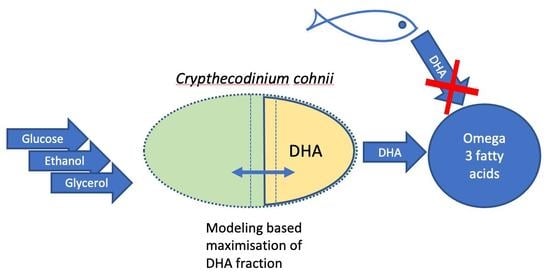
Kinetic and Stoichiometric Modeling-Based Analysis of Docosahexaenoic Acid (DHA) Production Potential by Crypthecodinium cohnii from Glycerol, Glucose and Ethanol
Abstract
1. Introduction
2. Results
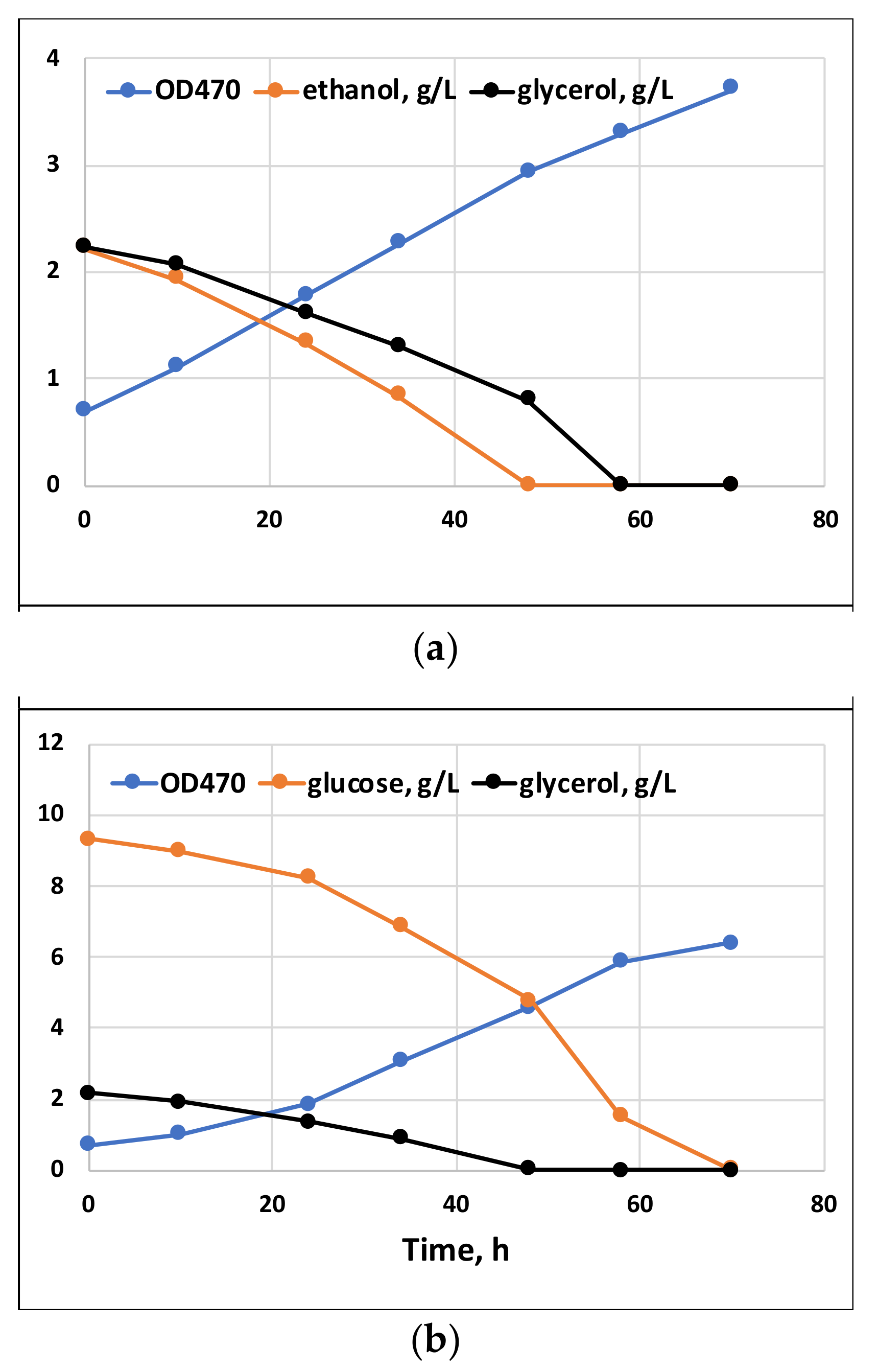
2.1. Comparison of Growth, Substrate Consumption, and Accumulation of PUFAs with Glucose, Ethanol and Glycerol
2.2. Pathway-Scale Kinetic Model of Substrate Uptake
2.2.1. Structure of the Model
2.2.2. Parameter Estimation Results
2.2.3. Simulation Results
2.3. Medium-Scale Stoichiometric Model of DHA Production
2.3.1. Validation of the Model
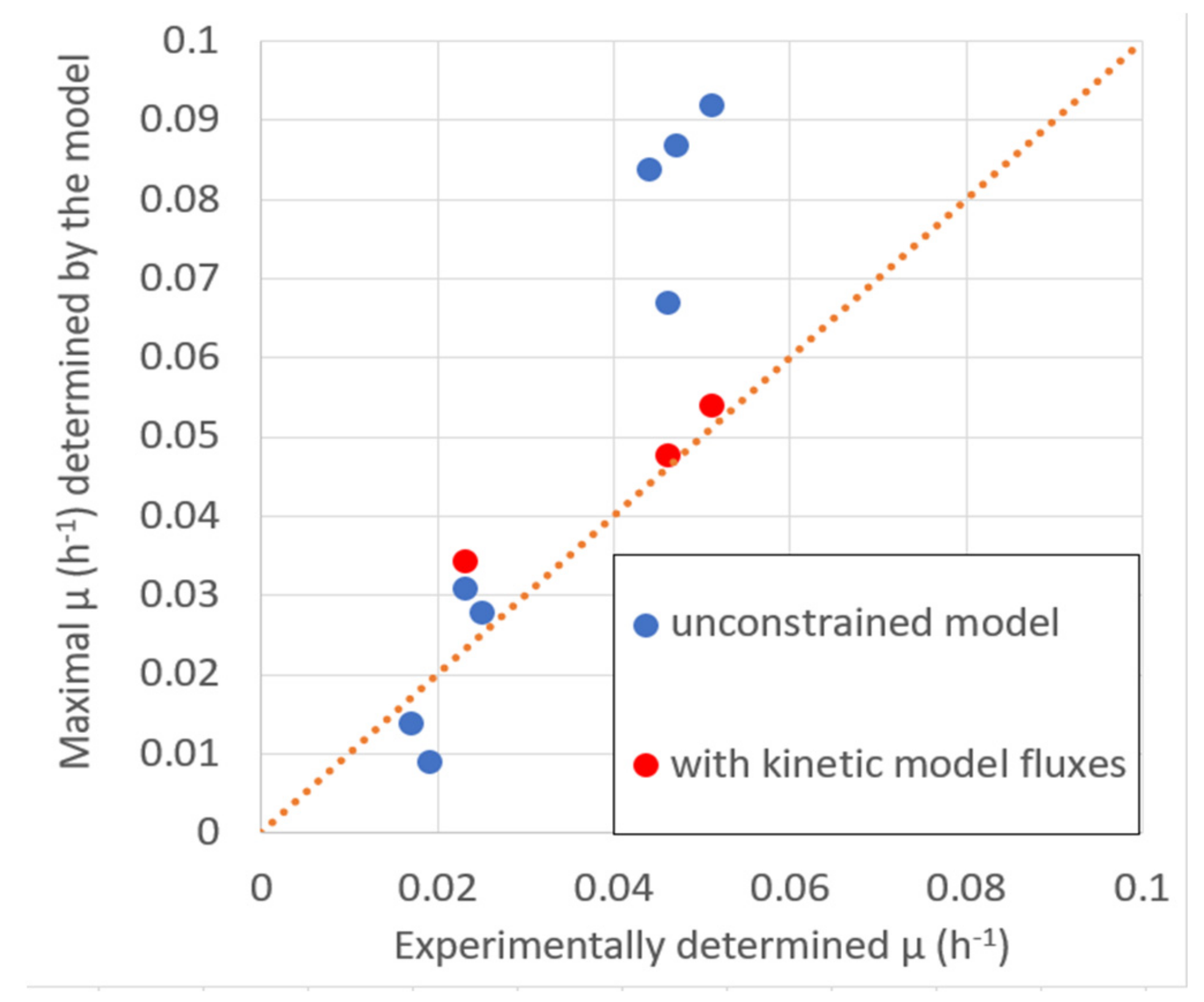
2.3.2. Validation of Steady-State Fluxes of the Kinetic Model
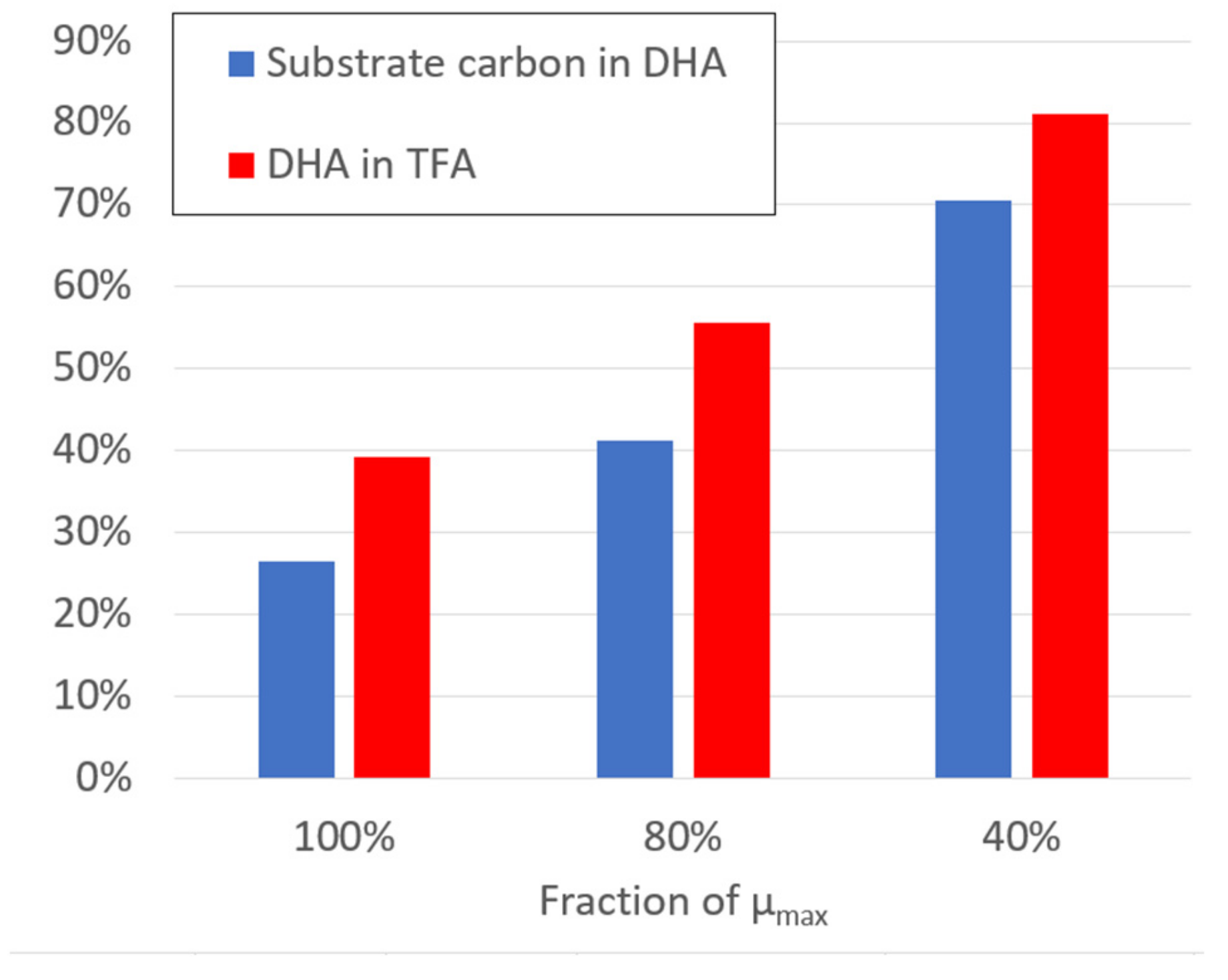
2.4. Model-Based Determination of DHA Production Potential
3. Discussion
3.1. Combining Kinetic and Stoichiometric Models
3.2. Analysis of Substrate-Specific Functioning of Central Metabolism by Experimental and Modeling Analysis
4. Materials and Methods
4.1. Experimental Materials and Methods
4.2. Development of a Pathway-Scale Kinetic Model
4.3. Development of the Constraint-Based Medium-Scale Stoichiometric Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Adarme-Vega, T.C.; Thomas-Hall, S.R.; Schenk, P.M. Towards Sustainable Sources for Omega-3 Fatty Acids Production. Curr. Opin. Biotechnol. 2014, 26, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Oliver, L.; Dietrich, T.; Marañón, I.; Villarán, M.C.; Barrio, R.J. Producing Omega-3 Polyunsaturated Fatty Acids: A Review of Sustainable Sources and Future Trends for the EPA and DHA Market. Resources 2020, 9, 148. [Google Scholar] [CrossRef]
- Ji, X.-J.; Ren, L.-J.; Huang, H. Omega-3 Biotechnology: A Green and Sustainable Process for Omega-3 Fatty Acids Production. Front. Bioeng. Biotechnol. 2015, 3, 1301–1315. [Google Scholar] [CrossRef] [PubMed]
- Graham, I.A.; Larson, T.; Napier, J.A. Rational Metabolic Engineering of Transgenic Plants for Biosynthesis of Omega-3 Polyunsaturates. Curr. Opin. Biotechnol. 2007, 18, 142–147. [Google Scholar] [CrossRef]
- Santos-Sánchez, N.F.; Valadez-Blanco, R.; Hernández-Carlos, B.; Torres-Ariño, A.; Guadarrama-Mendoza, P.C.; Salas-Coronado, R. Lipids Rich in ω-3 Polyunsaturated Fatty Acids from Microalgae. Appl. Microbiol. Biotechnol. 2016, 100, 8667–8684. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.; Reis, A.; Vasconcelos, R.; Guerra, P.; Lopes Da Silva, T. Crypthecodinium Cohnii with Emphasis on DHA Production: A Review. J. Appl. Phycol. 2009, 21, 199–214. [Google Scholar] [CrossRef]
- Sijtsma, L.; de Swaaf, M.E. Biotechnological Production and Applications of the ω-3 Polyunsaturated Fatty Acid Docosahexaenoic Acid. Appl. Microbiol. Biotechnol. 2004, 64, 146–153. [Google Scholar] [CrossRef] [PubMed]
- de Swaaf, M.E.; de Rijk, T.C.; Eggink, G.; Sijtsma, L. Optimisation of Docosahexaenoic Acid Production in Batch Cultivations by Crypthecodinium Cohnii. Prog. Ind. Microbiol. 1999, 35, 185–192. [Google Scholar] [CrossRef]
- Sijtsma, L.; Anderson, A.J.; Ratledge, C. Alternative Carbon Sources for Heterotrophic Production of Docosahexaenoic Acid by the Marine Alga Crypthecodinium Cohnii. In Single Cell Oils; Elsevier: Amsterdam, The Netherlands, 2010; pp. 131–149. [Google Scholar]
- Safdar, W.; Zan, X.; Song, Y. Synergistic Effects of PH, Temperature and Agitation on Growth Kinetics and Docosahexaenoic Acid Production of C. Cohnii Cultured on Different Carbon Sources. Int. J. Res. Agric. Sci. 2017, 4, 94–101. [Google Scholar]
- Moniz, P.; Silva, C.; Oliveira, A.C.; Reis, A.; Lopes da Silva, T. Raw Glycerol Based Medium for Dha and Lipids Production, Using the Marine Heterotrophic Microalga Crypthecodinium Cohnii. Processes 2021, 9, 2005. [Google Scholar] [CrossRef]
- Taborda, T.; Moniz, P.; Reis, A.; da Silva, T.L. Evaluating Low-Cost Substrates for Crypthecodinium Cohnii Lipids and DHA Production, by Flow Cytometry. J. Appl. Phycol. 2021, 33, 263–274. [Google Scholar] [CrossRef]
- Pentjuss, A.; Kalnenieks, U. Assessment of Zymomonas Mobilis Biotechnological Potential in Ethanol Production by Flux Variability Analysis. Biosyst. Inf. Technol. 2014, 3, 1–5. [Google Scholar] [CrossRef]
- Pentjuss, A.; Stalidzans, E.; Liepins, J.; Kokina, A.; Martynova, J.; Zikmanis, P.; Mozga, I.; Scherbaka, R.; Hartman, H.; Poolman, M.G.; et al. Model-Based Biotechnological Potential Analysis of Kluyveromyces Marxianus Central Metabolism. J. Ind. Microbiol. Biotechnol. 2017, 44, 1177–1190. [Google Scholar] [CrossRef] [PubMed]
- Otero, J.M.; Nielsen, J. Industrial Systems Biology. Biotechnol. Bioeng. 2010, 105, 439–460. [Google Scholar] [CrossRef] [PubMed]
- Palsson, B.O. Metabolic Systems Biology. FEBS Lett. 2009, 583, 3900–3904. [Google Scholar] [CrossRef] [PubMed]
- Cvijovic, M.; Höfer, T.; Aćimović, J.; Alberghina, L.; Almaas, E.; Besozzi, D.; Blomberg, A.; Bretschneider, T.; Cascante, M.; Collin, O.; et al. Strategies for Structuring Interdisciplinary Education in Systems Biology: An European Perspective. NPJ Syst. Biol. Appl. 2016, 2, 16011. [Google Scholar] [CrossRef] [PubMed]
- Stelling, J. Mathematical Models in Microbial Systems Biology. Curr. Opin. Microbiol. 2004, 7, 513–518. [Google Scholar] [CrossRef]
- Stalidzans, E.; Seiman, A.; Peebo, K.; Komasilovs, V.; Pentjuss, A. Model-Based Metabolism Design: Constraints for Kinetic and Stoichiometric Models. Biochem. Soc. Trans. 2018, 46, 261–267. [Google Scholar] [CrossRef]
- de Swaaf, M.E.; Pronk, J.T.; Sijtsma, L. Fed-Batch Cultivation of the Docosahexaenoic-Acid-Producing Marine Alga Crypthecodinium Cohnii on Ethanol. Appl. Microbiol. Biotechnol. 2003, 61, 40–43. [Google Scholar] [CrossRef]
- Didrihsone, E.; Dubencovs, K.; Grube, M.; Shvirksts, K.; Suleiko, A.; Suleiko, A.; Vanags, J. Crypthecodinium Cohnii Growth and Omega Fatty Acid Production in Mediums Supplemented with Extract from Recycled Bio-Mass. Mar. Drugs 2022, 20, 68. [Google Scholar] [CrossRef] [PubMed]
- Ripoche, A.; Guillard, A.S. Determination of Fatty Acid Composition of Pork Fat by Fourier Transform Infrared Spectroscopy. Meat Sci. 2001, 58, 299–304. [Google Scholar] [CrossRef]
- Ami, D.; Posteri, R.; Mereghetti, P.; Porro, D.; Doglia, S.M.; Branduardi, P. Fourier Transform Infrared Spectroscopy as a Method to Study Lipid Accumulation in Oleaginous Yeasts. Biotechnol. Biofuels 2014, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.; Lourenço, S.; Lopes, A.; Andrade, C.; Câmara, J.S.; Castilho, P.; Perestrelo, R. Evaluation of Fatty Acids Profile as a Useful Tool towards Valorization of By-Products of Agri-Food Industry. Foods 2021, 10, 2867. [Google Scholar] [CrossRef]
- Yoshida, S.; Yoshida, H. Noninvasive Analyses of Polyunsaturated Fatty Acids in Human Oral Mucosain Vivo by Fourier-Transform Infrared Spectroscopy. Biopolymers 2004, 74, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.; Guerra, P.; Madeira, V.; Ruano, F.; Lopes da Silva, T.; Reis, A. Study of Docosahexaenoic Acid Production by the Heterotrophic Microalga Crypthecodinium Cohnii CCMP 316 Using Carob Pulp as a Promising Carbon Source. World J. Microbiol. Biotechnol. 2007, 23, 1209–1215. [Google Scholar] [CrossRef]
- Chalima, A.; Taxeidis, G.; Topakas, E. Optimization of the Production of Docosahexaenoic Fatty Acid by the Heterotrophic Microalga Crypthecodinium Cohnii Utilizing a Dark Fermentation Effluent. Renew. Energy 2020, 152, 102–109. [Google Scholar] [CrossRef]
- Diao, J.; Li, X.; Pei, G.; Liu, L.; Chen, L. Comparative Metabolomic Analysis of Crypthecodinium Cohnii in Response to Different Dissolved Oxygen Levels during Docosahexaenoic Acid Fermentation. Biochem. Biophys. Res. Commun. 2018, 499, 941–947. [Google Scholar] [CrossRef]
- Lopes da Silva, T.; Reis, A. The Use of Multi-Parameter Flow Cytometry to Study the Impact of n-Dodecane Additions to Marine Dinoflagellate Microalga Crypthecodinium Cohnii Batch Fermentations and DHA Production. J. Ind. Microbiol. Biotechnol. 2008, 35, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Strazdina, I.; Klavins, L.; Galinina, N.; Shvirksts, K.; Grube, M.; Stalidzans, E.; Kalnenieks, U. Syntrophy of Crypthecodinium Cohnii and Immobilized Zymomonas Mobilis for Docosahexaenoic Acid Production from Sucrose-Containing Substrates. J. Biotechnol. 2021, 338, 63–70. [Google Scholar] [CrossRef]
- Pei, G.; Li, X.; Liu, L.; Liu, J.; Wang, F.; Chen, L.; Zhang, W. De Novo Transcriptomic and Metabolomic Analysis of Docosahexaenoic Acid (DHA)-Producing Crypthecodinium Cohnii during Fed-Batch Fermentation. Algal Res. 2017, 26, 380–391. [Google Scholar] [CrossRef]
- Cui, J.; Diao, J.; Sun, T.; Shi, M.; Liu, L.; Wang, F.; Chen, L.; Zhang, W. 13C Metabolic Flux Analysis of Enhanced Lipid Accumulation Modulated by Ethanolamine in Crypthecodinium Cohnii. Front. Microbiol. 2018, 9, 956. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR Core Data Resource in 2021: New Developments and Updates. Nucleic Acids Res. 2021, 49, D498–D508. [Google Scholar] [CrossRef] [PubMed]
- Wittig, U.; Kania, R.; Golebiewski, M.; Rey, M.; Shi, L.; Jong, L.; Algaa, E.; Weidemann, A.; Sauer-Danzwith, H.; Mir, S.; et al. SABIO-RK--Database for Biochemical Reaction Kinetics. Nucleic Acids Res. 2012, 40, D790–D796. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; O’Donovan, C.; Magrane, M.; Alpi, E.; Antunes, R.; Bely, B.; Bingley, M.; Bonilla, C.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Singh, V.K.; Ghosh, I. Kinetic Modeling of Tricarboxylic Acid Cycle and Glyoxylate Bypass in Mycobacterium Tuberculosis, and Its Application to Assessment of Drug Targets. Theor. Biol. Med. Model. 2006, 3, 27. [Google Scholar] [CrossRef][Green Version]
- Flamholz, A.; Noor, E.; Bar-Even, A.; Milo, R. Equilibrator—The Biochemical Thermodynamics Calculator. Nucleic Acids Res. 2012, 40, D770–D775. [Google Scholar] [CrossRef]
- Malik-Sheriff, R.S.; Glont, M.; Nguyen, T.V.N.; Tiwari, K.; Roberts, M.G.; Xavier, A.; Vu, M.T.; Men, J.; Maire, M.; Kananathan, S.; et al. BioModels-15 Years of Sharing Computational Models in Life Science. Nucleic Acids Res. 2020, 48, D407–D415. [Google Scholar] [CrossRef]
- Feist, A.M.; Palsson, B.O. The Biomass Objective Function. Curr. Opin. Microbiol. 2010, 13, 344–349. [Google Scholar] [CrossRef]
- Shoguchi, E.; Shinzato, C.; Kawashima, T.; Gyoja, F.; Mungpakdee, S.; Koyanagi, R.; Takeuchi, T.; Hisata, K.; Tanaka, M.; Fujiwara, M.; et al. Draft Assembly of the Symbiodinium Minutum Nuclear Genome Reveals Dinoflagellate Gene Structure. Curr. Biol. 2013, 23, 1399–1408. [Google Scholar] [CrossRef]
- Almquist, J.; Cvijovic, M.; Hatzimanikatis, V.; Nielsen, J.; Jirstrand, M. Kinetic Models in Industrial Biotechnology—Improving Cell Factory Performance. Metab. Eng. 2014, 24, 38–60. [Google Scholar] [CrossRef]
- Price, N.D.; Papin, J.A.; Schilling, C.H.; Palsson, B.O. Genome-Scale Microbial in Silico Models: The Constraints-Based Approach. Trends Biotechnol. 2003, 21, 162–169. [Google Scholar] [CrossRef]
- Thiele, I.; Palsson, B.O. A Protocol for Generating a High-Quality Genome-Scale Metabolic Reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef]
- Kalnenieks, U.; Pentjuss, A.; Rutkis, R.; Stalidzans, E.; Fell, D.A. Modeling of Zymomonas Mobilis Central Metabolism for Novel Metabolic Engineering Strategies. Front. Microbiol. 2014, 5, 42. [Google Scholar] [CrossRef][Green Version]
- Schrenk, D.F.; Bisswanger, H. Measurements of Electron Spin Resonance with the Pyruvate Dehydrogenase Complex from Escherichia Coli. Eur. J. Biochem. 1984, 143, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Pelley, J.W. Glycolysis and Pyruvate Oxidation. In Elsevier’s Integrated Biochemistry; Elsevier: Amsterdam, The Netherlands, 2007; pp. 47–53. [Google Scholar]
- Nielsen, J. Metabolic Engineering: Techniques for Analysis of Targets for Genetic Manipulations. Biotechnol. Bioeng. 1998, 58, 125–132. [Google Scholar] [CrossRef]
- Strazdina, I.; Balodite, E.; Lasa, Z.; Rutkis, R.; Galinina, N.; Kalnenieks, U. Aerobic Catabolism and Respiratory Lactate Bypass in Ndh-Negative Zymomonas Mobilis. Metab. Eng. Commun. 2018, 7, e00081. [Google Scholar] [CrossRef]
- Susi, H.; Byler, D.M. Resolution-Enhanced Fourier Transform Infrared Spectroscopy of Enzymes. Methods Enzymol. 1986, 130, 290–311. [Google Scholar] [CrossRef] [PubMed]
- Hoops, S.; Sahle, S.; Gauges, R.; Lee, C.; Pahle, J.; Simus, N.; Singhal, M.; Xu, L.; Mendes, P.; Kummer, U. COPASI—A complex pathway simulator. Bioinformatics 2006, 22, 3067–3074. [Google Scholar] [CrossRef]
- Mendes, P.; Hoops, S.; Sahle, S.; Gauges, R.; Dada, J.O.; Kummer, U. Computational Modeling of Biochemical Networks Using COPASI. In Methods in Molecular Biology, Systems Biology; Maly, I.V., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 500, pp. 17–59. ISBN 978-1-934115-64-0. [Google Scholar]
- Kostromins, A.; Mozga, I.; Stalidzans, E. ConvAn: A Convergence Analyzing Tool for Optimization of Biochemical Networks. Biosystems 2012, 108, 73–77. [Google Scholar] [CrossRef]
- Elsts, A.; Pentjuss, A.; Stalidzans, E. SpaceScanner: COPASI Wrapper for Automated Management of Global Stochastic Optimization Experiments. Bioinformatics 2017, 33, 2966–2967. [Google Scholar] [CrossRef]
- Stalidzans, E.; Landmane, K.; Sulins, J.; Sahle, S. Misinterpretation Risks of Global Stochastic Optimisation of Kinetic Models Revealed by Multiple Optimisation Runs. Math. Biosci. 2019, 307, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Park, J.O.; Rubin, S.A.; Xu, Y.-F.; Amador-Noguez, D.; Fan, J.; Shlomi, T.; Rabinowitz, J.D. Metabolite Concentrations, Fluxes and Free Energies Imply Efficient Enzyme Usage. Nat. Chem. Biol. 2016, 12, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Uchida, M.; Sun, Y.; McDermott, G.; Knoechel, C.; le Gros, M.A.; Parkinson, D.; Drubin, D.G.; Larabell, C.A. Quantitative Analysis of Yeast Internal Architecture Using Soft X-ray Tomography. Yeast 2011, 28, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Bordbar, A.; Monk, J.M.; King, Z.A.; Palsson, B.O. Constraint-Based Models Predict Metabolic and Associated Cellular Functions. Nat. Rev. Genet. 2014, 15, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Heirendt, L.; Arreckx, S.; Pfau, T.; Mendoza, S.N.; Richelle, A.; Heinken, A.; Haraldsdóttir, H.S.; Wachowiak, J.; Keating, S.M.; Vlasov, V.; et al. Creation and Analysis of Biochemical Constraint-Based Models Using the COBRA Toolbox v.3.0. Nat. Protoc. 2019, 14, 639–702. [Google Scholar] [CrossRef]
- Wang, H.; Marcišauskas, S.; Sánchez, B.J.; Domenzain, I.; Hermansson, D.; Agren, R.; Nielsen, J.; Kerkhoven, E.J. RAVEN 2.0: A Versatile Toolbox for Metabolic Network Reconstruction and a Case Study on Streptomyces Coelicolor. PLoS Comput. Biol. 2018, 14, e1006541. [Google Scholar] [CrossRef]
- Kostromins, A.; Stalidzans, E. Paint4Net: COBRA Toolbox Extension for Visualization of Stoichiometric Models of Metabolism. Biosystems 2012, 109, 233–239. [Google Scholar] [CrossRef] [PubMed]
- King, Z.A.; Dräger, A.; Ebrahim, A.; Sonnenschein, N.; Lewis, N.E.; Palsson, B.O. Escher: A Web Application for Building, Sharing, and Embedding Data-Rich Visualizations of Biological Pathways. PLoS Comput. Biol. 2015, 11, e1004321. [Google Scholar] [CrossRef]
- Petrovs, R.; Stalidzans, E.; Pentjuss, A. IMFLer: A Web Application for Interactive Metabolic Flux Analysis and Visualization. J. Comput. Biol. 2021, 28, 1021–1032. [Google Scholar] [CrossRef]
- Stalidzans, E.; Dace, E. Sustainable Metabolic Engineering for Sustainability Optimisation of Industrial Biotechnology. Comput. Struct. Biotechnol. J. 2021, 19, 4770–4776. [Google Scholar] [CrossRef] [PubMed]




| Experimental Data | Substrate Concentration mmoL·L−1 | Substrate Uptake mmoL·min−1·L−1 | Single Carbon (C1) Uptake mmoL·min−1·L−1 | Krebs Cycle Flux mmoL·min−1·L−1 | ACL EC 2.3.3.8 Flux mmoL·min−1·L−1 | Specific Growth Rate μ h−1 |
|---|---|---|---|---|---|---|
| Cui et.al. 2018 [32] | Glucose, up to 50 | 3.58 | 21.46 | 2.43 | 3.87 | 0.051 |
| This study | Glycerol, up to 130 | 2.42 | 7.27 | 0.90 | 1.44 | 0.023 |
| This study | Ethanol, up to 32 | 7.76 | 15.52 | 3.00 | 4.76 | 0.046 |
| Reference | Consumption mmoL·gDW−1·h−1 | Specific Growth Rate μ h−1 |
|---|---|---|
| Cui et.al. 2018 [32] | Glucose 0.65 | 0.051 |
| Cui et.al. 2018 with ETA [32] | Glucose 0.61 | 0.047 |
| This study | Glucose 0.59 | 0.044 |
| Taborda et al. 2021 [12] | Glucose 0.37 | 0.017 |
| This study | Glycerol 0.44 | 0.023 |
| Taborda et al. 2021 [12] | Glycerol 0.43 | 0.019 |
| This study | Ethanol 1.41 | 0.046 |
| Taborda et al. 2021 [12] | Acetate 0.60 | 0.025 |
| Experimental Data | Substrate Uptake mmoL·gDW−1·h−1 | Carbon (C1) Uptake mmoL·gDW−1·h−1 | Experimental | Optimized by Stoichiometric Modeling | ||
|---|---|---|---|---|---|---|
| μ h−1 | Carbon C1 per gDW Biomass mmoL·gDW−1 | μmax h−1 | Carbon C1 per gDW Biomass mmoL·gDW−1 | |||
| Cui et.al. 2018 [32] | Glucose 0.65 (=3.58 mmol·min−1·L−1) | 3.9 | 0.051 | 76.5 | 0.092 | 42.4 |
| This study | Glycerol 0.44 (=2.42 mmol·min−1·L−1) | 1.32 | 0.023 | 57.4 | 0.031 | 42.6 |
| This study | Ethanol 1.41 (=7.76 mmol·min−1·L−1) | 2.82 | 0.046 | 61.3 | 0.067 | 42.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berzins, K.; Muiznieks, R.; Baumanis, M.R.; Strazdina, I.; Shvirksts, K.; Prikule, S.; Galvanauskas, V.; Pleissner, D.; Pentjuss, A.; Grube, M.; et al. Kinetic and Stoichiometric Modeling-Based Analysis of Docosahexaenoic Acid (DHA) Production Potential by Crypthecodinium cohnii from Glycerol, Glucose and Ethanol. Mar. Drugs 2022, 20, 115. https://doi.org/10.3390/md20020115
Berzins K, Muiznieks R, Baumanis MR, Strazdina I, Shvirksts K, Prikule S, Galvanauskas V, Pleissner D, Pentjuss A, Grube M, et al. Kinetic and Stoichiometric Modeling-Based Analysis of Docosahexaenoic Acid (DHA) Production Potential by Crypthecodinium cohnii from Glycerol, Glucose and Ethanol. Marine Drugs. 2022; 20(2):115. https://doi.org/10.3390/md20020115
Chicago/Turabian StyleBerzins, Kristaps, Reinis Muiznieks, Matiss R. Baumanis, Inese Strazdina, Karlis Shvirksts, Santa Prikule, Vytautas Galvanauskas, Daniel Pleissner, Agris Pentjuss, Mara Grube, and et al. 2022. "Kinetic and Stoichiometric Modeling-Based Analysis of Docosahexaenoic Acid (DHA) Production Potential by Crypthecodinium cohnii from Glycerol, Glucose and Ethanol" Marine Drugs 20, no. 2: 115. https://doi.org/10.3390/md20020115
APA StyleBerzins, K., Muiznieks, R., Baumanis, M. R., Strazdina, I., Shvirksts, K., Prikule, S., Galvanauskas, V., Pleissner, D., Pentjuss, A., Grube, M., Kalnenieks, U., & Stalidzans, E. (2022). Kinetic and Stoichiometric Modeling-Based Analysis of Docosahexaenoic Acid (DHA) Production Potential by Crypthecodinium cohnii from Glycerol, Glucose and Ethanol. Marine Drugs, 20(2), 115. https://doi.org/10.3390/md20020115