Microalgae as an Efficient Vehicle for the Production and Targeted Delivery of Therapeutic Glycoproteins against SARS-CoV-2 Variants
Abstract
1. Introduction
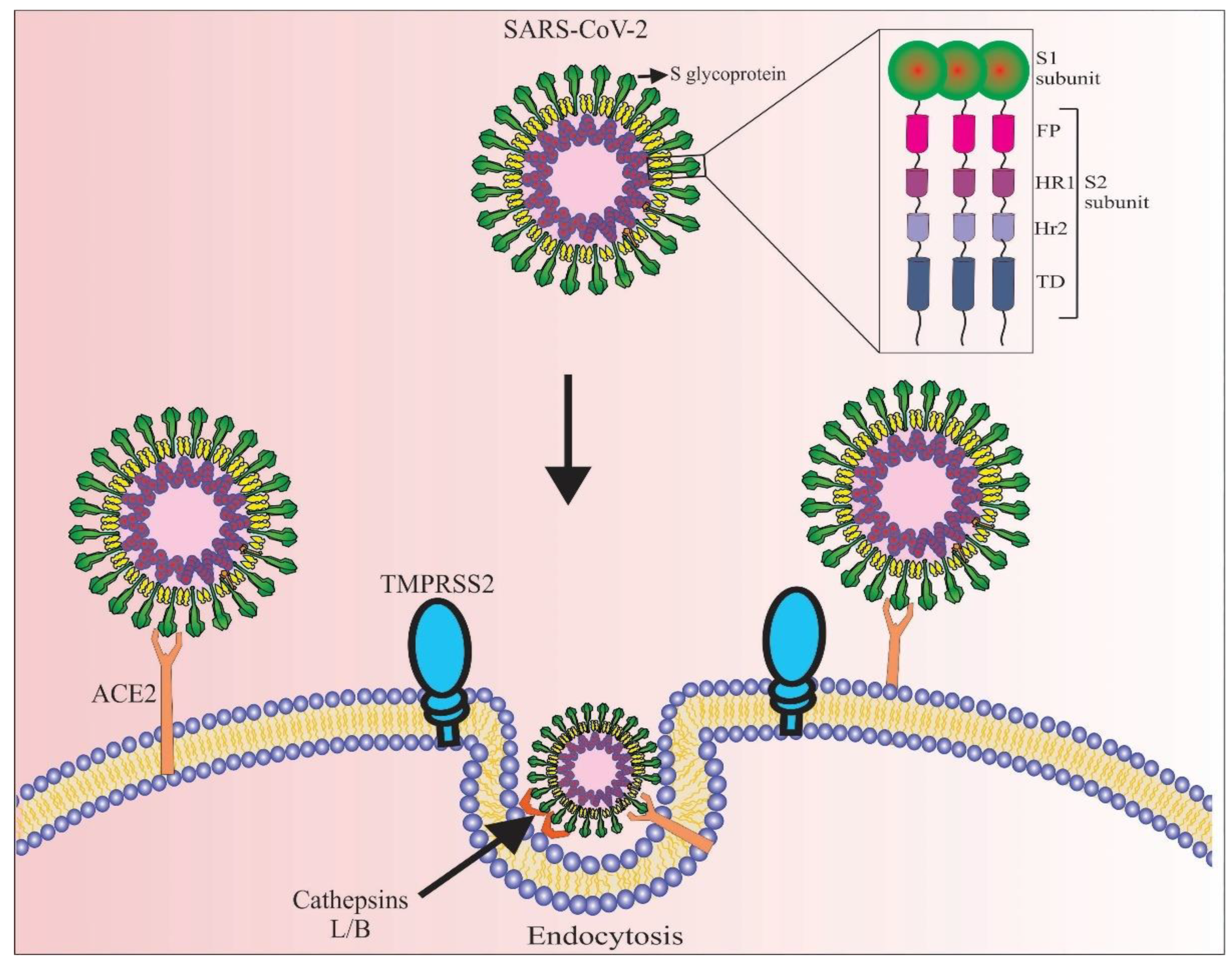
2. SARS-CoV-2 Virus and Its Pathogenic Mechanism
3. Therapeutic Potential of SARS-CoV-2 S-Glycoprotein
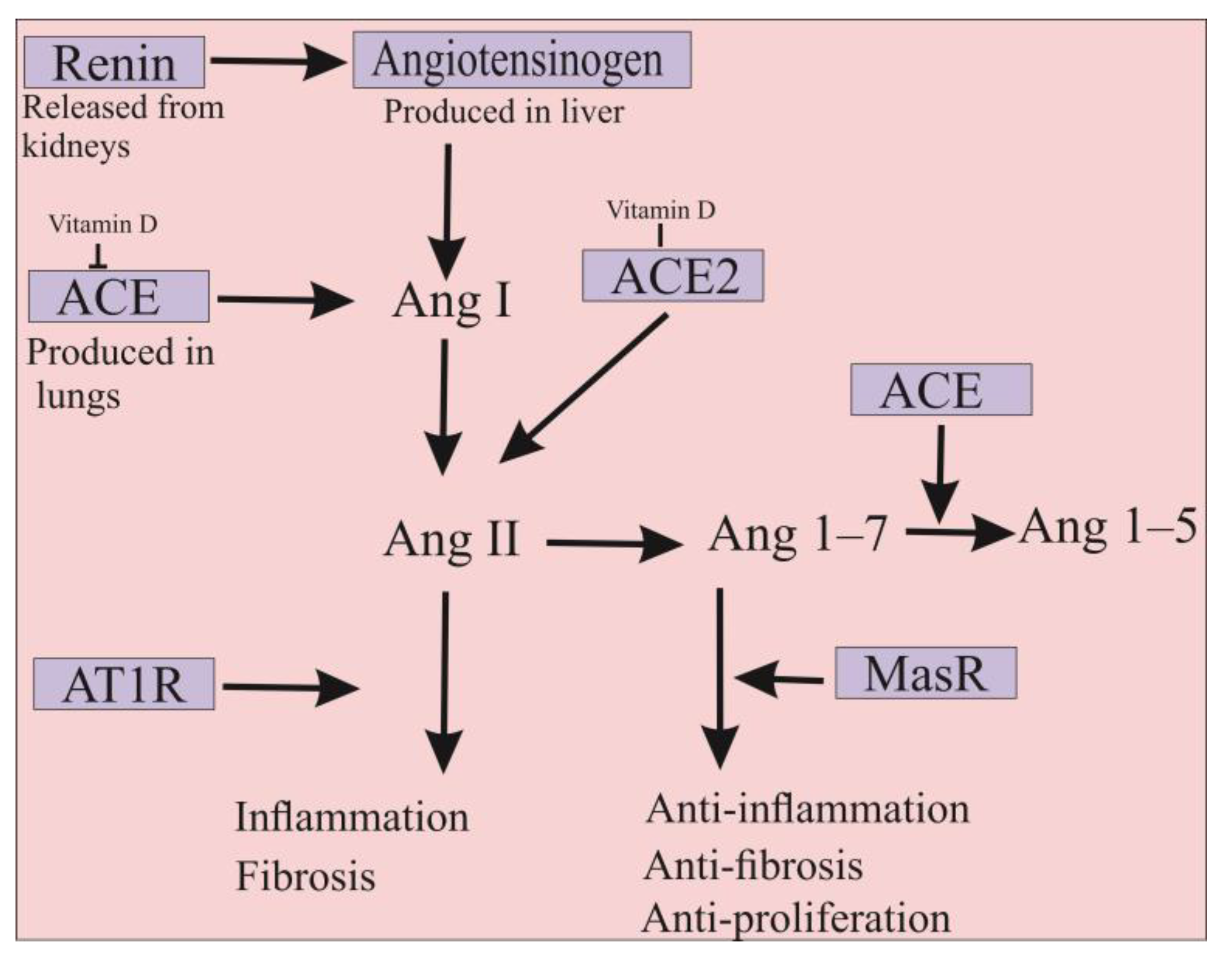
4. ACE2 Is Also a Promising Approach for the Treatment of COVID-19 Infection
5. Potential of Microalgae for the Production of Recombinant S-Glycoprotein and ACE2
6. Edible Microalgae as a Biosystem for Production and Delivery of S-Glycoprotein and Human ACE2
6.1. Edible Microalgae-Based Vaccine against SARS-CoV-2
6.2. Targeted Delivery of Soluble rhACE2 by Microalgae
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, L.; Wang, Y.; Ye, D.; Liu, Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int. J. Antimicrob. 2020, 55, 105948. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Sempowski, G.D.; Saunders, K.O.; Acharya, P.; Haynes, B.F. SARS-CoV-2 Neutralizing Antibodies for COVID-19 Prevention and Treatment. Annu. Rev. Med. 2022, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- South, A.M.; Tomlinson, L.; Edmonston, D.; Hiremath, S.; Sparks, M.A. Controversies of renin-angiotensin system inhibition during the COVID-19 pandemic. Nat. Rev. Nephrol. 2020, 16, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Du, R.H.; Liang, L.R.; Yang, C.Q.; Wang, W.; Cao, T.Z.; Li, M.; Guo, G.Y.; Du, J.; Zheng, C.L.; Zhu, Q.; et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: A prospective cohort study. Eur. Respir. J. 2020, 55, 2002961. [Google Scholar] [CrossRef]
- Rothan, H.A.; Byrareddy, S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020, 109, 102433. [Google Scholar] [CrossRef]
- Jin, X.; Lian, J.S.; Hu, J.H.; Gao, J.; Zheng, L.; Zhang, Y.M.; Hao, S.R.; Jia, H.Y.; Cai, H.; Zhang, X.L.; et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 2020, 69, 1002–1009. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, L.; Deng, Q.; Zhang, G.; Wu, K.; Ni, L.; Yang, Y.; Liu, B.; Wang, W.; Wei, C.; et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020, 92, 833–840. [Google Scholar] [CrossRef]
- Arons, M.M.; Hatfield, K.M.; Reddy, S.C.; Kimball, A.; James, A.; Jacobs, J.R.; Taylor, J.; Spicer, K.; Bardossy, A.C.; Oakley, L.P.; et al. Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility. N. Engl. J. Med. 2020, 382, 2081–2090. [Google Scholar] [CrossRef]
- DosSantos, M.F.; Devalle, S.; Aran, V.; Capra, D.; Roque, N.R.; Coelho-Aguiar, J.M.; Spohr, T.; Subilhaga, J.G.; Pereira, C.M.; D‘Andrea Meira, I.; et al. Neuromechanisms of SARS-CoV-2: A Review. Front. Neuroanat. 2020, 14, 37. [Google Scholar] [CrossRef]
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.L.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.D.; et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J. Pathol. 2020, 251, 228–248. [Google Scholar] [CrossRef]
- Pourseif, M.M.; Parvizpour, S.; Jafari, B.; Dehghani, J.; Naghili, B.; Omidi, Y. Prophylactic domain-based vaccine against SARSCoV-2, causative agent of COVID-19 pandemic. BioImpacts 2020, 11, 65–84. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Detailed Dissection and Critical Evaluation of the Pfizer/BioNTech and Moderna mRNA Vaccines. Vaccines 2021, 9, 734. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Flores, D.; Zepeda-Cervantes, J.; Cruz-Reséndiz, A.; Aguirre-Sampieri, S.; Sampieri, A.; Vaca, L. SARS-CoV-2 Vaccines Based on the Spike Glycoprotein and Implications of New Viral Variants. Front. Immunol. 2021, 12, 701501. [Google Scholar] [CrossRef] [PubMed]
- Pillet, S.; Arunachalam, P.S.; Andreani, G.; Golden, N.; Fontenot, J.; Aye, P.P.; Röltgen, K.; Lehmicke, G.; Gobeil, P.; Dubé, C.; et al. Safety, immunogenicity, and protection provided by unadjuvanted and adjuvanted formulations of a recombinant plant-derived virus-like particle vaccine candidate for COVID-19 in nonhuman primates. Cell. Mol. Immunol. 2022, 19, 222–233. [Google Scholar] [CrossRef]
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M.; et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020, 369, 1014–1018. [Google Scholar] [CrossRef]
- Cameroni, E.; Saliba, C.; Bowen, J.E.; Rosen, L.E.; Culap, K.; Pinto, D.; VanBlargan, L.A.; De Marco, A.; Zepeda, S.K.; Iulio, J.D.; et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022, 602, 664–670. [Google Scholar] [CrossRef]
- Baral, P.; Bhattarai, N.; Hossen, M.L.; Stebliankin, V.; Gerstman, B.S.; Narasimhan, G.; Chapagain, P.P. Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 Delta variant B.1.617.2 and implications for immune evasion. Biochem. Biophys. Res. Commun. 2021, 574, 14–19. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef]
- Akash, K.; Sharma, A.; Kumar, D.; Singh, S.K.; Gupta, G.; Chellappan, D.K.; Dua, K.; Nagraik, R. Molecular aspects of Omicron, vaccine development, and recombinant strain XE: A review. J. Med. Virol. 2022, 94, 4628–4643. [Google Scholar] [CrossRef]
- Ikemura, N.; Taminishi, S.; Inaba, T.; Arimori, T.; Motooka, D.; Katoh, K.; Kirita, Y.; Higuchi, Y.; Li, S.; Suzuki, T.; et al. An engineered ACE2 decoy neutralizes the SARS-CoV-2 Omicron variant and confers protection against infection in vivo. Sci. Transl. Med. 2022, 14, eabn7737. [Google Scholar] [CrossRef]
- Dehghani, J.; Adibkia, K.; Movafeghi, A.; Maleki-Kakelar, H.; Saeedi, N.; Omidi, Y. Towards a new avenue for producing therapeutic proteins: Microalgae as a tempting green biofactory. Biotechnol. Adv. 2020, 40, 107499. [Google Scholar] [CrossRef] [PubMed]
- Rasala, B.A.; Lee, P.A.; Shen, Z.; Briggs, S.P.; Mendez, M.; Mayfield, S.P. Robust expression and secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. PLoS ONE 2012, 7, e43349. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Almehdi, A.M.; Khoder, G.; Alchakee, A.S.; Alsayyid, A.T.; Sarg, N.H.; Soliman, S.S.M. SARS-CoV-2 spike protein: Pathogenesis, vaccines, and potential therapies. Infection 2021, 49, 855–876. [Google Scholar] [CrossRef]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care. Med. 2020, 46, 586–590. [Google Scholar] [CrossRef]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef]
- Wu, K.; Peng, G.; Wilken, M.; Geraghty, R.J.; Li, F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012, 287, 8904–8911. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Pawłowski, P. SARS-CoV-2 variant Omicron (B.1.1.529) is in a rising trend of mutations increasing the positive electric charge in crucial regions of the spike protein S. Acta Biochim. Pol. 2021, 69, 263–264. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Zhang, J.; Kreutzberger, A.J.B.; Eaton, A.; Edwards, R.J.; Jing, C.; Dai, H.Q.; Sempowski, G.D.; Cronin, K.; Parks, R.; et al. An Antibody from Single Human V(H)-rearranging Mouse Neutralizes All SARS-CoV-2 Variants Through BA.5 by Inhibiting Membrane Fusion. Sci. Immunol. 2022, 7, eadd5446. [Google Scholar] [CrossRef]
- Mannar, D.; Saville, J.W.; Sun, Z.; Zhu, X.; Marti, M.M.; Srivastava, S.S.; Berezuk, A.M.; Zhou, S.; Tuttle, K.S.; Sobolewski, M.D.; et al. SARS-CoV-2 variants of concern: Spike protein mutational analysis and epitope for broad neutralization. Nat. Commun. 2022, 13, 4696. [Google Scholar] [CrossRef]
- Shah, V.K.; Firmal, P.; Alam, A.; Ganguly, D.; Chattopadhyay, S. Overview of Immune Response During SARS-CoV-2 Infection: Lessons from the Past. Front. Immunol. 2020, 11, 1949. [Google Scholar] [CrossRef]
- Singh, L.; Bajaj, S.; Gadewar, M.; Verma, N.; Ansari, M.N.; Saeedan, A.S.; Kaithwas, G.; Singh, M. Modulation of Host Immune Response Is an Alternative Strategy to Combat SARS-CoV-2 Pathogenesis. Front. Immunol. 2021, 12, 660632. [Google Scholar] [CrossRef]
- Xia, X. Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design. Viruses 2021, 13, 109. [Google Scholar] [CrossRef]
- Gong, Y.; Qin, S.; Dai, L.; Tian, Z. The glycosylation in SARS-CoV-2 and its receptor ACE2. Signal Transduct. Target. Ther. 2021, 6, 396. [Google Scholar] [CrossRef]
- Wang, N.; Shang, J.; Jiang, S.; Du, L. Subunit Vaccines Against Emerging Pathogenic Human Coronaviruses. Front. Microbiol. 2020, 11, 298. [Google Scholar] [CrossRef]
- Kam, Y.W.; Kien, F.; Roberts, A.; Cheung, Y.C.; Lamirande, E.W.; Vogel, L.; Chu, S.L.; Tse, J.; Guarner, J.; Zaki, S.R.; et al. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into B cells in vitro. Vaccine 2007, 25, 729–740. [Google Scholar] [CrossRef]
- Di Domenico, M.; De Rosa, A.; Boccellino, M. Detection of SARS-COV-2 Proteins Using an ELISA Test. Diagnostics 2021, 11, 698. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Lin, R.T.P.; Renia, L.; Ng, L.F.P. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol. 2020, 11, 879. [Google Scholar] [CrossRef] [PubMed]
- Mathuria, J.P.; Yadav, R.; Rajkumar. Laboratory diagnosis of SARS-CoV-2—A review of current methods. J. Infect. Public Health 2020, 13, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Sakagami, H.; Miwa, N. ACE2: The key Molecule for Understanding the Pathophysiology of Severe and Critical Conditions of COVID-19: Demon or Angel? Viruses 2020, 12, 491. [Google Scholar] [CrossRef]
- Imai, Y.; Kuba, K.; Rao, S.; Huan, Y.; Guo, F.; Guan, B.; Yang, P.; Sarao, R.; Wada, T.; Leong-Poi, H.; et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 2005, 436, 112–116. [Google Scholar] [CrossRef]
- Roca-Ho, H.; Riera, M.; Palau, V.; Pascual, J.; Soler, M.J. Characterization of ACE and ACE2 Expression within Different Organs of the NOD Mouse. Int. J. Mol. Sci. 2017, 18, 563. [Google Scholar] [CrossRef]
- Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000, 275, 33238–33243. [Google Scholar] [CrossRef]
- Sanchis-Gomar, F.; Lavie, C.J.; Perez-Quilis, C.; Henry, B.M.; Lippi, G. Angiotensin-Converting Enzyme 2 and Antihypertensives (Angiotensin Receptor Blockers and Angiotensin-Converting Enzyme Inhibitors) in Coronavirus Disease 2019. Mayo. Clin. Proc. 2020, 95, 1222–1230. [Google Scholar] [CrossRef]
- Hamming, I.; Cooper, M.E.; Haagmans, B.L.; Hooper, N.M.; Korstanje, R.; Osterhaus, A.D.; Timens, W.; Turner, A.J.; Navis, G.; van Goor, H. The emerging role of ACE2 in physiology and disease. J. Pathol. 2007, 212, 1–11. [Google Scholar] [CrossRef]
- Hamming, I.; van Goor, H.; Turner, A.J.; Rushworth, C.A.; Michaud, A.A.; Corvol, P.; Navis, G. Differential regulation of renal angiotensin-converting enzyme (ACE) and ACE2 during ACE inhibition and dietary sodium restriction in healthy rats. Exp. Physiol. 2008, 93, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Timens, W.; Bulthuis, M.L.; Lely, A.T.; Navis, G.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [PubMed]
- Kim, J.; Mukherjee, A.; Nelson, D.; Jozic, A.; Sahay, G. Rapid generation of circulating and mucosal decoy ACE2 using mRNA nanotherapeutics for the potential treatment of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Yang, X.H.; Deng, W.; Tong, Z.; Liu, Y.X.; Zhang, L.F.; Zhu, H.; Gao, H.; Huang, L.; Liu, Y.L.; Ma, C.M.; et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007, 57, 450–459. [Google Scholar]
- Zhang, L.; Dutta, S.; Xiong, S.; Chan, M.; Chan, K.K.; Fan, T.M.; Bailey, K.L.; Lindeblad, M.; Cooper, L.M.; Rong, L.; et al. Engineered ACE2 decoy mitigates lung injury and death induced by SARS-CoV-2 variants. Nat. Chem. Biol. 2022, 18, 342–351. [Google Scholar] [CrossRef]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913.e7. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Pang, X.; Cui, Y.; Zhu, Y. Recombinant human ACE2: Potential therapeutics of SARS-CoV-2 infection and its complication. Acta. Pharmacol. Sin. 2020, 41, 1255–1257. [Google Scholar] [CrossRef]
- Haschke, M.; Schuster, M.; Poglitsch, M.; Loibner, H.; Salzberg, M.; Bruggisser, M.; Penninger, J.; Krähenbühl, S. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin-converting enzyme 2 in healthy human subjects. Clin. Pharmacokinet. 2013, 52, 783–792. [Google Scholar] [CrossRef]
- Khan, A.; Benthin, C.; Zeno, B.; Albertson, T.E.; Boyd, J.; Christie, J.D.; Hall, R.; Poirier, G.; Ronco, J.J.; Tidswell, M.; et al. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit. Care 2017, 21, 234. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wysocki, J.; Souma, T.; Ye, M.; Ramirez, V.; Zhou, B.; Wilsbacher, L.D.; Quaggin, S.E.; Batlle, D.; Jin, J. Novel ACE2-Fc chimeric fusion provides long-lasting hypertension control and organ protection in mouse models of systemic renin angiotensin system activation. Kidney Int. 2018, 94, 114–125. [Google Scholar] [CrossRef]
- Zhang, Z.; Zeng, E.; Zhang, L.; Wang, W.; Jin, Y.; Sun, J.; Huang, S.; Yin, W.; Dai, J.; Zhuang, Z.; et al. Potent prophylactic and therapeutic efficacy of recombinant human ACE2-Fc against SARS-CoV-2 infection in vivo. Cell Discov. 2021, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, J.; Adibkia, K.; Movafeghi, A.; Pourseif, M.M.; Omidi, Y. Designing a new generation of expression toolkits for engineering of green microalgae; robust production of human interleukin-2. Bioimpacts 2020, 10, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Lau, J.; Klingl, A.; Maier, U.G. Algae as protein factories: Expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PLoS ONE 2011, 6, e28424. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Maier, U.G. An engineered diatom acting like a plasma cell secreting human IgG antibodies with high efficiency. Microb. Cell Factories 2012, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Vanier, G.; Hempel, F.; Chan, P.; Rodamer, M.; Vaudry, D.; Maier, U.G.; Lerouge, P.; Bardor, M. Biochemical Characterization of Human Anti-Hepatitis B Monoclonal Antibody Produced in the Microalgae Phaeodactylum tricornutum. PLoS ONE 2015, 10, e0139282. [Google Scholar] [CrossRef]
- Vanier, G.; Stelter, S.; Vanier, J.; Hempel, F.; Maier, U.G.; Lerouge, P.; Ma, J.; Bardor, M. Alga-Made Anti-Hepatitis B Antibody Binds to Human Fcγ Receptors. Biotechnol. J. 2018, 13, e1700496. [Google Scholar] [CrossRef]
- Mathieu-Rivet, E.; Mati-Baouche, N.; Walet-Balieu, M.L.; Lerouge, P.; Bardor, M. N- and O-Glycosylation Pathways in the Microalgae Polyphyletic Group. Front. Plant Sci. 2020, 11, 609993. [Google Scholar] [CrossRef]
- Grama, S.B.; Liu, Z.; Li, J. Emerging Trends in Genetic Engineering of Microalgae for Commercial Applications. Mar. Drugs 2022, 20, 285. [Google Scholar] [CrossRef]
- Slattery, S.S.; Giguere, D.J.; Stuckless, E.E.; Shrestha, A.; Briere, L.K.; Galbraith, A.; Reaume, S.; Boyko, X.; Say, H.H.; Browne, T.S.; et al. Phosphate-regulated expression of the SARS-CoV-2 receptor-binding domain in the diatom Phaeodactylum tricornutum for pandemic diagnostics. Sci. Rep. 2022, 12, 7010. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Maurer, M.; Brockmann, B.; Mayer, C.; Biedenkopf, N.; Kelterbaum, A.; Becker, S.; Maier, U.G. From hybridomas to a robust microalgal-based production platform: Molecular design of a diatom secreting monoclonal antibodies directed against the Marburg virus nucleoprotein. Microb. Cell Factories 2017, 16, 131. [Google Scholar] [CrossRef] [PubMed]
- Eichler-Stahlberg, A.; Weisheit, W.; Ruecker, O.; Heitzer, M. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 2009, 229, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, Y.T.; Cho, J.J.; Bae, J.H.; Hur, S.B.; Hwang, I.; Choi, T.J. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar. Biotechnol. 2002, 4, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Ratha, S.K.; Renuka, N.; Rawat, I.; Bux, F. Prospective options of algae-derived nutraceuticals as supplements to combat COVID-19 and human coronavirus diseases. Nutrition 2021, 83, 111089. [Google Scholar] [CrossRef] [PubMed]
- Wan Afifudeen, C.L.; Teh, K.Y.; Cha, T.S. Bioprospecting of microalgae metabolites against cytokine storm syndrome during COVID-19. Mol. Biol. Rep. 2022, 49, 1475–1490. [Google Scholar] [CrossRef]
- Wannathong, T.; Waterhouse, J.C.; Young, R.E.; Economou, C.K.; Purton, S. New tools for chloroplast genetic engineering allow the synthesis of human growth hormone in the green alga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2016, 100, 5467–5477. [Google Scholar] [CrossRef]
- Geng, D.; Wang, Y.; Wang, P.; Li, W.; Sun, Y. Stable expression of hepatitis B surface antigen gene in Dunaliella salina (Chlorophyta). J. Appl. Phycol. 2003, 15, 451–456. [Google Scholar] [CrossRef]
- Commault, A.S.; Kaur Walia, N.; Fabris, M.; Barolo, L.; Siboni, N.; Adriaans, J.; Ralph, P.J.; Pernice, M. Effect of biphasic temperature regime on therapeutic recombinant protein production in the green alga Chlamydomonas reinhardtii. Algal Res. 2020, 50, 101997. [Google Scholar] [CrossRef]
- Kiefer, A.; Niemeyer, J.; Probst, A.; Erkel, G.; Schroda, M. Production and secretion of functional full-length SARS-CoV-2 spike protein in Chlamydomonas reinhardtii. Front. Plant. Sci. 2021, 20, 988870. [Google Scholar]
- Berndt, A.J.; Smalley, T.N.; Ren, B.; Simkovsky, R.; Badary, A.; Sproles, A.E.; Fields, F.J.; Torres-Tiji, Y.; Heredia, V.; Mayfield, S.P. Recombinant production of a functional SARS-CoV-2 spike receptor binding domain in the green algae Chlamydomonas reinhardtii. PLoS ONE 2021, 16, e0257089. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ramírez, J.; Wong-Arce, A.; González-Ortega, O.; Rosales-Mendoza, S. Expression in algae of a chimeric protein carrying several epitopes from tumor associated antigens. Int. J. Biol. Macromol. 2020, 147, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Jarquín-Cordero, M.; Chávez, M.N.; Centeno-Cerdas, C.; Bohne, A.V.; Hopfner, U.; Machens, H.G.; Egaña, J.T.; Nickelsen, J. Towards a biotechnological platform for the production of human pro-angiogenic growth factors in the green alga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2020, 104, 725–739. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhang, H.; Zhan, M.; Jiang, J.; Yin, H.; Dauphars, D.J.; Li, S.Y.; Li, Y.; He, Y.W. Preventing Mortality in COVID-19 Patients: Which Cytokine to Target in a Raging Storm? Front. Cell Dev. Biol. 2020, 8, 677. [Google Scholar] [CrossRef]
- Ávila-Román, J.; Talero, E.; de Los Reyes, C.; García-Mauriño, S.; Motilva, V. Microalgae-derived oxylipins decrease inflammatory mediators by regulating the subcellular location of NFκB and PPAR-γ. Pharmacol. Res. 2018, 128, 220–230. [Google Scholar] [CrossRef]
- Talukdar, J.; Bhadra, B.; Dattaroy, T.; Nagle, V.; Dasgupta, S. Potential of natural astaxanthin in alleviating the risk of cytokine storm in COVID-19. Biomed. Pharmacother. 2020, 132, 110886. [Google Scholar] [CrossRef]
- Tzachor, A.; Rozen, O.; Khatib, S.; Jensen, S.; Avni, D. Photosynthetically Controlled Spirulina, but Not Solar Spirulina, Inhibits TNF-α Secretion: Potential Implications for COVID-19-Related Cytokine Storm Therapy. Mar. Biotechnol. 2021, 23, 149–155. [Google Scholar] [CrossRef]
- Chia, W.Y.; Kok, H.; Chew, K.W.; Low, S.S.; Show, P.L. Can algae contribute to the war with Covid-19? Bioengineered 2021, 12, 1226–1237. [Google Scholar] [CrossRef]
- Khavari, F.; Saidijam, M.; Taheri, M.; Nouri, F. Microalgae: Therapeutic potentials and applications. Mol. Biol. Rep. 2021, 48, 4757–4765. [Google Scholar] [CrossRef]
- Guzmán, S.; Gato, A.; Lamela, M.; Freire-Garabal, M.; Calleja, J.M. Anti-inflammatory and immunomodulatory activities of polysaccharide from Chlorella stigmatophora and Phaeodactylum tricornutum. Phytother. Res. 2003, 17, 665–670. [Google Scholar] [CrossRef]
- Kwon, P.S.; Oh, H.; Kwon, S.J.; Jin, W.; Zhang, F.; Fraser, K.; Hong, J.J.; Linhardt, R.J.; Dordick, J.S. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020, 6, 50. [Google Scholar] [CrossRef] [PubMed]
- Esposito, D.; Mehalko, J.; Drew, M.; Snead, K.; Wall, V.; Taylor, T.; Frank, P.; Denson, J.P.; Hong, M.; Gulten, G.; et al. Optimizing high-yield production of SARS-CoV-2 soluble spike trimers for serology assays. Protein Expr. Purif. 2020, 174, 105686. [Google Scholar] [CrossRef] [PubMed]
- Mouffak, S.; Shubbar, Q.; Saleh, E.; El-Awady, R. Recent advances in management of COVID-19: A review. Biomed. Pharmacother. 2021, 143, 112107. [Google Scholar] [CrossRef] [PubMed]
- Monreal-Escalante, E.; Ramos-Vega, A.; Angulo, C.; Bañuelos-Hernández, B. Plant-Based Vaccines: Antigen Design, Diversity, and Strategies for High Level Production. Vaccines 2022, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, S.; Hefferon, K.; Makhzoum, A.; Abouhaidar, M. Combating Human Viral Diseases: Will Plant-Based Vaccines Be the Answer? Vaccines 2021, 9, 761. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.J.; Gobeil, P.; Séguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.Y.; Couture, M.; D‘Aoust, M.A.; Dhaliwall, J.; Finkle, C.; et al. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 2021, 27, 1071–1078. [Google Scholar] [CrossRef]
- Burnett, M.J.B.; Burnett, A.C. Therapeutic recombinant protein production in plants: Challenges and opportunities. Plants People Planet 2020, 2, 121–132. [Google Scholar] [CrossRef]
- Balieu, J.; Jung, J.-W.; Chan, P.; Lomonossoff, G.P.; Lerouge, P.; Bardor, M. Investigation of the N-Glycosylation of the SARS-CoV-2 S Protein Contained in VLPs Produced in Nicotiana benthamiana. Molecules 2022, 27, 5119. [Google Scholar] [CrossRef]
- McGuire, B.E.; Mela, J.E.; Thompson, V.C.; Cucksey, L.R.; Stevens, C.E.; McWhinnie, R.L.; Winkler, D.F.H.; Pelech, S.; Nano, F.E. Escherichia coli recombinant expression of SARS-CoV-2 protein fragments. Microb. Cell Factories 2022, 21, 21. [Google Scholar] [CrossRef]
- Argentinian AntiCovid Consortium. Structural and functional comparison of SARS-CoV-2-spike receptor binding domain produced in Pichia pastoris and mammalian cells. Sci. Rep. 2020, 10, 21779. [Google Scholar] [CrossRef]
- Toustou, C.; Walet-Balieu, M.L.; Kiefer-Meyer, M.C.; Houdou, M.; Lerouge, P.; Foulquier, F.; Bardor, M. Towards understanding the extensive diversity of protein N-glycan structures in eukaryotes. Biol. Rev. Camb. Philos. Soc. 2022, 97, 732–748. [Google Scholar] [CrossRef] [PubMed]
- Malla, A.; Rosales-Mendoza, S.; Phoolcharoen, W.; Vimolmangkang, S. Efficient Transient Expression of Recombinant Proteins Using DNA Viral Vectors in Freshwater Microalgal Species. Front. Plant. Sci. 2021, 12, 650820. [Google Scholar] [CrossRef] [PubMed]
- Siriwattananon, K.; Manopwisedjaroen, S.; Kanjanasirirat, P.; Budi Purwono, P.; Rattanapisit, K.; Shanmugaraj, B.; Smith, D.R.; Borwornpinyo, S.; Thitithanyanont, A.; Phoolcharoen, W. Development of Plant-Produced Recombinant ACE2-Fc Fusion Protein as a Potential Therapeutic Agent Against SARS-CoV-2. Front. Plant. Sci. 2020, 11, 604663. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Du, H.; Chen, J.; Aslam, M.; Wang, W.; Chen, W.; Li, P.; Du, H.; Liu, X. Global Profiling of N-Glycoproteins and N-Glycans in the Diatom Phaeodactylum tricornutum. Front. Plant Sci. 2021, 12, 779307. [Google Scholar] [CrossRef] [PubMed]
- Baïet, B.; Burel, C.; Saint-Jean, B.; Louvet, R.; Menu-Bouaouiche, L.; Kiefer-Meyer, M.C.; Mathieu-Rivet, E.; Lefebvre, T.; Castel, H.; Carlier, A.; et al. N-glycans of Phaeodactylum tricornutum diatom and functional characterization of its N-acetylglucosaminyltransferase I enzyme. J. Biol. Chem. 2011, 286, 6152–6164. [Google Scholar] [CrossRef]
- Dumontier, R.; Loutelier-Bourhis, C.; Walet-Balieu, M.L.; Burel, C.; Mareck, A.; Afonso, C.; Lerouge, P.; Bardor, M. Identification of N-glycan oligomannoside isomers in the diatom Phaeodactylum tricornutum. Carbohydr. Polym. 2021, 259, 117660. [Google Scholar] [CrossRef]
- Stiefvatter, L.; Neumann, U.; Rings, A.; Frick, K.; Schmid-Staiger, U.; Bischoff, S.C. The Microalgae Phaeodactylum tricornutum Is Well Suited as a Food with Positive Effects on the Intestinal Microbiota and the Generation of SCFA: Results from a Pre-Clinical Study. Nutrients 2022, 14, 2504. [Google Scholar] [CrossRef]
- Domozych, D.S.; Ciancia, M.; Fangel, J.U.; Mikkelsen, M.D.; Ulvskov, P.; Willats, W.G. The Cell Walls of Green Algae: A Journey through Evolution and Diversity. Front. Plant Sci. 2012, 3, 82. [Google Scholar] [CrossRef]
- Imam, S.H.; Snell, W.J. The Chlamydomonas cell wall degrading enzyme, lysin, acts on two substrates within the framework of the wall. J. Cell Biol. 1988, 106, 2211–2221. [Google Scholar] [CrossRef]
- Gerken, H.G.; Donohoe, B.; Knoshaug, E.P. Enzymatic cell wall degradation of Chlorella vulgaris and other microalgae for biofuels production. Planta 2013, 237, 239–253. [Google Scholar] [CrossRef]
- Le Costaouëc, T.; Unamunzaga, C.; Mantecon, L.; Helbert, W. New structural insights into the cell-wall polysaccharide of the diatom Phaeodactylum tricornutum. Algal Res. 2017, 26, 172–179. [Google Scholar] [CrossRef]
- Vela Ramirez, J.E.; Sharpe, L.A.; Peppas, N.A. Current state and challenges in developing oral vaccines. Adv. Drug Deliv. Rev. 2017, 114, 116–131. [Google Scholar] [CrossRef] [PubMed]
- Riccio, G.; Lauritano, C. Microalgae with Immunomodulatory Activities. Mar. Drugs 2019, 18, 2. [Google Scholar] [CrossRef] [PubMed]
- Specht, E.A.; Mayfield, S.P. Algae-based oral recombinant vaccines. Front. Microbiol. 2014, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, B.; Gothandam, K.M. A review on edible vaccines and their prospects. Braz. J. Med. Biol. Res. 2020, 53, e8749. [Google Scholar] [CrossRef] [PubMed]
- Naik, P. Chapter 17—Edible vaccines: Current scenario and future prospects. In Future Foods; Bhat, R., Ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 305–313. [Google Scholar]
- Chan, H.T.; Daniell, H. Plant-made oral vaccines against human infectious diseases-Are we there yet? Plant Biotechnol. J. 2015, 13, 1056–1070. [Google Scholar] [CrossRef]
- Dreesen, I.A.; Charpin-El Hamri, G.; Fussenegger, M. Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection. J. Biotechnol. 2010, 145, 273–280. [Google Scholar] [CrossRef]
- Sami, N.; Ahmad, R.; Fatma, T. Exploring algae and cyanobacteria as a promising natural source of antiviral drug against SARS-CoV-2. Biomed. J. 2021, 44, 54–62. [Google Scholar] [CrossRef]
- Ramos-Vega, A.; Angulo, C.; Bañuelos-Hernández, B.; Monreal-Escalante, E. Microalgae-made vaccines against infectious diseases. Algal Res. 2021, 58, 102408. [Google Scholar] [CrossRef]
- Jia, H.; Neptune, E.; Cui, H. Targeting ACE2 for COVID-19 Therapy: Opportunities and Challenges. Am. J. Respir. Cell Mol. Biol. 2021, 64, 416–425. [Google Scholar] [CrossRef]
- Donoghue, M.; Hsieh, F.; Baronas, E.; Godbout, K.; Gosselin, M.; Stagliano, N.; Donovan, M.; Woolf, B.; Robison, K.; Jeyaseelan, R.; et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000, 87, E1–E9. [Google Scholar] [CrossRef] [PubMed]
- Mehdipour, A.R.; Hummer, G. Dual nature of human ACE2 glycosylation in binding to SARS-CoV-2 spike. Proc. Natl. Acad. Sci. USA 2021, 118, e2100425118. [Google Scholar] [CrossRef] [PubMed]
- Hati, S.; Bhattacharyya, S. Impact of Thiol-Disulfide Balance on the Binding of Covid-19 Spike Protein with Angiotensin-Converting Enzyme 2 Receptor. ACS Omega 2020, 5, 16292–16298. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, J.; Adibkia, K.; Movafeghi, A.; Barzegari, A.; Pourseif, M.M.; Maleki Kakelar, H.; Golchin, A.; Omidi, Y. Stable transformation of Spirulina (Arthrospira) platensis: A promising microalga for production of edible vaccines. Appl. Microbiol. Biotechnol. 2018, 102, 9267–9278. [Google Scholar] [CrossRef]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef]
- Li, L.Y.; Wu, W.; Chen, S.; Gu, J.W.; Li, X.L.; Song, H.J.; Du, F.; Wang, G.; Zhong, C.Q.; Wang, X.Y.; et al. Digestive system involvement of novel coronavirus infection: Prevention and control infection from a gastroenterology perspective. J. Dig. Dis. 2020, 21, 199–204. [Google Scholar] [CrossRef]
- Malek Mahdavi, A. A brief review of interplay between vitamin D and angiotensin-converting enzyme 2: Implications for a potential treatment for COVID-19. J. Med. Virol. 2020, 30, e2119. [Google Scholar] [CrossRef]
- Jäpelt, R.B.; Jakobsen, J. Vitamin D in plants: A review of occurrence, analysis, and biosynthesis. Front. Plant Sci. 2013, 4, 136. [Google Scholar] [CrossRef]
- Del Mondo, A.; Smerilli, A.; Sané, E.; Sansone, C.; Brunet, C. Challenging microalgal vitamins for human health. Microb. Cell Factories 2020, 19, 201. [Google Scholar] [CrossRef]
- Schroda, M. Good News for Nuclear Transgene Expression in Chlamydomonas. Cells 2019, 8, 1534. [Google Scholar] [CrossRef]
- Einhaus, A.; Baier, T.; Rosenstengel, M.; Freudenberg, R.A.; Kruse, O. Rational Promoter Engineering Enables Robust Terpene Production in Microalgae. ACS Synth. Biol. 2021, 10, 847–856. [Google Scholar] [CrossRef]
- Baier, T.; Wichmann, J.; Kruse, O.; Lauersen, K.J. Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii. Nucleic Acids Res. 2018, 46, 6909–6919. [Google Scholar] [CrossRef]
- Baier, T.; Jacobebbinghaus, N.; Einhaus, A.; Lauersen, K.J.; Kruse, O. Introns mediate post-transcriptional enhancement of nuclear gene expression in the green microalga Chlamydomonas reinhardtii. PLoS Genet. 2020, 16, e1008944. [Google Scholar] [CrossRef]
| Recombinant Protein | Expression Host | Targeted Disease | Biological Activity | References |
|---|---|---|---|---|
| Human interleukin 2 | C. reinhardtii, C. vulgaris, and D. salina | Cancer | Yes | [64] |
| Human Anti-Hepatitis B surface antigen antibody (CL4mAb) | P. tricornutum | Hepatitis B | Yes | [65,66] |
| Hepatitis B surface antigen | P. tricornutum | Hepatitis B | Yes | [65] |
| RBD of SARS-CoV-2 | P. tricornutum | COVID-19 | Yes | [71] |
| Human growth hormone | C. reinhardtii | Turner syndrome | Yes | [77] |
| Hepatitis B surface antigen | D. salina | Hepatitis B | Yes | [78] |
| Human interferon alpha 2a | C. reinhardtii | Cancer | Yes | [79] |
| Spike glycoprotein of SARS-CoV-2 | C. reinhardtii | COVID-19 | Yes | [80] |
| RBD of SARS-CoV-2 | C. reinhardtii | COVID-19 | Yes | [81] |
| BCB; a multi-epitope protein | Schizochytrium sp. | Breast cancer | Yes | [82] |
| Human VEGF-165 | C. reinhardtii | Wound healing | Yes | [83] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dehghani, J.; Movafeghi, A.; Mathieu-Rivet, E.; Mati-Baouche, N.; Calbo, S.; Lerouge, P.; Bardor, M. Microalgae as an Efficient Vehicle for the Production and Targeted Delivery of Therapeutic Glycoproteins against SARS-CoV-2 Variants. Mar. Drugs 2022, 20, 657. https://doi.org/10.3390/md20110657
Dehghani J, Movafeghi A, Mathieu-Rivet E, Mati-Baouche N, Calbo S, Lerouge P, Bardor M. Microalgae as an Efficient Vehicle for the Production and Targeted Delivery of Therapeutic Glycoproteins against SARS-CoV-2 Variants. Marine Drugs. 2022; 20(11):657. https://doi.org/10.3390/md20110657
Chicago/Turabian StyleDehghani, Jaber, Ali Movafeghi, Elodie Mathieu-Rivet, Narimane Mati-Baouche, Sébastien Calbo, Patrice Lerouge, and Muriel Bardor. 2022. "Microalgae as an Efficient Vehicle for the Production and Targeted Delivery of Therapeutic Glycoproteins against SARS-CoV-2 Variants" Marine Drugs 20, no. 11: 657. https://doi.org/10.3390/md20110657
APA StyleDehghani, J., Movafeghi, A., Mathieu-Rivet, E., Mati-Baouche, N., Calbo, S., Lerouge, P., & Bardor, M. (2022). Microalgae as an Efficient Vehicle for the Production and Targeted Delivery of Therapeutic Glycoproteins against SARS-CoV-2 Variants. Marine Drugs, 20(11), 657. https://doi.org/10.3390/md20110657







