-West Pacific Conus (Gastropoda: Conidae). J. Biogeogr. 2005, 32, 1429–1439. [Google Scholar] [CrossRef]
- Duda, T.F.J.; Kohn, A.J. Species-level phylogeography and evolutionary history of the hyperdiverse marine gastropod genus Conus. Mol. Phylogenet. Evol. 2005, 34, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Bouchet, P.; Duda, T.F.J.; Kauferstein, S.; Kohn, A.J.; Olivera, B.M.; Watkins, M.; Meyer, C. Molecular phylogeny and evolution of the cone snails (Gastropoda, Conoidea). Mol. Phylogenet. Evol. 2014, 78, 290–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abalde, S.; Tenorio, M.J.; Uribe, J.E.; Zardoya, R. Conidae phylogenomics and evolution. Zool. Scr. 2019, 48, 194–214. [Google Scholar] [CrossRef]
- Stanley, S.M. An analysis of the history of marine animal diversity. Paleobiology 2007, 33, 1–55. [Google Scholar] [CrossRef]
- Puillandre, N.; Koua, D.; Favreau, P.; Olivera, B.M.; Stöcklin, R. Molecular phylogeny, classification and evolution of conopeptides. J. Mol. Evol. 2012, 74, 297–309. [Google Scholar] [CrossRef] [Green Version]
- Abdelkrim, J.; Aznar-Cormano, L.; Fedosov, A.E.; Kantor, Y.I.; Lozouet, P.; Phuong, M.A.; Zaharias, P.; Puillandre, N. Exon-capture-based phylogeny and diversification of the venomous gastropods (Neogastropoda, Conoidea). Mol. Biol. Evol. 2018, 35, 2355–2374. [Google Scholar] [CrossRef]
- Fassio, G.; Modica, M.V.; Mary, L.; Zaharias, P.; Fedosov, A.E.; Gorson, J.; Kantor, Y.I.; Holford, M.; Puillandre, N. Venom Diversity and evolution in the most divergent cone snail genus Profundiconus. Toxins 2019, 11, 623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, A.W.; Duda, T.F.J. Reticulate evolution in conidae: Evidence of nuclear and mitochondrial introgression. Mol. Phylogenetics Evol. 2021, 161, 107182. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Griffin, J.; Lewis, R.J. Phyla Molluska: The venom apparatus of cone snails. In Marine and Freshwater Toxins; Gopalakrishnakone, P., Haddad, V., Jr., Tubaro, A., Kim, E., Kem, W.R., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 327–340. [Google Scholar]
- Duda, T.F.J.; Lee, T. Ecological release and venom evolution of a predatory marine snail at Easter Island. PLoS ONE 2009, 4, e5558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duda, T.F.J.; Bolin, M.B.; Meyer, C.P.; Kohn, A.J. Hidden diversity in a hyperdiverse gastropod genus: Discovery of previously unidentified members of a Conus species complex. Mol. Phylogenet. Evol. 2008, 49, 867–876. [Google Scholar]
- Lawler, A.J.; Duda, T.F.J. Molecular and morphometric data suggest the presence of a neglected species in the marine gastropod family Conidae. Mol. Phylogenet. Evol. 2017, 109, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Cerda, P.A.; Hewitt, T.L.; Haponski, A.E.; Duda, T.F.J. Unraveling cryptic morphological diversity in a marine snail species complex using nuclear genomic data. Am. Malacol. Bull. 2020, 37, 45–52. [Google Scholar] [CrossRef]
- Barghi, N.; Concepcion, G.P.; Olivera, B.M.; Lluisma, A.O. High conopeptide diversity in Conus tribblei revealed through analysis of venom duct transcriptome using two high-throughput sequencing platforms. Mar. Biotech. 2015, 17, 81–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Barghi, N.; Lu, A.; Fedosov, A.E.; Bandyopadhyay, P.K.; Lluisma, A.O.; Concepcion, G.P.; Yandell, M.; Olivera, B.M.; Safavi-Hemami, H. Divergence of the venom exogene repertoire in two sister species of Turriconus. Genome Biol. Evol. 2017, 9, 2211–2225. [Google Scholar] [CrossRef] [Green Version]
- Pardos-Blas, J.R.; Irisarri, I.; Abalde, S.; Tenorio, M.J.; Zardoya, R. Conotoxin diversity in the venom gland transcriptome of the magician’s cone, pionoconus magus. Mar. Drugs 2019, 17, 553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Himaya, S.W.A.; Jin, A.H.; Hamilton, B.; Rai, S.K.; Alewood, P.; Lewis, R.J. Venom duct origins of prey capture and defensive conotoxins in piscivorous Conus striatus. Sci. Rep. 2021, 11, 13282. [Google Scholar] [CrossRef] [PubMed]
- Dutt, M.; Dutertre, S.; Jin, A.H.; Lavergne, V.; Alewood, P.F.; Lewis, R.J. Venomics reveals venom complexity of the piscivorous cone snail, Conus tulipa. Mar. Drugs 2019, 17, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivera, B.M.; McIntosh, J.M.; Cruz, L.J.; Luque, F.A.; Gray, W.R. Purification and sequence of a presynaptic peptide toxin from Conus geographus venom. Biochemistry 1984, 23, 5087–5090. [Google Scholar] [CrossRef]
- Jin, A.H.; Muttenthaler, M.; Dutertre, S.; Himaya, S.W.A.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Conotoxins: Chemistry and biology. Chem. Rev. 2019, 119, 11510–11549. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.H.; Dekan, Z.; Smout, M.J.; Wilson, D.; Dutertre, S.; Vetter, I.; Lewis, R.J.; Loukas, A.; Daly, N.L.; Alewood, P.F. Conotoxin Φ-MiXXVIIA from the superfamily G2 employs a novel cysteine framework that mimics granulin and displays anti-apoptotic activity. Angew. Chem. Int. Ed. 2017, 56, 14973–14976. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.H.; Kaas, Q.; Jones, A.; Alewood, P.F.; Lewis, R.J. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell Proteom. 2013, 12, 312–329. [Google Scholar] [CrossRef] [Green Version]
- Jin, A.H.; Dutertre, S.; Kaas, Q.; Lavergne, V.; Kubala, P.; Lewis, R.J.; Alewood, P.F. Transcriptomic messiness in the venom duct of Conus miles contributes to conotoxin diversity. Mol. Cell Proteom. 2013, 12, 3824–3833. [Google Scholar] [CrossRef] [Green Version]
- Akondi, K.B.; Muttenthaler, M.; Dutertre, S.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Discovery, synthesis, and structure–activity relationships of conotoxins. Chem. Rev. 2014, 114, 5815–5847. [Google Scholar] [CrossRef] [PubMed]
- Buczek, O.; Bulaj, G.; Olivera, B.M. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol. Life Sci. 2005, 62, 3067–3079. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.H.; Vetter, I.; Himaya, S.W.A.; Alewood, P.F.; Lewis, R.J.; Dutertre, S. Transcriptome and proteome of Conus planorbis identify the nicotinic receptors as primary target for the defensive venom. Proteomics 2015, 15, 4030–4040. [Google Scholar] [CrossRef] [PubMed]
- Jin, A.H.; Dutertre, S.; Dutt, M.; Lavergne, V.; Jones, A.; Lewis, R.J.; Alewood, P.F. Transcriptomic-proteomic correlation in the predation-evoked venom of the cone snail, Conus imperialis. Mar. Drugs 2019, 17, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lluisma, A.O.; Milash, B.A.; Moore, B.; Olivera, B.M.; Bandyopadhyay, P.K. Novel venom peptides from the cone snail Conus pulicarius discovered through next-generation sequencing of its venom duct transcriptome. Mar. Genom. 2012, 5, 43–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phuong, M.A.; Mahardika, G.N. Targeted sequencing of venom genes from cone snail genomes reveals coupling between dietary breadth and conotoxin diversity. Biorxiv 2017, 1, 107672. [Google Scholar]
- Zamora-Bustillos, R.; Martínez-Núñez, M.A.; Aguilar, M.B.; Collí-Dula, R.C.; Brito-Domínguez, D.A. Identification of novel conotoxin precursors from the cone snail Conus spurius by high-throughput RNA sequencing. Mar. Drugs 2021, 19, 547. [Google Scholar] [CrossRef] [PubMed]
- Monnier, E.; Limpalaër, L.; Robin, A.; Roux, C.A. Taxonomic Iconography of Living Conidae; ConchBooks: Bonn, Germany, 2018; Volume 1–2. [Google Scholar]
- Dutertre, S.; Jin, A.H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J.; et al. Evolution of separate predation-and defence-evoked venoms in carnivorous cone snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prashanth, J.R.; Dutertre, S.; Jin, A.H.; Lavergne, V.; Hamilton, B.; Cardoso, F.C.; Griffin, J.; Venter, D.J.; Alewood, P.F.; Lewis, R.J. The role of defensive ecological interactions in the evolution of conotoxins. Mol. Ecol. 2016, 25, 598–615. [Google Scholar] [CrossRef] [PubMed]
- Abalde, S.; Tenorio, M.J.; Afonso, C.M.; Uribe, J.E.; Echeverry, A.M.; Zardoya, R. Phylogenetic relationships of cone snails endemic to Cabo Verde based on mitochondrial genomes. BMC Evol. Biol. 2017, 17, 231. [Google Scholar] [CrossRef] [Green Version]
- Abalde, S.; Tenorio, M.J.; Afonso, C.M.; Zardoya, R. Comparative transcriptomics of the venoms of continental and insular radiations of West African cones. Proc. R. Soc. B 2020, 287, 20200794. [Google Scholar] [CrossRef] [PubMed]
- Himaya, S.W.A.; Jin, A.H.; Dutertre, S.; Giacomotto, J.; Mohialdeen, H.; Vetter, I.; Alewood, P.F.; Lewis, R.J. Comparative venomics reveals the complex prey capture strategy of the piscivorous cone snail Conus catus. J. Prot. Res. 2015, 14, 4372–4381. [Google Scholar] [CrossRef] [PubMed]
- Himaya, S.W.A.; Rai, S.K.; Pamfili, G.; Jin, A.H.; Alewood, P.F.; Lewis, R.J. Venomic interrogation reveals the complexity of Conus striolatus venom. Aust. J. Chem. 2020, 73, 357–365. [Google Scholar] [CrossRef]
- Duda, T.F.J.; Chang, D.; Lewis, B.D.; Lee, T. Geographic variation in venom allelic composition and diets of the widespread predatory marine gastropod Conus ebraeus. PLoS ONE 2009, 4, e6245. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.R.; McArthur, J.R.; Brust, A.; Bhola, R.F.; Rosengren, K.J.; Ragnarsson, L.; Dutertre, S.; Alewood, P.F.; Christie, M.J.; Adams, D.J.; et al. Novel analgesic ω-conotoxins from the vermivorous cone snail Conus moncuri provide new insights into the evolution of conopeptides. Sci. Rep. 2018, 8, 13397. [Google Scholar] [CrossRef] [PubMed]
- Grandal, M.; Hoggard, M.; Neely, B.; Davis, W.C.; Marí, F. Proteogenomic assessment of intraspecific venom variability: Molecular adaptations in the venom arsenal of conus purpurascens. Mol. Cell Prot. 2021, 20, 100100. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, E.C.; Watkins, M.; Juszczak, L.J.; Cruz, L.J.; Olivera, B.M. Contryphans from Conus textile venom ducts. Toxicon 2001, 39, 803–808. [Google Scholar] [CrossRef]
- Sabareesh, V.; Gowd, K.H.; Ramasamy, P.; Sudarslal, S.; Krishnan, K.S.; Sikdar, S.K.; Balaram, P. Characterization of contryphans from Conus loroisii and Conus amadis that target calcium channels. Peptides 2006, 27, 2647–2654. [Google Scholar] [CrossRef] [PubMed]
- Franklin, J.B.; Rajesh, R.P.; Vinithkumar, N.V.; Kirubagaran, R. Identification of short single disulfide-containing contryphans from the venom of cone snails using de novo mass spectrometry-based sequencing methods. Toxicon 2017, 132, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Duda, T.F.J.; Remigio, E.A. Variation and evolution of toxin gene expression patterns of six closely related venomous marine snails. Mol. Ecol. 2008, 17, 3018–3032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavergne, V.; Dutertre, S.; Jin, A.H.; Lewis, R.J.; Taft, R.J.; Alewood, P.F. Systematic interrogation of the Conus marmoreus venom duct transcriptome with ConoSorter reveals 158 novel conotoxins and 13 new gene superfamilies. BMC Genom. 2013, 14, 708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaas, Q.; Yu, R.; Jin, A.H.; Dutertre, S.; Craik, D.J. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012, 40, D325–D330. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, J.R.; Lewis, R.J. An efficient transcriptome analysis pipeline to accelerate venom peptide discovery and characterisation. Toxicon 2015, 107, 282–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hale, J.E.; Butler, J.P.; Gelfanova, V.; You, J.S.; Knierman, M.D. A simplified procedure for the reduction and alkylation of cysteine residues in proteins prior to proteolytic digestion and mass spectral analysis. Anal. Biochem. 2004, 333, 174–181. [Google Scholar] [CrossRef] [PubMed]
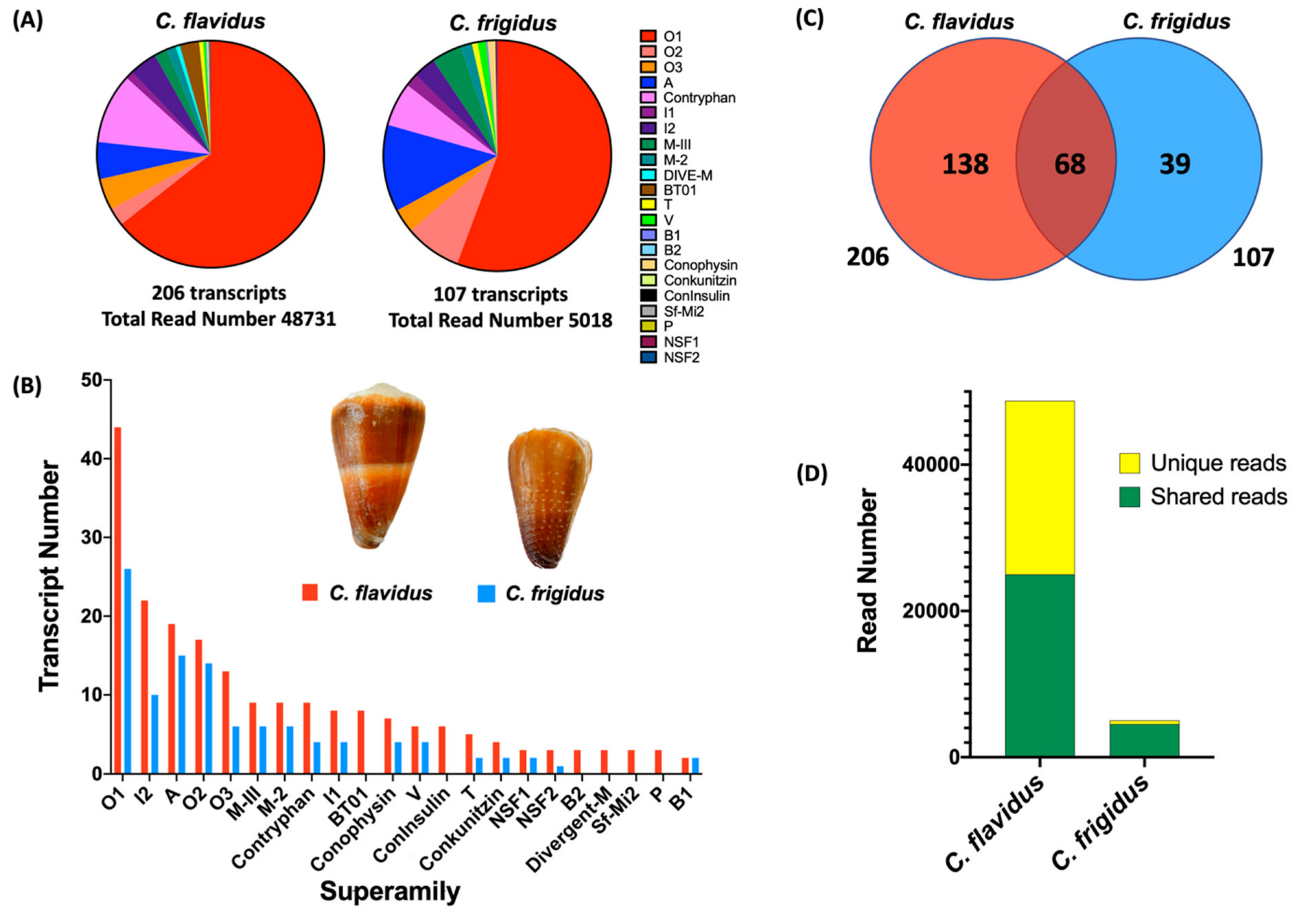

| Unique C. flavidus Transcripts | Unique C. frigidus Transcripts | Common Transcripts | All Transcripts | |||||
|---|---|---|---|---|---|---|---|---|
| Superfamily | Number | Read Number (Relative Expression) | Number | Read Number (Relative Expression) | Number | C. flavidus Read Number (Relative Expression) | C. frigidus Read Number (Relative Expression) | Number |
| O1 | 29 | 14,539 (39.4) | 11 | 241 (4.9) | 15 | 16,836 (39.2) | 2561 (51.9) | 55 |
| O2 | 8 | 320 (0.4) | 5 | 52 (1.1) | 9 | 919 (1.2) | 350 (7.1) | 22 |
| O3 | 8 | 959 (1.2) | 1 | 18 (0.4) | 5 | 1250 (1.6) | 152 (3.1) | 14 |
| I2 | 19 | 1781 (2.2) | 6 | 95 (1.9) | 4 | 140 (0.2) | 63 (1.3) | 28 |
| I1 | 5 | 291 (0.4) | 1 | 9 (0.2) | 3 | 258 (0.3) | 84 (1.7) | 9 |
| A | 11 | 635 (0.8) | 6 | 56 (1.1) | 8 | 1930 (2.4) | 462 (9.4) | 25 |
| M | 9 | 417 (0.52) | 3 | 8 (0.2) | 9 | 1135 (1.4) | 289 (5.9) | 21 |
| Divergent M | 3 | 291 (0.6) | 0 | 0 | 0 | 0 | 0 | 3 |
| Contryphans | 4 | 2846 (3.6) | 0 | 0 | 4 | 2038 (2.6) | 310 (6.3) | 9 |
| Conophysin | 5 | 21 (0.03) | 2 | 21 (0.4) | 2 | 25 (0.03) | 27 (0.6) | 9 |
| T | 3 | 149 (1.2) | 0 | 0 | 2 | 132 (0.2) | 41 (0.8) | 5 |
| V | 2 | 42 (0.05) | 0 | 0 | 4 | 143 (0.2) | 60 (1.2) | 6 |
| B1 | 2 | 37 (0.05) | 2 | 11 (0.2) | 0 | 0 | 0 | 4 |
| B2 | 3 | 57 (0.07) | 0 | 0 | 0 | 0 | 0 | 3 |
| Conkunitzin | 3 | 15 (0.02) | 1 | 3 (0.06) | 1 | 24 (0.03) | 4 (0.08) | 5 |
| NSF1 | 2 | 12 (0.02) | 1 | 2 (0.04) | 1 | 30 (0.04) | 9 (02) | 4 |
| NSF2 | 2 | 25 (0.03) | 0 | 0 | 1 | 22 (0.03) | 4 (0.08) | 3 |
| bt01 | 8 | 1349 (1.7) | 0 | 0 | 0 | 0 | 0 | 8 |
| ConoInsulin | 6 | 20 (0.03) | 0 | 0 | 0 | 0 | 0 | 6 |
| P | 3 | 17 (0.02) | 0 | 0 | 0 | 0 | 0 | 3 |
| SF-Mi2 | 3 | 19 (0.02) | 0 | 0 | 0 | 0 | 0 | 3 |
| Total | 138 | 23,844 (48.93) | 39 | 516 (10.3) | 68 | 24,887 (51.07) | 4502 (89.7) | 245 |
| Species/Group | C. frigidus | C. flavidus | Sub-Group 1 | Sub-Group 2 |
|---|---|---|---|---|
| Shell (In the native collected form) |  |  |  |  |
| Shell length | 3.5–4 cm | 4.5–5 cm | 4.5–5 cm | 4–4.5 cm |
| Syphon color | Yellow and black stripes | White and black stripes | White and black stripes | White and black stripes |
| Crown height | 45–50 mm | 30–35 mm | 40–45 mm | 10–15 mm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Himaya, S.W.A.; Arkhipov, A.; Yum, W.Y.; Lewis, R.J. Comparative Venomics of C. flavidus and C. frigidus and Closely Related Vermivorous Cone Snails. Mar. Drugs 2022, 20, 209. https://doi.org/10.3390/md20030209
Himaya SWA, Arkhipov A, Yum WY, Lewis RJ. Comparative Venomics of C. flavidus and C. frigidus and Closely Related Vermivorous Cone Snails. Marine Drugs. 2022; 20(3):209. https://doi.org/10.3390/md20030209
Chicago/Turabian StyleHimaya, S. W. A., Alexander Arkhipov, Wai Ying Yum, and Richard J. Lewis. 2022. "Comparative Venomics of C. flavidus and C. frigidus and Closely Related Vermivorous Cone Snails" Marine Drugs 20, no. 3: 209. https://doi.org/10.3390/md20030209
APA StyleHimaya, S. W. A., Arkhipov, A., Yum, W. Y., & Lewis, R. J. (2022). Comparative Venomics of C. flavidus and C. frigidus and Closely Related Vermivorous Cone Snails. Marine Drugs, 20(3), 209. https://doi.org/10.3390/md20030209






