Biochemical Characterization and Cold-Adaption Mechanism of a PL-17 Family Alginate Lyase Aly23 from Marine Bacterium Pseudoalteromonas sp. ASY5 and Its Application for Oligosaccharides Production
Abstract
:1. Introduction
2. Results and Discussion
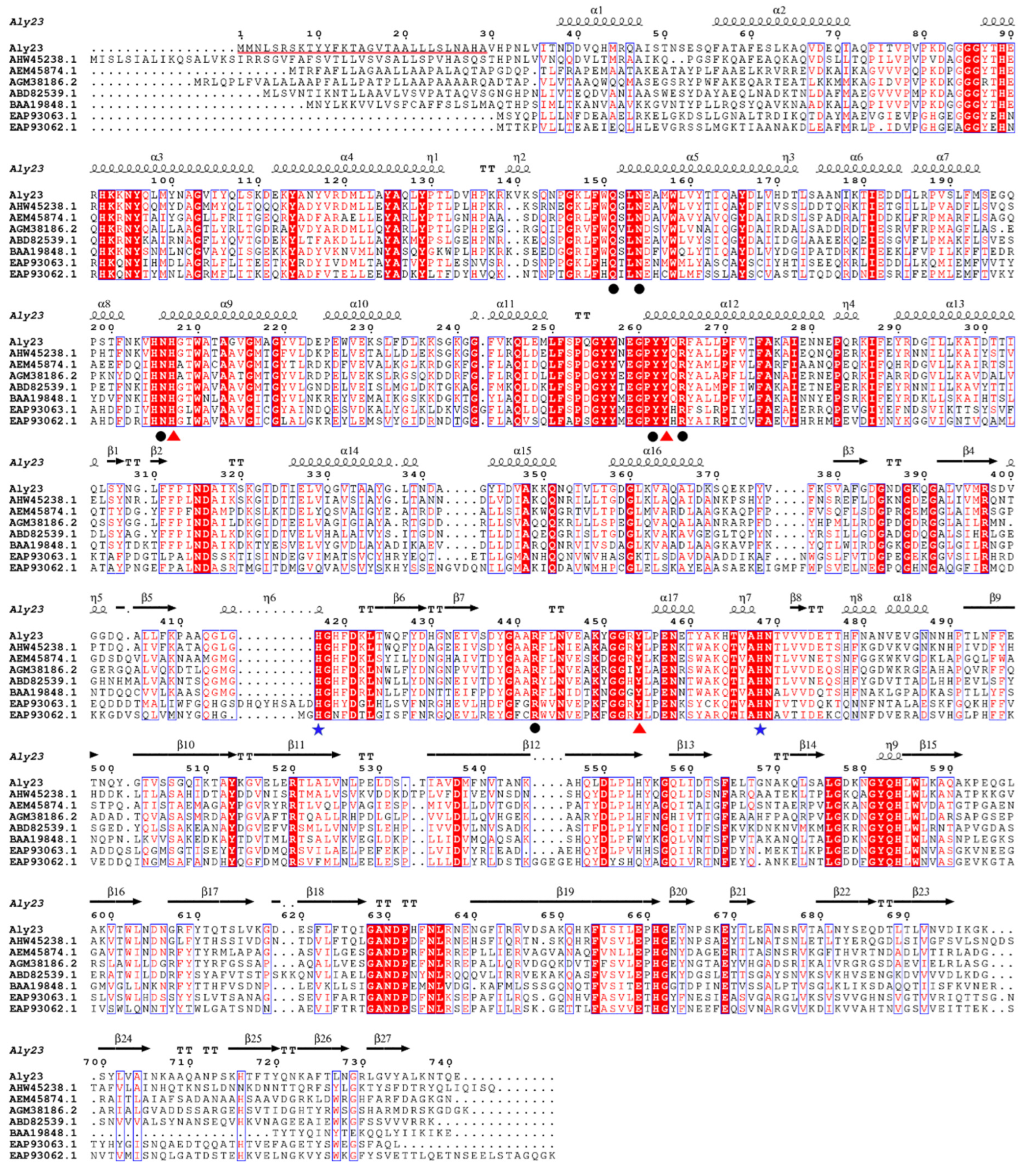
2.1. Cloning and Sequence Analysis of Aly23
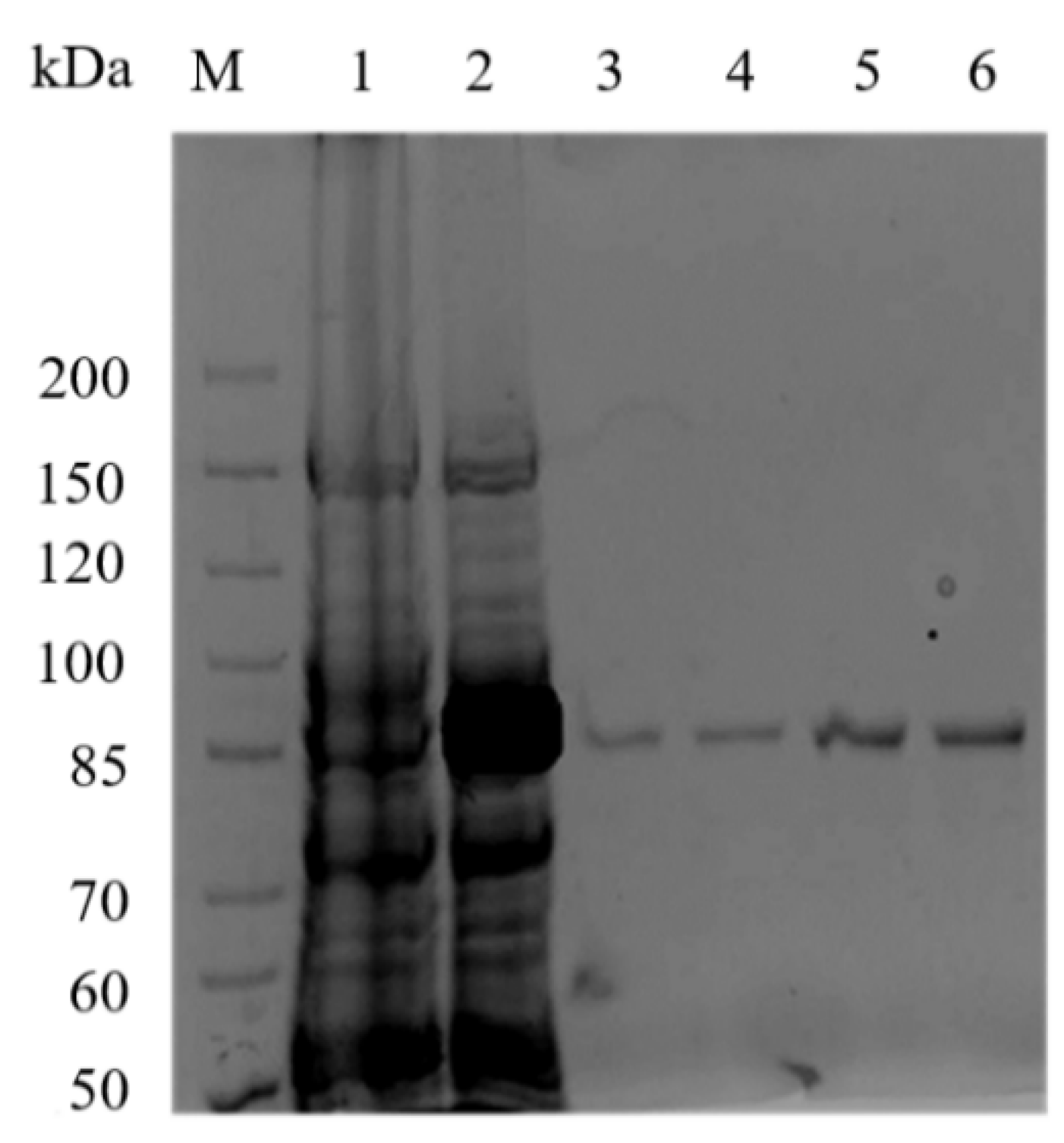
2.2. Expression and Purification of Recombinant Aly23
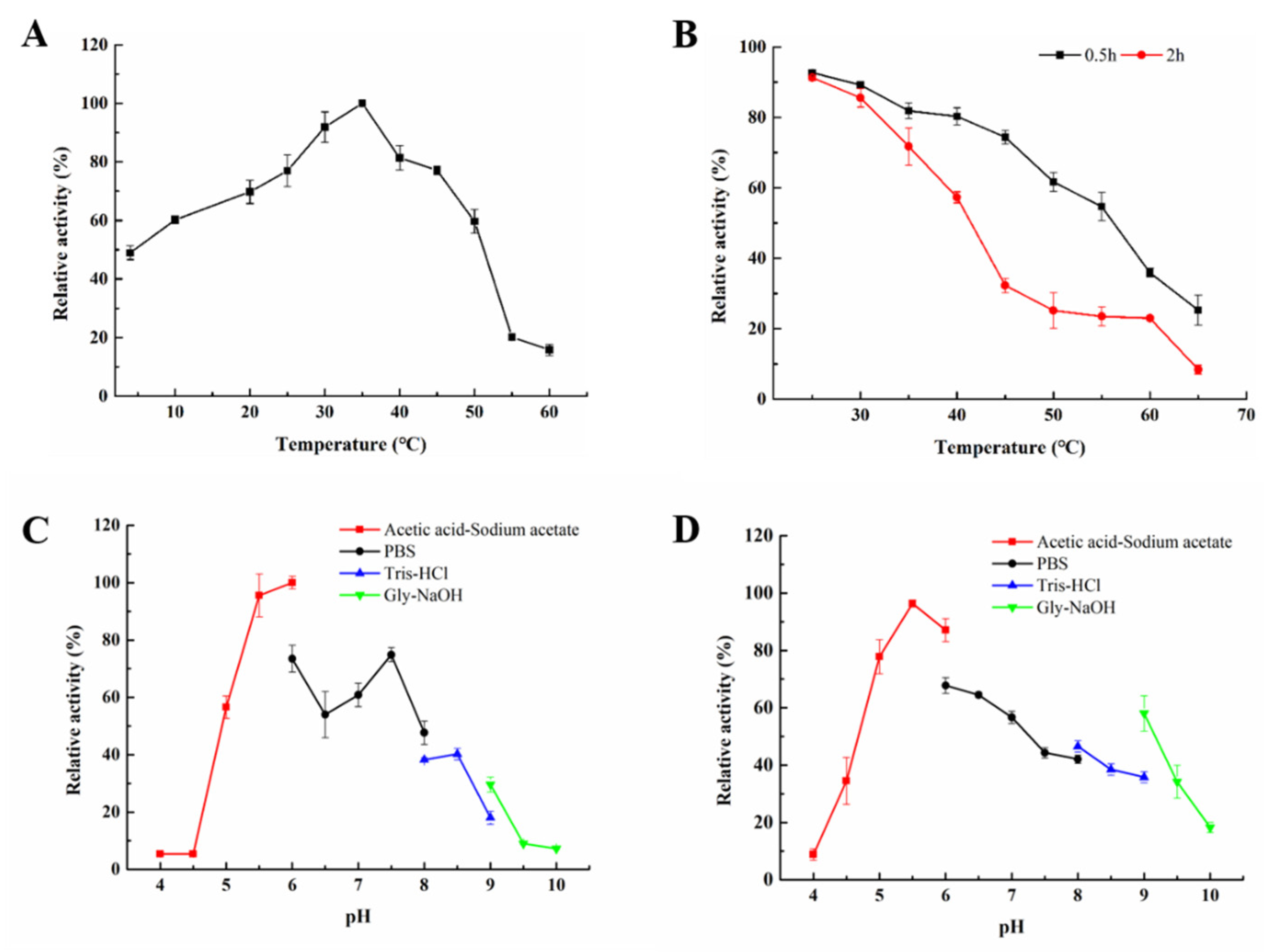
2.3. Biochemical Characterization of Aly23
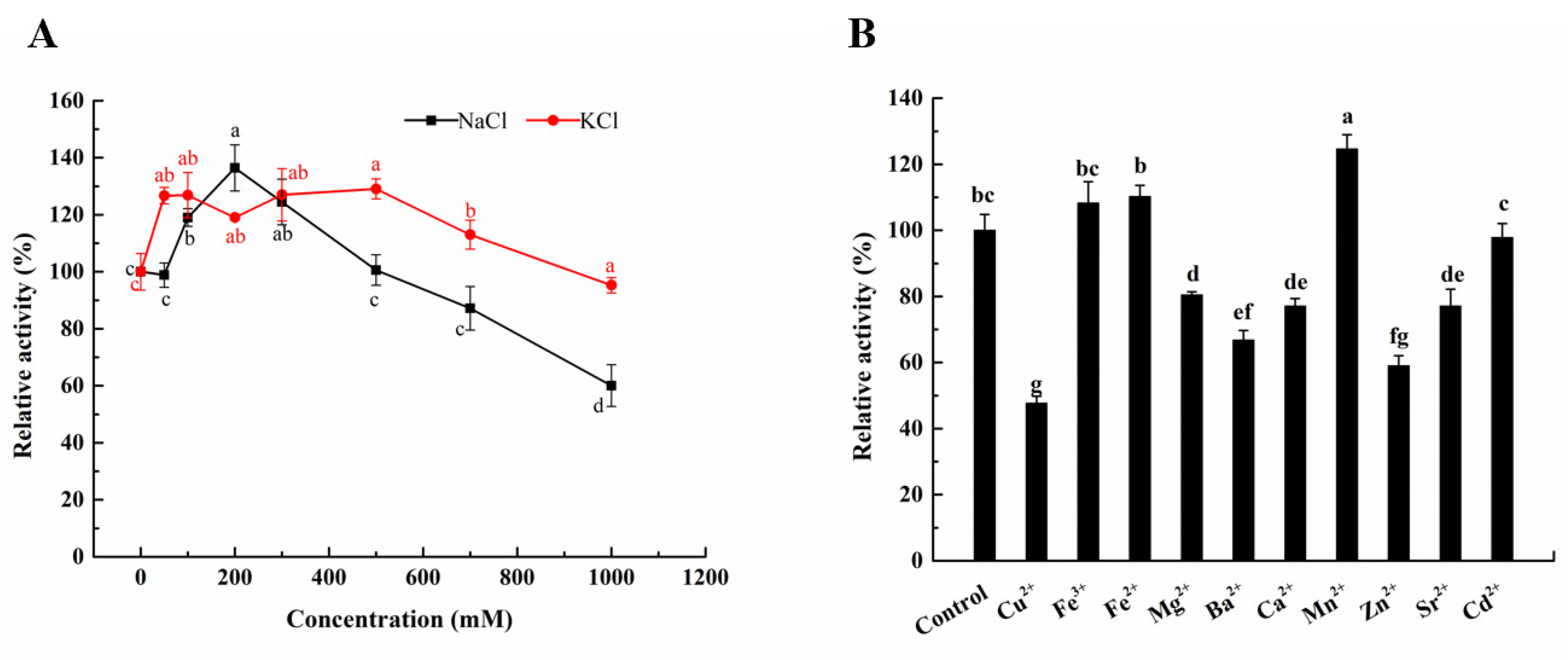
2.4. Effects of Ions on the Activity of Aly23
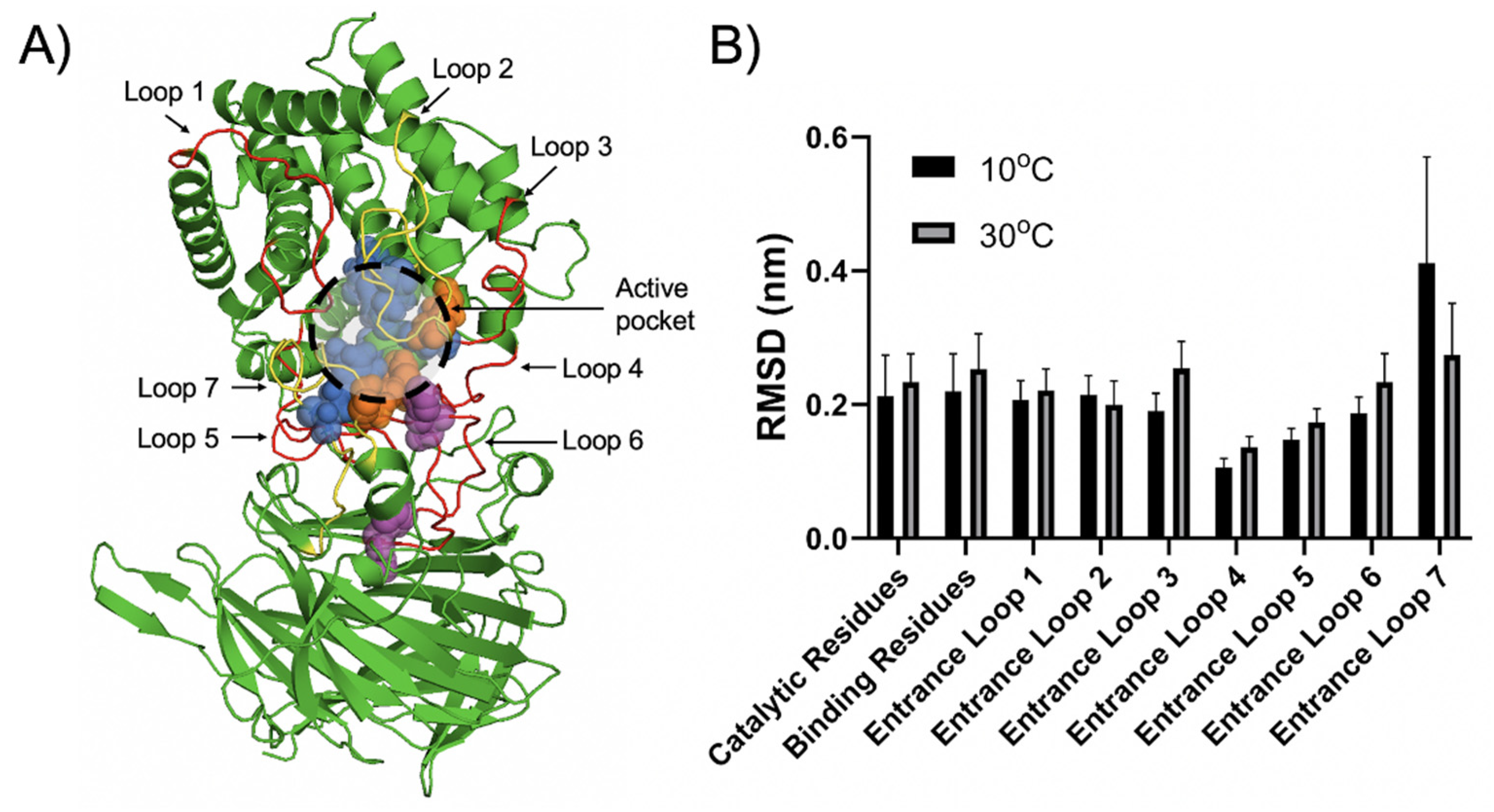
2.5. Structural Insight into the Cold-Adaptation Property of Aly23
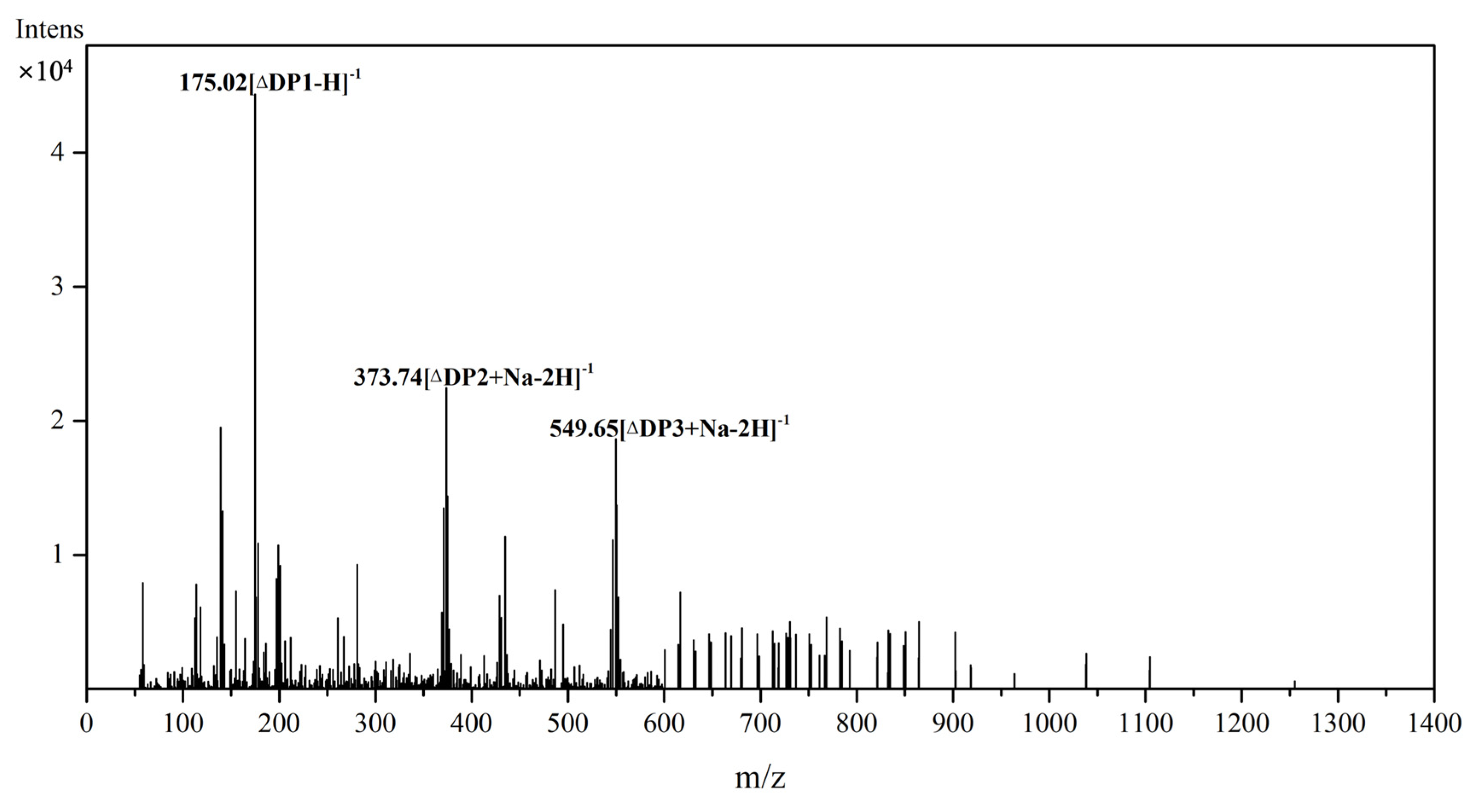
2.6. Substrate Specificity and Degradation Products of Aly23
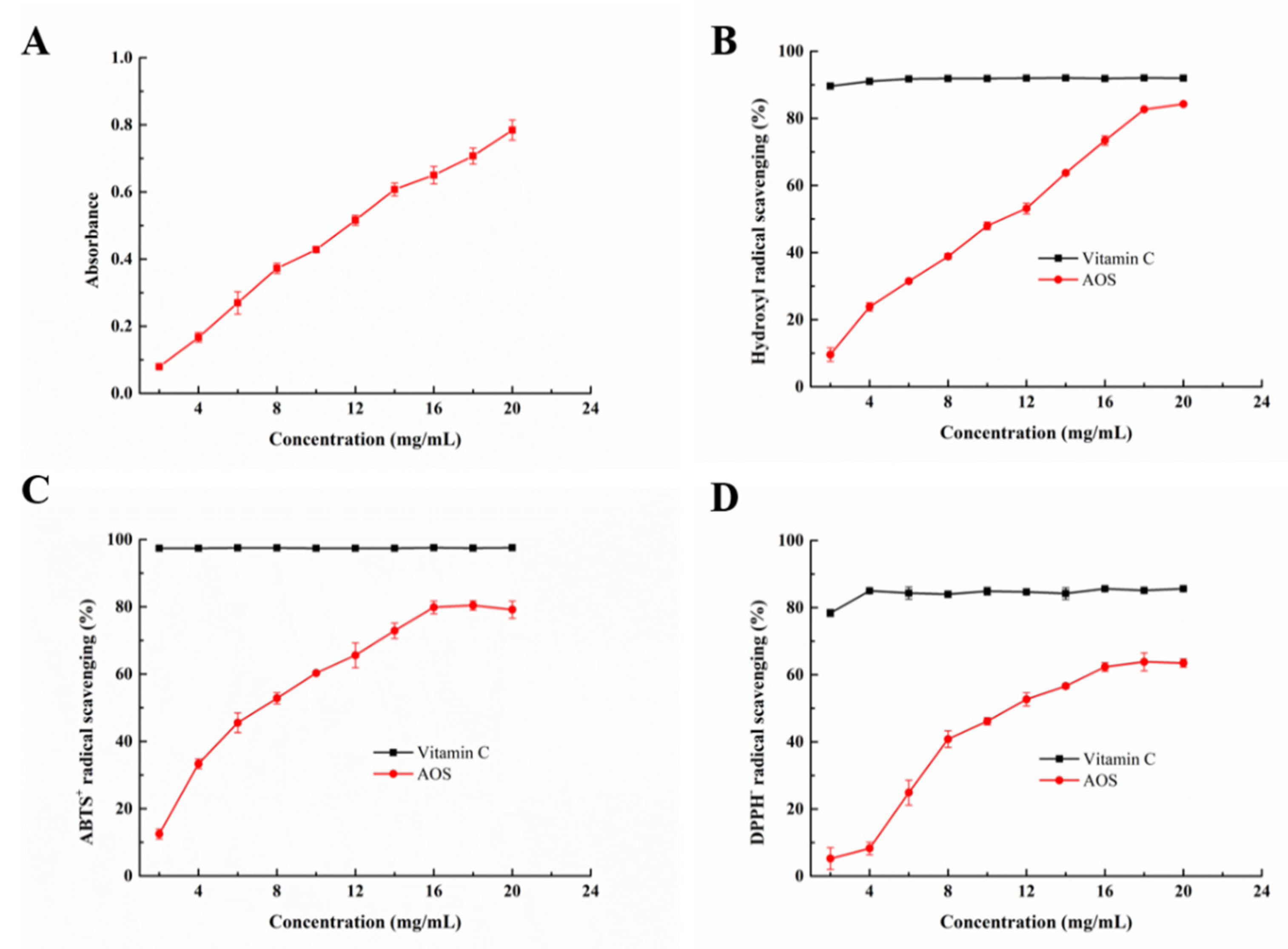
2.7. Antioxidant Function of the Degradation Products of Aly23
3. Materials and Methods
3.1. Materials
3.2. Construction of Recombinant E. coli and Sequence Analysis of Aly23
3.3. Expression and Purification of Aly23
3.4. Enzymatic Activity Assay
3.5. Determination of Enzymatic Kinetics of Recombinant Alginate Lyases
3.6. Biochemical Characterization of Aly23
3.7. Comparative Study by MD Simulation
3.8. Substrates Specificity of Aly23
3.9. ESI-MS Analysis of the Degradation Products of Aly23
3.10. Electrospray Ionization–Mass Spectrometry (ESI-MS) Analysis of the Degradation Products of Aly23
3.10.1. Hydroxyl Radical Scavenging
3.10.2. DPPH Radical Scavenging
3.10.3. Ferric Reducing Power
3.10.4. ABTS Radical Scavenging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Yan, J.; Chen, P.; Zeng, Y.; Men, Y.; Mu, S.; Zhu, Y.; Chen, Y.; Sun, Y. The characterization and modification of a novel bifunctional and robust alginate lyase derived from Marinimicrobium sp. H1. Mar. Drugs 2019, 17, 545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badur, A.H.; Jagtap, S.S.; Yalamanchili, G.; Lee, J.K.; Zhao, H.; Rao, C.V. Alginate lyases from alginate-degrading Vibrio splendidus 12B01 are endolytic. Appl. Environ. Microbiol. 2015, 81, 1865–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Li, S.; Yu, W.; Gong, Q. Cloning, overexpression and characterization of a new oligoalginate lyase from a marine bacterium, Shewanella sp. Biotechnol. Lett. 2015, 37, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. Alginate lyase: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, Z.; Li, S.; Su, H.; Tang, L.; Tan, Y.; Yu, W.; Han, F. Characterization of a new endo-type polysaccharide lyase (PL) family 6 alginate lyase with cold-adapted and metal ions-resisted property. Int. J. Biol. Macromol. 2018, 120, 729–735. [Google Scholar] [CrossRef]
- Singh, A.; Nigam, P.S.; Murphy, J.D. Renewable fuels from algae: An answer to debatable land based fuels. Bioresour Technol. 2011, 102, 10–16. [Google Scholar] [CrossRef]
- Åqvist, J.; Isaksen, G.V.; Brandsdal, B.O. Computation of enzyme cold adaptation. Nat. Rev. Chem. 2017, 1, 1–14. [Google Scholar] [CrossRef]
- Bose, S.K.; Howlader, P.; Jia, X.; Wang, W.; Yin, H. Alginate oligosaccharide postharvest treatment preserve fruit quality and increase storage life via abscisic acid signaling in strawberry. Food Chem. 2019, 283, 665–674. [Google Scholar] [CrossRef]
- Powell, L.C.; Sowedan, A.; Khan, S.; Wright, C.J.; Hawkins, K.; Onsoyen, E.; Myrvold, R.; Hill, K.E.; Thomas, D.W. The effect of alginate oligosaccharides on the mechanical properties of Gram-negative biofilms. Biofouling 2013, 29, 413–421. [Google Scholar] [CrossRef]
- Falkeborg, M.; Cheong, L.Z.; Gianfico, C.; Sztukiel, K.M.; Kristensen, K.; Glasius, M.; Xu, X.; Guo, Z. Alginate oligosaccharides: Enzymatic preparation and antioxidant property evaluation. Food Chem. 2014, 164, 185–194. [Google Scholar] [CrossRef]
- Wang, Y.; Han, F.; Hu, B.; Li, J.B.; Yu, W.G. In vivo prebiotic properties of alginate oligosaccharides prepared through enzymatic hydrolysis of alginate. Nutr. Res. 2006, 26, 597–603. [Google Scholar] [CrossRef]
- Fang, W.S.; Bi, D.C.; Zheng, R.J.; Cai, N.; Xu, H.; Zhou, R.; Lu, J.; Wan, M.; Xu, X. Identification and activation of TLR4-mediated signalling pathways by alginate-derived guluronate oligosaccharide in RAW264.7 macrophages. Sci. Rep.-UK 2017, 7, 1663. [Google Scholar] [CrossRef] [Green Version]
- Hao, J.J.; Hao, C.; Zhang, L.J.; Liu, X.; Zhou, X.L.; Dun, Y.L.; Li, H.H.; Li, G.S.; Zhao, X.L.; An, Y.Y.; et al. OM2, a Novel Oligomannuronate-Chromium (III) Complex, Promotes Mitochondrial Biogenesis and Lipid Metabolism in 3T3-L1 Adipocytes via the AMPK-PGC1 alpha Pathway. PLoS ONE 2015, 10, e0131930. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Dong, S.; Li, F.L.; Ma, X.Q. Structural basis for the exolytic activity of polysaccharide lyase family 6 alginate lyase BcAlyPL6 from human gut microbe Bacteroides clarus. Biochem. Biophys. Res. Commun. 2021, 547, 111–117. [Google Scholar] [CrossRef]
- Inoue, A.; Anraku, M.; Nakagawa, S.; Ojima, T. Discovery of a novel alginate lyase from Nitratiruptor sp. SB155-2 thriving at deep-sea hydrothermal vents and identification of the residues responsible for its heat stability. J. Biol. Chem. 2016, 291, 15551–15563. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.W.; Yin, H. Alginate lyase: Review of major sources and classification, properties, structure-function analysis and applications. Bioengineered 2015, 6, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Lim, H.G.; Kwak, D.H.; Park, S.; Woo, S.; Yang, J.S.; Kang, C.W.; Kim, B.; Noh, M.H.; Seo, S.W.; Jung, G.Y. Vibrio sp. dhg as a platform for the biorefinery of brown macroalgae. Nat. Commun. 2019, 10, 2486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.H.; Kam, N.; Lee, E.Y.; Kim, H.S. Cloning and Characterization of a Novel Oligoalginate Lyase from a Newly Isolated Bacterium Sphingomonas sp MJ-3. Mar. Biotechnol. 2012, 14, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Wang, Y.; Gao, S.; Wu, H.; Wang, D.; Yu, W.; Han, F. Biochemical characteristics and molecular mechanism of an exo-type alginate lyase VxAly7D and its use for the preparation of unsaturated monosaccharides. Biotechnol. Biofuels 2020, 13, 99. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Guo, M.; Zhang, X.; Chen, K.; Yan, J.; Irbis, C. Purification and characterization of alginate lyase from Sphingomonas sp. ZH0. J. Biosci. Bioeng. 2018, 126, 310–316. [Google Scholar] [CrossRef]
- Yang, J.; Cui, D.; Ma, S.; Chen, W.; Chen, D.; Shen, H. Characterization of a novel PL 17 family alginate lyase with exolytic and endolytic cleavage activity from marine bacterium Microbulbifer sp. SH-1. Int. J. Biol. Macromol. 2021, 169, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Belik, A.; Silchenko, A.; Malyarenko, O.; Rasin, A.; Kiseleva, M.; Kusaykin, M.; Ermakova, S. Two new alginate lyases of pl7 and pl6 families from polysaccharide-degrading bacterium Formosa algae KMM 3553(T): Structure, properties, and products analysis. Mar. Drugs 2020, 18, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, S.; Yang, J.; Zhang, X.Y.; Shi, M.; Song, X.Y.; Chen, X.L.; Zhang, Y.Z. Cultivable alginate lyase-excreting bacteria associated with the Arctic brown alga Laminaria. Mar. Drugs 2012, 10, 2481–2491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.L.; Dong, S.; Xu, F.; Dong, F.; Li, P.Y.; Zhang, X.Y.; Zhou, B.C.; Zhang, Y.Z.; Xie, B.B. Characterization of a new cold-adapted and salt-activated polysaccharide lyase family 7 alginate lyase from Pseudoalteromonas sp. SM0524. Front. Microbiol. 2016, 7, 1120. [Google Scholar] [CrossRef] [Green Version]
- Narsico, J.; Inoue, A.; Oka, S.; Ojima, T. Production of a novel dimeric 4-deoxy-L-erythro-5-hexoseulose uronic acid by a PL-17 exolytic alginate lyase from Hydrogenophaga sp. UMI-18. Biochem. Biophys. Res. Commun. 2020, 525, 982–988. [Google Scholar] [CrossRef]
- Park, D.; Jagtap, S.; Nair, S.K. Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J. Biol. Chem. 2014, 289, 8645–8655. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.X.; Xu, S.S.; Yin, X.J.; Wang, F.L.; Li, Y. Characterization of a new bifunctional and cold-adapted polysaccharide lyase (PL) family 7 alginate lyase from Flavobacterium sp. Mar. Drugs 2020, 18, 388. [Google Scholar] [CrossRef]
- Jiang, Z.; Guo, Y.; Wang, X.; Li, H.; Ni, H.; Li, L.; Xiao, A.; Zhu, Y. Molecular cloning and characterization of AlgL17, a new exo-oligoalginate lyase from Microbulbifer sp. ALW1. Protein Expr. Purif. 2019, 161, 17–27. [Google Scholar] [CrossRef]
- Yu, Z.; Zhu, B.; Wang, W.; Tan, H.; Yin, H. Characterization of a new oligoalginate lyase from marine bacterium Vibrio sp. Int. J. Biol. Macromol. 2018, 112, 937–942. [Google Scholar] [CrossRef]
- Kim, H.T.; Chung, J.H.; Wang, D.; Lee, J.; Woo, H.C.; Choi, I.G.; Kim, K.H. Depolymerization of alginate into a monomeric sugar acid using Alg17C, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2-40. Appl. Microbiol. Biotechnol. 2012, 93, 2233–2239. [Google Scholar] [CrossRef]
- Jagtap, S.S.; Hehemann, J.H.; Polz, M.F.; Lee, J.K.; Zhao, H. Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Appl. Environ. Microbiol. 2014, 80, 4207–4214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Wang, L.; Jung, S.; Lee, B.S.; He, N.; Lee, M.S. Biochemical characterization of a new oligoalginate lyase and its biotechnological application in Laminaria japonica degradation. Front. Microbiol. 2020, 11, 316. [Google Scholar] [CrossRef]
- Huang, G.; Wen, S.; Liao, S.; Wang, Q.; Pan, S.; Zhang, R.; Lei, F.; Liao, W.; Feng, J.; Huang, S. Characterization of a bifunctional alginate lyase as a new member of the polysaccharide lyase family 17 from a marine strain BP-2. Biotechnol. Lett. 2019, 41, 1187–1200. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.M.; Li, S.Y.; Wu, Y.; Yu, W.G.; Han, F. Cloning and characterization of two thermo- and salt-tolerant oligoalginate lyases from marine bacterium Halomonas sp. FEMS Microbiol. Lett. 2016, 363, fnw079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, B.; Sun, Y.; Ni, F.; Ning, L.; Yao, Z. Characterization of a new endo-type alginate lyase from Vibrio sp. NJU-03. Int. J. Biol. Macromol. 2018, 108, 1140–1147. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, L.; Hao, J.; Xing, M.; Sun, J.; Sun, M. Purification and Characterization of a New Alginate Lyase from Marine Bacterium Vibrio sp. SY08. Mar. Drugs 2016, 15, 1. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Zhu, Y.; Men, Y.; Zeng, Y.; Sun, Y. Purification and characterization of a novel alginate lyase from the marine bacterium Bacillus sp. Alg07. Mar. Drugs 2018, 16, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.P.; Cao, M.; Li, B.; Ji, X.F.; Zhang, X.Y.; Zhang, Y.Q.; Wang, H.Y. Cloning, secretory expression and characterization of a unique pH-stable and cold-adapted alginate lyase. Mar. Drugs 2020, 18, 189. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.D.; Yuan, S.; Li, L.; Chen, S.; Xu, J.H.; Zhou, J. Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates. Proc. Natl. Acad. Sci. USA 2014, 111, 15717–15722. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.H.; Shao, Y.; Jiao, C.; Yang, Q.M.; Weng, H.F.; Xiao, A.F. Characterization and application of an alginate lyase, Aly1281 from marine bacterium Pseudoalteromonas carrageenovora ASY5. Mar. Drugs 2020, 18, 95. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Wang, W.; Zhao, X.; Wang, H.; Yin, H. Preparation of alginate oligosaccharides and their biological activities in plants: A review. Carbohydr. Res. 2020, 494, 108056. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Song, X.N.; Lin, Y.; Xiao, Q.; Du, X.P.; Chen, Y.H.; Xiao, A.F. Antioxidant capacity and prebiotic effects of Gracilaria neoagaro oligosaccharides prepared by agarase hydrolysis. Int. J. Biol. Macromol. 2019, 137, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Q.; Zhang, J.; Zhou, X.; Lyu, F.; Zhao, P.; Ding, Y. Preparation, composition analysis and antioxidant activities of konjac oligo-glucomannan. Carbohydr. Polym. 2015, 130, 398–404. [Google Scholar] [CrossRef]
- Ramírez-Jiménez, A.; Guerra-Hernández, E.; García-Villanova, B. Evolution of non-enzymatic browning during storage of infant rice cereal. Food Chem. 2003, 83, 219–225. [Google Scholar] [CrossRef]
- Liu, M.; Liu, L.; Zhang, H.-f.; Yi, B.; Everaert, N. Alginate oligosaccharides preparation, biological activities and their application in livestock and poultry. J. Integrative Agric. 2021, 20, 24–34. [Google Scholar] [CrossRef]
- Hernandez-Marin, E.; Martinez, A. Carbohydrates and Their Free Radical Scavenging Capability: A Theoretical Study. J. Phys. Chem. B 2012, 116, 9668–9675. [Google Scholar] [CrossRef] [PubMed]
- Kang, O.L.; Ghani, M.; Hassan, O.; Rahmati, S.; Ramli, N. Novel agaro-oligosaccharide production through enzymatic hydrolysis: Physicochemical properties and antioxidant activities. Food Hydrocolloids 2014, 42, 304–308. [Google Scholar] [CrossRef]
- Gulcin, I.; Bursal, E.; Sehitoglu, M.H.; Bilsel, M.; Goren, A.C. Polyphenol contents and antioxidant activity of lyophilized aqueous extract of propolis from Erzurum, Turkey. Food Chem. Toxicol. 2010, 48, 2227–2238. [Google Scholar] [CrossRef]
- Zeng, J.; An, D.; Jiao, C.; Xiao, Q.; Weng, H.; Yang, Q.; Xiao, A. Cloning, expression, and characterization of a new pH- and heat-stable alginate lyase from Pseudoalteromonas carrageenovora ASY5. J. Food Biochem. 2019, 43, e12886. [Google Scholar] [CrossRef]


| Enzyme | Source | Optimal Temperature | Optimal pH | Product DP | Ref. |
|---|---|---|---|---|---|
| Aly23 | Pseudoaltermonas sp. ASY5 | 35 °C | 6 | 1–3 | This study |
| oalS17 | Shewanella sp. Kz7 | 50 °C | 6.2 | 1 | [3] |
| HyAly-I | Hydrogenopahaga sp. UMI-18 | 40 °C | 6 | 1–3 | [25] |
| algL17 | Microbulbifer sp. ALW1 | 35 °C | 8 | 1–4 | [28] |
| AlgSH17 | Microbulbifer sp. SH-1 | 30 °C | 7 | 1–6 | [21] |
| Oal17A | Vibrio sp. W13 | 30 °C | 7 | 1 | [29] |
| Alg17C | Saccharophagus degradans 2-40 | 40 °C | 6 | 1 | [30] |
| OalB | Vibrio.splendidus 12B01 | 30 °C | 7 | — | [31] |
| OalC | Vibrio.splendidus 12B01 | 35 °C | 7.5 | — | [31] |
| OalV17 | Vibrio sp. SY01 | 40 °C | 7.2 | 1 | [32] |
| Alg17B | strain BP-2 | 40–45 °C | 7.5–8.0 | 1–6 | [33] |
| OalY1 | Halomonas sp. QY114 | 45 °C | 7.05 | 1 | [34] |
| OalY2 | Halomonas sp. QY114 | 50 °C | 6.6 | 1 | [34] |
| Parameter a | Simulation Temperature | |
|---|---|---|
| 10 °C | 30 °C | |
| RMSD (nm) | 0.314 ± 0.052 | 0.322 ± 0.52 |
| RMSF (nm) | 0.131 ± 0.064 | 0.127 ± 0.055 |
| Radius of gyrate (nm) | 2.882 ± 0.013 | 2.864 ± 0.020 |
| SASA (Å2) | 33130 ± 283 | 32473 ± 443 |
| Salt bridge | 35 ± 3 | 34 ± 3 |
| Hbond p-p b | 514 ± 11 | 506 ± 11 |
| Hbond p-w b | 1497 ± 24 | 1454 ± 29 |
| Hbond s-s b | 85 ± 5 | 87 ± 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, X.; Jiao, C.; Wei, Y.; Zhuang, X.-Y.; Xiao, Q.; Chen, J.; Chen, F.-Q.; Yang, Q.-M.; Weng, H.-F.; Fang, B.-S.; et al. Biochemical Characterization and Cold-Adaption Mechanism of a PL-17 Family Alginate Lyase Aly23 from Marine Bacterium Pseudoalteromonas sp. ASY5 and Its Application for Oligosaccharides Production. Mar. Drugs 2022, 20, 126. https://doi.org/10.3390/md20020126
Tang X, Jiao C, Wei Y, Zhuang X-Y, Xiao Q, Chen J, Chen F-Q, Yang Q-M, Weng H-F, Fang B-S, et al. Biochemical Characterization and Cold-Adaption Mechanism of a PL-17 Family Alginate Lyase Aly23 from Marine Bacterium Pseudoalteromonas sp. ASY5 and Its Application for Oligosaccharides Production. Marine Drugs. 2022; 20(2):126. https://doi.org/10.3390/md20020126
Chicago/Turabian StyleTang, Xiang, Chao Jiao, Yi Wei, Xiao-Yan Zhuang, Qiong Xiao, Jun Chen, Fu-Quan Chen, Qiu-Ming Yang, Hui-Fen Weng, Bai-Shan Fang, and et al. 2022. "Biochemical Characterization and Cold-Adaption Mechanism of a PL-17 Family Alginate Lyase Aly23 from Marine Bacterium Pseudoalteromonas sp. ASY5 and Its Application for Oligosaccharides Production" Marine Drugs 20, no. 2: 126. https://doi.org/10.3390/md20020126
APA StyleTang, X., Jiao, C., Wei, Y., Zhuang, X.-Y., Xiao, Q., Chen, J., Chen, F.-Q., Yang, Q.-M., Weng, H.-F., Fang, B.-S., Zhang, Y.-H., & Xiao, A.-F. (2022). Biochemical Characterization and Cold-Adaption Mechanism of a PL-17 Family Alginate Lyase Aly23 from Marine Bacterium Pseudoalteromonas sp. ASY5 and Its Application for Oligosaccharides Production. Marine Drugs, 20(2), 126. https://doi.org/10.3390/md20020126






